Abstract
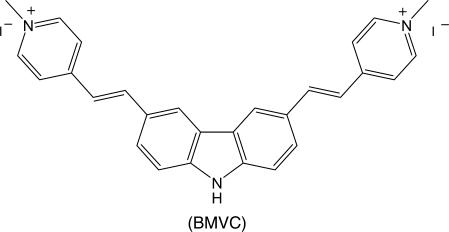
We have introduced a G-quadruplex-binding ligand, 3,6-bis(1-methyl-4-vinylpyridinium)carbazole diiodide (BMVC), to verify the major structure of d(T2AG3)4 (H24) in potassium solution and examine the structural conversion of H24 in sodium solution upon potassium titration. The studies of circular dichroism, induced circular dichroism, spectral titration and gel competition have allowed us to determine the binding mode and binding ratio of BMVC to the H24 in solution and eliminate the parallel form as the major G-quadruplex structure. Although the mixed-type form could not be eliminated as a main component, the basket and chair forms are more likely the main components of H24 in potassium solution. In addition, the circular dichroism spectra and the job plots reveal that a longer telomeric sequence d(T2AG3)13 (H78) could form two units of G4 structure both in sodium or potassium solutions. Of particular interest is that no appreciable change on the induced circular dichroism spectra of BMVC is found during the change of the circular dichroism patterns of H24 upon potassium titration. Considering similar spectral conversion detected for H24 and a long sequence H78 together with the G4 structure stabilized by BMVC, it is therefore unlikely that the rapid spectral conversion of H24 and H78 is due to structural change between different types of the G4 structures. With reference to the circular dichroism spectra of d(GAA)7 and d(GAAA)5, we suggest that the spectral conversion of H24 upon potassium titration is attributed to fast ion exchange resulting in different loop base interaction and various hydrogen bonding effects.
INTRODUCTION
A very challenging question in determining the G-quadruplex (G4) structures of the telomeric repeats d(T2AG3)4 (H24) in potassium solutions has currently received extensive attention (1–7). This is because the 3′-overhang G-rich single strand with 50–200 bases could adopt G-quadruplex structures under physiological conditions. Since the folding of telomeric DNA into G4 structure has been shown to inhibit telomerase activity in vitro (8,9), molecules that stabilize G4 structures have the potential to interfere with telomere replication and possibly to serve as anti-cancer agents (10–12). Knowledge of the telomeric structure is critical for the drug design of structure-specific DNA-binding ligands.
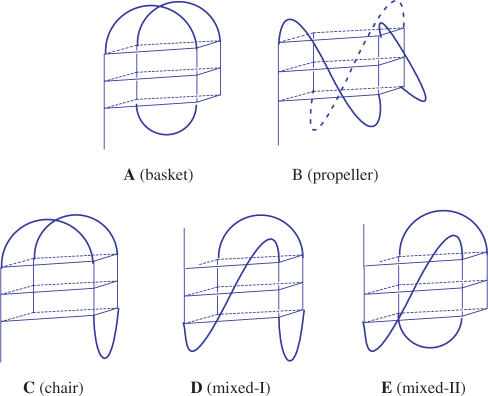
Two intramolecular G4 structures have been determined for human telomeric sequence d[AG3(T2AG3)3] (H22), the NMR structure in Na+ solution (Scheme IA) determined by Wang and Patel (13) and the crystal structure in the presence of K+ (Scheme IB) revealed by Neidle et al. (14). However, the NMR spectrum of the H22 in K+ solution showed a broad envelop with some fine lines, implying the presence of multiple conformational isomers (1,15). Bolton et al. (2) suggested that the propeller G4 structure of H24 occurs in K+ solution. In contrast, the sedimentation and fluorescence studies suggested that the crytal structure of H22 cannot be the major structure in K+ solution (3). Analysis of the fine NMR structures suggested the presence of the mixed-I-type structure (Scheme ID) (1). The platinum cross-linking studies suggested that the basket-type structure (Scheme IA) coexists with other G4 structures in both Na+ and K+ solutions (4). The 125I-radioprobing studies suggested that a chair-type structure (Scheme IC) is a major structure in K+ solution (5). Sugiyama et al. (6) suggested that a mixture of chair-type (Scheme IC) and mixed-I-type (Scheme ID) structures coexist for the H22 in K+ solution. Up to now, determination of the major component on the G4 structure of H24 in K+ solution still remains open.
Scheme I.
In addition, a spectral conversion from the G4 structure in Na+ solution to that in K+ solution was observed upon K+ titration (1,6). Considering the basket-type form of H22 in Na+ solution and the mixed-I-type form in K+ solution, Yang et al. (1) proposed a mechanism for the spectral conversion resulting from structural conversion via the switch of the orientation of the 5′ end GGG strand from the basket- type form to a mixed-I-type form. Sugiyama et al. (6) suggested a very similar mechanism to describe the structural conversion between the chair-type form and the mixed-I-type form. Considering the possible correlation of different G4 structures to their biological roles (3), investigation of structural conversion among various G4 structures deserves further studies and more evidences are necessary in verifying the proposed structural conversion (1,6).
Since different G4 structures could have different binding sites to the ligand, one may distinguish the structural isomers of quadruplexes by monitoring the signal from ligand binding. Particularly, if the ligand has high binding affinity to the G4 structure of H24, one could then examine the structural conversion under K+ titration. In addition, it would be interesting to examine if the ligand binding could inhibit spectral conversion. We have synthesized a novel molecule, 3,6-bis(1-methyl-4-vinylpyridinium)carbazole diiodide (BMVC), to stabilize the G4 structure of H24 (16,17). In addition, significant increase of fluorescence yield and distinctive fluorescence properties of BMVC upon binding to various DNA structures have allowed us to map the G4 structure in human metaphase chromosomes (18,19). Here spectral titration, induced circular dichroism and gel competition are applied to determine the binding modes and compare the binding properties of BMVC to H24 in Na+ and in K+ solutions for verifying the major G4 structure in K+ solution. In addition, circular dichroism (CD) spectra of DNA and induced CD spectra of BMVC together with the melting temperature are used to study the spectral conversion upon K+ titration for examining the structural conversion of the G4 structures of H24. Furthermore, we have applied BMVC to determine the units of G4 structure for a long telomeric sequence, d(T2AG3)13 (H78), by spectral titration and examine its spectral conversion upon K+ titration. Table 1 lists the DNA sequences studied in this work.
Scheme II.
Table 1.
Oligonucleotides used in this work
| Sequence | ε260 (M−1 cm−1) | Abbreviation | |
|---|---|---|---|
| 1 | 5′-TTAGGGTTAGGGTTAGGGTTAGGG | 244 600 | H24 |
| 2 | 5′-TTAGGGTTTGGGTTAGGGTTAGGG | 238 700 | H24-T9 |
| 3 | 5′-TTAGGGTTAGGGTTTGGGTTAGGG | 238 700 | H24-T15 |
| 4 | 5′-TTAGGGTTAGGGTTGGGTTAGGG | 230 600 | M23 |
| 5 | 5′-TTAGGGTTAGGGTTTTGGGTTAGGG | 246 800 | M25 |
| 6 | 5′-GGGTTAGGGTTAGGGTTAGGG | 215 000 | H21 |
| 7 | 5′-TTAGGGTTAGGG | 122 400 | H12 |
| 8 | 5′-(TTAGGG)9 | 550 100 | H54 |
| 9 | 5′-(TTAGGG)13 | 794 500 | H78 |
| 10 | 5′-AGGGTTAGGGTTAGGGTTAGGG | 228 500 | H22 |
| 11 | 5′-AGGGTTAGGGTTAGGGTTAGGGTT | 245 100 | H24-B |
| 12 | 5′-TTAGGGTTAGGGTTAGGGTTAGGGTT | 261 200 | H26 |
MATERIALS AND METHODS
Chemicals
Synthesis of the BMVC molecule from 3,6-dibromocarbazole has been described elsewhere (16,17). All oligonucleotides were purchased from Applied Biosystems. Solutions of 10 mM Tris-HCl (pH 7.5) and 150 mM NaCl or KCl mixed with each DNA were heated to 90°C for 2 min, cooled slowly to room temperature, and then stored for >2 days at 4°C before use. The molar concentration of DNA was determined by monitoring the 260 nm absorbance. The calculated molar extinction coefficient from mononucleotide and dinucleotide by using the nearest neighbor method (20) are listed in Table 1. We further assumed that the G-rich sequence is a single strand at 90°C, but forms a G-quadruplex structure at room temperature. Indeed, the 295 nm positive CD band of each sequence H24, H54 and H78 detected at 25°C is negligible at 90°C. Their absorption spectra could be found in the Supplementary Data. The extinction coefficient of the folded structures of H24, H54 and H78 are estimated to be 2.28 × 105, 4.73 × 105 and 6.91 × 105 M−1 cm−1 at 260 nm based on the ratio of the sample absorbance at 25 and 90°C, respectively.
PAGE
The PAGE was conducted in 10 mM Tris-HCl and 150 mM NaCl or KCl (pH 7.5) with 20% native gels. Electrophoresis gels were carried out at 100 V/cm for 15 h at 4°C. The pre-stained gels were normally conducted by incubated 20 μM of BMVC with 20 μM of DNA for 10 min before running the gels. After photographing with UV shadowing, gels were post-stained by 10 μM of BMVC for 10 s at room temperature, rinsed with distilled water, and then photographed under UV light at 254 nm by a digital camera.
Absorption, fluorescence and CD spectra
Absorption spectra were taken on a Hitachi U3200 UV-visible spectrophotometer and fluorescence spectra were recorded on a Hitachi F4010 spectrofluorimeter with a 2 nm band pass in a 1- or 0.1-cm cell length at room temperature. The CD spectra were averages of 10 scans on a Jasco J-715 spectropolarimeter with a 2 nm bandwidth at room temperature. The scan speed was 50 nm/min with a 0.2 nm step resolution.
RESULTS
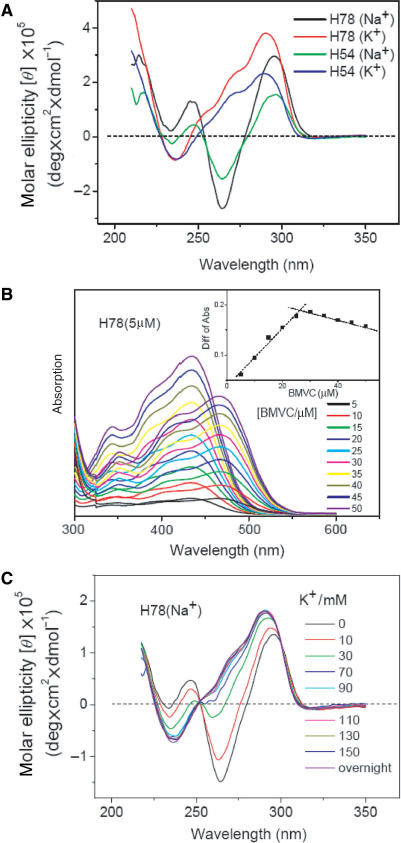
CD spectral conversion of d(T2AG3)4 (H24)
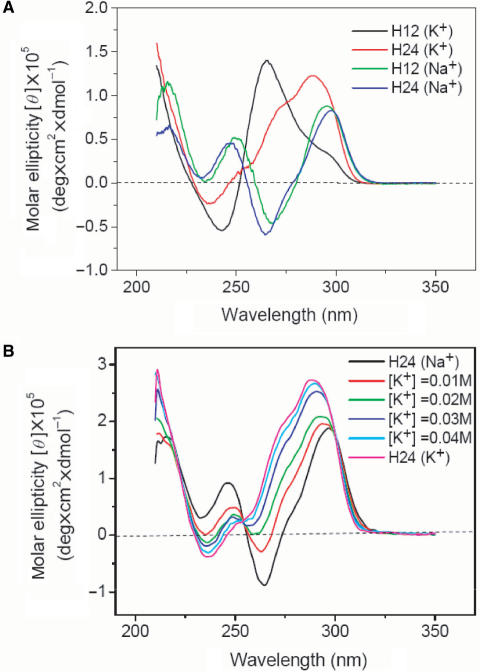
CD spectra have been extensively applied to study the G4 structures (1–3,21–25). It is well known that linear parallel G4 structures, such as propeller form, give a positive band ∼265 nm and a negative band ∼240 nm, while antiparallel G4 structures, such as basket and chair forms, show two positive bands ∼295 and 240 nm and a negative band ∼265 nm. These spectral features are mainly attributed to the specific guanine stacking in various G4 structures. Figure 1A shows CD spectra of H12 and H24 in solutions containing 150 mM of Na+ or K+ cation. The CD patterns of H12 and H24 are similar to each other in Na+ solution, but quite different in K+ solution. The CD pattern detected in Na+ solution indicates the presence of alternative anti/syn glycosidic conformations from guanines of adjacent G-tetrads (26). The 265-nm positive CD band associated with a 295-nm positive shoulder of H12 is probably due to coexistence of dimeric parallel and antiparallel G4 structures in K+ solution. This is because the CD pattern of H12 is similar to the CD pattern of d(TAGGGTTAGGGT) (H12-B) (7) and NMR analysis has revealed the coexistence of the dimeric antiparallel and parallel G4 structures of H12-B in K+ solution (15). According to the finding of multiple conformations from NMR analysis (1,15), the positive CD band ∼290 nm associated with a positive shoulder ∼270 nm in the CD spectrum of H24 in K+ solution can be attributed to a combination of several components.
Figure 1.
(A) CD spectra of 10 μM H12 and H24 DNA in the solutions containing 150 mM of Na+ or K+ cation. (B) CD spectra of 20 μM H24 in 150 mM Na+ solution upon K+ titration. Each CD spectrum was recorded right after the K+ titration.
Figure 1B shows CD spectra of H24 in 150 mM Na+ solution upon K+ titration. Each CD spectrum was recorded right after the K+ titration. Our data show a rapid conversion from the spectral pattern of H24 in Na+ solution to that in K+ solution. In contrast, we have not found any appreciable difference in the CD spectra of H24 in K+ solution by adding Na+, even after incubation overnight (data not shown). It appears that the G4 structure of H24 in K+ solution is more stable than that in Na+ solution. Indeed the melting temperature of the G4 structure of H24 is ∼65°C in K+ solution, higher than the ∼55°C in Na+ solution. Similar spectral conversion of H22 upon K+ titration has been described by a structural conversion via the switch of the orientation of the 5′ end of the GGG strand from the basket form (Scheme IA) to the mixed-I form (Scheme ID) (1). However, we consider that the rapid spectral conversion due to the drastically structural conversion deserves more study.
Spectral titration for binding characters of BMVC to H24
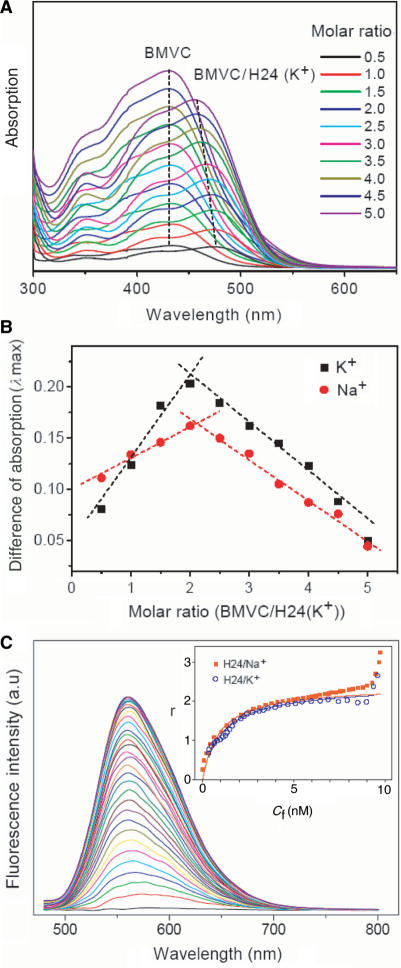
We have conducted spectral titrations to compare the binding ratio and binding affinity of BMVC to H24 in Na+ and in K+ solutions. Figure 2A shows the absorption spectra of free BMVC and its complexes with H24 in K+ solution as a function of BMVC concentration. The absorption maximum at ∼435 nm for free BMVC is red-shifted to ∼475 nm for bound BMVC at 1:2 molar ratio of BMVC to H24 and then gradually blue-shifted upon adding more BMVC. To determine the binding ratio of BMVC to H24, Figure 2B shows the job plots of BMVC to H24 in Na+ and in K+ solutions. A job plot is obtained by taking the intensity difference between the free and bound BMVC to the molar ratio of BMVC–H24. Our results reveal a ∼2:1 binding ratio for BMVC to H24 both in Na+ or K+ solutions, implying that both ends of the G-quartet are binding sites of H24 for BMVC in both solutions.
Figure 2.
(A) Absorption spectra of free BMVC and its complexes with H24 in K+ solution upon BMVC titration. (B) The job plots of BMVC to the H24 in Na+ and in K+ solutions obtained from (A). (C) Fluorescence titration of 10 nM BMVC by adding H24 from 0.25 to 12 nM in K+ solution. The inset showed the binding plots of γ versus Cf for the titration. The fitting parameters to the equation, γ = nKCf/(1 + KCf), are K = 1.12 × 109 with n ∼ 2.4 in Na+ solution, and K = 1.07 × 109 mol−1 with n ∼ 2.3 in K+ solution.
To measure the binding affinity of BMVC to the G4 structure of H24, we have studied both absorption and fluorescence titration of 15 μM BMVC by adding H24 from 0.015 to 6 μM in K+ solution (data not shown). The strong enhancement of the BMVC fluorescence upon interaction with H24 allowed us to measure the binding affinity by using a low concentration in fluorescence titration. Figure 2C shows the fluorescence titration of 10 nM BMVC by adding H24 from 0.25 to 12 nM in K+ solution. The titration data applied to construct the binding plots of γ versus Cf are shown in the inset. The binding ratio γ is defined as Cb/CDNA, where Cf, Cb and CDNA are the molar concentrations of free BMVC, bound BMVC and DNA, respectively. The difference between Ct and Cb gives the magnitude of Cf, where Ct is the total concentration of BMVC. The curve of the binding plots again indicates that the binding is a complex process. Binding parameters can be obtained by fitting the plots with a multiple-equivalent-site model (27):
where K is the equilibrium binding constant, and n represents the average number of ligands bound per each DNA structure. Note that this equation is identical to the Scatchard equation, γ/Cf = K(n − γ). Using the binding plots of γ versus Cf, the question of slightly non-linear Scatchard plots for obtaining both K and n values becomes irrelevant (27). Here the K values for BMVC to the H24 are 1.12 × 109 and 1.07 × 109 mol−1 with n ∼ 2.4 and ∼2.3 in Na+ and K+ solutions, respectively. The large value of γ at very low concentrations of H24 is probably due to nonspecific binding.
Binding sites of the G4 structures
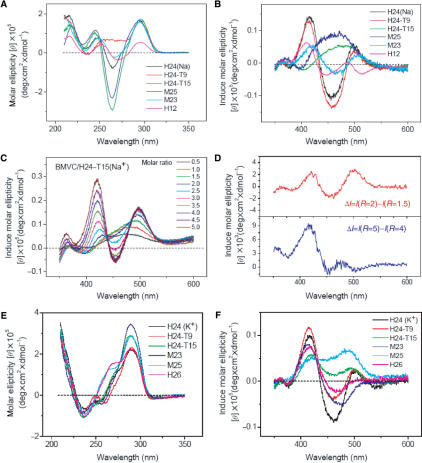
Since the loops and tails of the G4 structures could play an important role in ligand binding (23–25), substituting the base or varying the length in the loops might allow us to distinguish structural isomers and to determine the binding modes of different G4 structures. Figure 3A and B shows the CD spectra of H24, H24-T9, H24-T15, M23, M25 and H12 and the induced CD spectra of BMVC upon interaction with these G4 structures at their molar ratio 1:1 in Na+ solution, respectively. It is found that substitution of the A-base in a TTA lateral loop by a T-base could substantially change the CD spectra. Of particular interest is the region around the 265 nm CD band. Note that the CD pattern of H24-T21 is very similar to that of the H24-T9 (data not shown). On the other hand, both diagonal loops of the TTT in H24-T15 and the TTTT in M25 enhance the negative CD band at ∼265 nm. In addition, similar CD patterns of H24 and M23 with slight band shifts are observed. Considering the relative short TT loop, the M23 is likely to form a chair-type G4 structure with two antiparallel TTA lateral loops and a TT lateral loop at the other end of the G-quartet, whereas the H24 forms a basket-type G4 structure with two parallel TTA lateral loops and another TTA diagonal loop at the other end of the G-quartet. Our data clearly show no appreciable difference in the ∼295 nm positive CD band, but show some changes to the ∼265 and ∼240 nm CD bands upon loop modification.
Figure 3.
(A) CD spectra of H24, H24-T9, H24-T15, M25, M23 and H12 and (B) the corresponding induced CD spectra of 20 μM BMVC upon interaction with these G4 structures at their molar ratio 1:1 in Na+ solution. (C) Induced CD spectra of BMVC upon interaction with H24-T15 at molar ratio from 0.5 to 5.0 in Na+ solution. (D) Differential spectra obtained from the difference between the induced CD spectra at molar ratios of 2 and 1.5 (upper) and molar ratios of 5 and 4 (lower). (E) CD spectra of H24, H24-T9, H24-T15, M25, M23 and H26 and (F) the corresponding induced CD spectra of 20 μM BMVC upon interaction with these G4 structures at their molar ratio 1:1 in K+ solution.
In contrast to very different CD patterns between H24 and H24-T9, Figure 3B shows similar induced CD patterns of BMVC in H24 and H24-T9. It implies that the binding is not appreciably perturbed by substituting the A-base in the lateral TTA loops by a T-base. On the other hand, the induced CD pattern changes a lot in H24-T15 when substituting the A-base in the diagonal TTA loop by a T-base. The induced CD patterns of BMVC in H24-T15 and M25 characterized by a broad positive band ∼470 nm is mainly attributed to the interaction with the TTT or the TTTT diagonal loops, since similar induced CD pattern of BMVC has been also observed in the dimeric hairpin G4 structure of Oxytricha telomeric sequence [d(G4T4G4)]2 (Oxy12) (28,29) with a TTTT diagonal loop at each end of the G-quartet (data not shown). It appears that the induced CD pattern is dominated by the interaction of BMVC to the end of G-quartet with the diagonal loop. Considering the same parallel TTA lateral loops at the other end of the G-quartet in H24 and in H24-T15, different induced CD patterns of BMVC confirm that the end of G-quartet with the diagonal loop and a 5′-TTA tail of H24 is the major binding site for BMVC at the molar ratio of 1:1. On the other hand, the induced CD pattern of BMVC in H12 is quite different from the others, implying that the external stacking to the end of the G-quartet is not the major binding site of H12 for BMVC. It is important to elucidate the possible binding sites of H12 for the interaction with BMVC.
It would be useful if each binding site can be characterized by its induced CD pattern. Figure 3C shows the induced CD spectra of BMVC at different molar ratios with 20 μM H24-T15. Different induced CD patterns are found, for example, at molar ratios of 1:2, 2:1 and 5:1. Considering the binding preference of BMVC to H24-T15 at the end surface of G-quartet with a TTT diagonal loop and a 5′-TTA tail over the end surface of G-quartet with two lateral TTA loops, the contribution to spectral difference of the induced CD spectra between the molar ratio of 2:1 and 3:2 is mainly by external stacking to the end surface of G-quartet with two lateral TTA loops. Similarly, the spectral difference of the induced CD spectra between the molar ratio of 5:1 and 4:1 could be attributed to contribution from some minor binding sites with BMVC, such as groove interaction. These two different patterns are shown in Figure 3D. Surprisingly, the difference between the latter two induced CD spectra resulting from the minor binding sites is quite similar to the induced CD pattern of BMVC in H12, indicating that BMVC is not external stacking to the end surface of the G-quartet, but may externally interact with the groove of H12. Hence we have illustrated a simple approach for characterizing different binding sites of the G4 structures by the induced CD spectra. Moreover, it is clear why the binding strength of BMVC to H12 is much weaker than that of H24 (17).
It is of interest to compare the induced CD spectra of BMVC upon interaction with these G-rich sequences in K+ solution. Figure 3E and F shows the CD spectra of H24, H24-T9, H24-T15, M23, M25 and H26 and their induced CD spectra of BMVC at a 1:1 molar ratio in K+ solution, respectively. Among them, the mixed-I-type structure is suggested to be dominated in both H24 and H26 in K+ solution (1). To our knowledge, no structural information has been given to other sequences. It is found that the CD patterns are quite similar among them with a positive band ∼290 nm associated with a positive shoulder ∼265 nm and a negative band ∼240 nm in K+ solution, which are different from those in Na+ solution. Although the induced CD patterns of BMVC upon interaction with H24 and H24-T9 in K+ solution are similar to those in Na+ solution, the induced CD patterns of BMVC upon interaction with H24-T15, M25 and M23 in K+ solution are different from those in Na+ solution. Furthermore, the induced CD pattern of BMVC in H24-T9 is similar to that in H24, but slightly different from those in M25 and H24-T15, and quite different from those in H26 and M23 in K+ solution. It implies that the induced CD patterns of BMVC may be useful in distinguishing different local structures of the binding sites.
Structural conversion upon K+ titration examined by BMVC
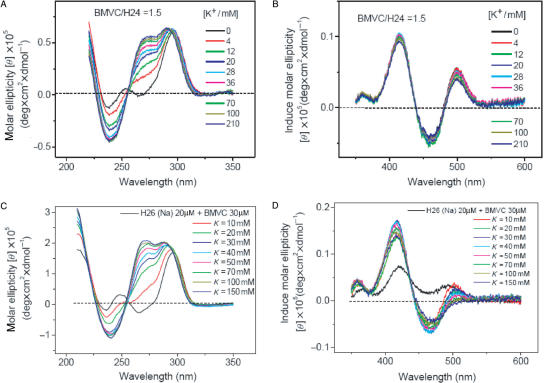
We now take advantage of a high binding affinity of the BMVC to the G4 structure of H24 to test the hypothesis of structural conversion from the basket-type form to the mixed-type form upon K+ titration. Figure 4A and B shows the CD spectra of H24 and the induced CD spectra of BMVC at a molar ratio 1.5 in Na+ solution upon K+ titration, respectively. The key finding is the detection of gradual changes on the CD spectra, but no appreciable difference on the induced CD spectra upon K+ titration. If the structural conversion from the basket form to the propeller form or the mixed form occurs upon K+ titration, one would expect to detect signal change in the induced CD spectra of the BMVC during K+ titration. It is possible that similar induced CD of BMVC in Na+ and in K+ solutions is not able to detect the structural conversion if the conversion between two different G4 structures is very rapid. Another possibility is that the rapid spectral conversion of H24 is not due to structural conversion via a switch in the orientation of the GGG strand upon K+ titration.
Figure 4.
(A) CD spectra of 10 μM H24 and (B) the corresponding induced CD spectra of BMVC at molar ratio 1:1.5 in Na+ solution upon K+ titration. (C) CD spectra of 20 μM H26 and (D) the corresponding induced CD spectra of BMVC at molar ratio 1:1.5 in Na+ solution upon K+ titration.
Considering different CD spectra of H26 together with different induced CD spectra of BMVC in Na+ and in K+ solutions, Figure 4C and D shows the CD spectra of H26 and the induced CD spectra of BMVC–H26 at a molar ratio 1.5 in Na+ solution upon K+ titration, respectively. Again, we have detected the CD spectral change. Of particular interest is that the induced CD spectrum shows spectral change right after the first 10 mM K+ titration and then slight change for the further K+ titration. Yang et al. (1) suggested that the mixed-I-type G4 structure is dominated for the H26 in K+ solution. Unfortunately, the major structure of H26 in Na+ solution has not been determined yet. Nevertheless, we have observed the spectral change of the induced CD pattern of BMVC. If we adopt the hypothesis that the CD spectral conversion of H26 is due to the structural conversion between different types of the G4 structures, the main question is whether it is possible to unfold and refold the G4 structures of H26 upon K+ titration for the structural conversion within few minutes.
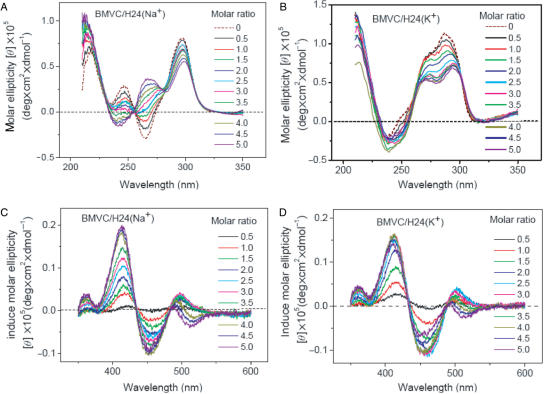
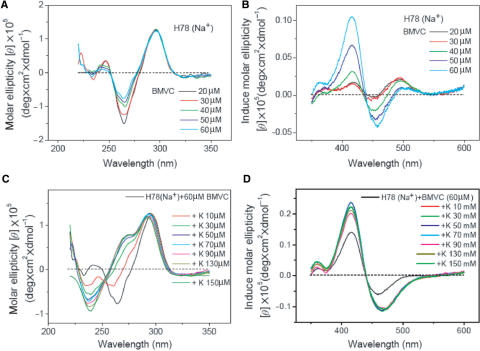
Since BMVC could not only bind to the two ends of G-quartet, but also interact with groove, we now examine the BMVC-binding effect to the G4 structure of H24 as a function of BMVC. Figure 5A–D shows the CD spectra of H24 and the induced CD spectra of BMVC in the presence of Na+ and K+ upon BMVC titration, respectively. The CD patterns are similar in K+ solution, but different in Na+ solution upon BMVC titration. In Na+ solution, it is found that the ∼265 nm negative CD band converts to a positive band and the ∼245 nm positive CD band converts to a negative band upon BMVC titration up to 100 μM. The change of the CD pattern clearly indicates that the BMVC binding could perturb the G4 structure. Under the same experimental conditions, the CD patterns of H24 in K+ solution show no appreciable difference upon BMVC titration. On the other hand, the induced CD patterns of BMVC upon interaction with H24 in Na+ and in K+ solutions are similar with each other during BMVC titration, implying that similar binding sites of H24 to the BMVC are involved in Na+ and in K+ solutions.
Figure 5.
(A and B) CD spectra of 10 μM H24 and (C and D) its induced CD spectra of BMVC in the presence of Na+ (A and C) or K+ (B and D) upon BMVC titration, respectively.
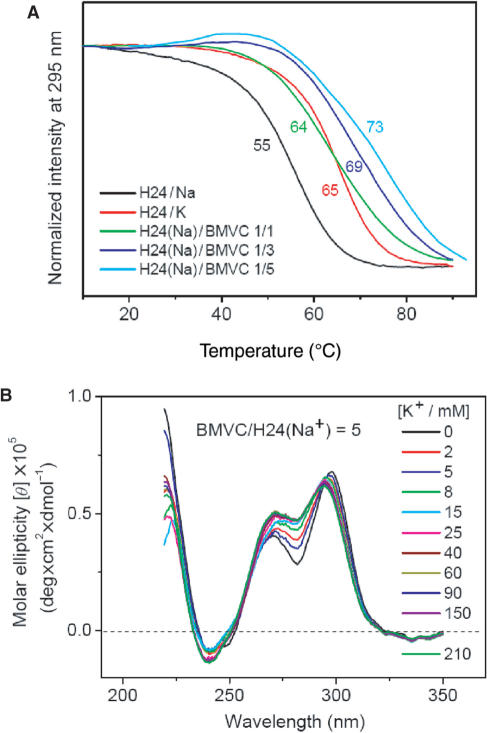
In addition, it is of interest to evaluate how the G4 structure of H24 can be stabilized by various BMVC concentrations. Figure 6A shows the CD intensity at 295 nm of H24 and its complexes with various BMVC concentrations as a function of temperature. It is found that the melting temperature of H24 increases as a function of BMVC. The change of melting temperature of the folded and unfolded structures upon interaction with BMVC provides evidence of thermal stabilization of the H24 structure.
Figure 6.
(A) CD signal at 295 nm for the measurement of melting temperature of H24 and its complexes of BMVC as a function of temperature. (B) CD spectra of 10 μM H24 mixed with 50 μM BMVC in Na+ solution upon K+ titration.
Since the melting temperature of H24 in Na+ solution increases by 18°C upon interaction with BMVC at molar ratio 5, the unfolding and refolding of the G4 structure should be more difficult at high concentrations of BMVC. Figure 6B shows the CD spectra of H24 mixed with BMVC at a molar ratio 5 in Na+ solution upon K+ titration. The change in the CD pattern at molar ratio 5 is less pronounced than that at molar ratio 1.5, implying that more BMVC could bind to H24 and may lower the interaction of K+ with H24.
G4 Structural units and spectral conversion of a longer sequence d(T2AG3)13 (H78)
Considering the 50–200 bases in a single-stranded telomeric sequence, it is of interest to examine the structural units of G4 in longer sequences d(T2AG3)9 (H54) and d(T2AG3)13 (H78), and the possible spectral conversion upon K+ titration. Figure 7A shows the CD spectra of H54 and H78 in Na+ and in K+ solutions. The CD patterns of H54 and H78 are almost identical to the patterns of H24 in Na+ or in K+ solutions, indicating the presence of G4 structure. In order to determine the unit number of the G4 structure in a longer sequence, Figure 7B shows the absorption spectra of free BMVC and its complexes with H78 in Na+ solution as a function of BMVC concentration. The job plots are shown in the inset. Our results reveal a binding ratio ∼5 for BMVC to H78 in Na+ solution. Although the detailed structure of the whole H78 is not clear at present, the CD spectra together with the job plots suggest that H78 could form two units of G4 structures in Na+ solution. This finding illustrates that a longer single- stranded telomeric sequence could form more than one unit of G4 structure.
Figure 7.
(A) CD spectra of 10 μM H54 and 10 μM H78 DNA in the solutions containing 150 mM of Na+ or K+ cation. (B) Absorption spectra of free BMVC and its complexes with H78 in Na+ solution upon BMVC titration. The inset shows the job plots to determine the binding ratio. (C) CD spectra of 5 μM H78 in 150 mM Na+ solution upon K+ titration.
Considering more complexes of local environment around the G4 structure of H78, we have examined if the spectral conversion of H78 occurs upon K+ titration. Figure 7C shows the CD spectra of H78 in 150 mM Na+ solution upon K+ titration. Surprisingly, the spectral conversion of H78 is similar to that of H24. Similar CD spectral conversion of H54 is also observed upon K+ titration, although the spectral titration reveals a binding ratio ∼3 for BMVC to H54 in both Na+ and K+ solutions (data not shown). Since there are two units of G4 structure in H78 and one unit of G4 structure with long tails in H54, it would be more difficult to unfold and refold the G-quadruplex for structural conversion of H54 and H78.
We have further investigated the BMVC-binding effect on the spectral conversion of H78. Figure 8A and B shows the CD spectra of H78 and the induced CD spectra of BMVC in Na+ solution upon BMVC titration. The ∼265-nm negative CD band changes toward a positive direction and the ∼245-nm positive CD band changes toward a negative direction. The CD spectral change of H78 is slower than that of H24 upon BMVC titration. This difference is probably due to the nonspecific binding of BMVC to the more complex structure of H78. An induced CD positive band of BMVC–H78 complexes at ∼415 nm increases associated with a relative weak positive band at ∼495 nm switches to a negative band at ∼460 nm upon BMVC titration up to molar ratio 12. Nevertheless, the induced CD patterns of BMVC–H78 at molar ratio ∼8 are similar to that of BMVC–H24 at molar ratio ∼2.
Figure 8.
(A) CD spectra of 5 μM H78 and (B) the corresponding induced CD spectra of BMVC in Na+ solution upon BMVC titration. (C) CD spectra of 5 μM H78 and (D) the induced CD spectra of BMVC–H78 at a molar ratio 12 in Na+ solution upon K+ titration.
Considering two units of the G4 structure in H78, we use a relative high molar ratio of BMVC with H78 to assure the two ends of each G4 structure of H78 bound by BMVC. Figure 8C and D show the CD spectra of H78 and the induced CD spectra of BMVC–H78 at a molar ratio ∼12 in Na+ solution upon K+ titration, respectively. Again, we have detected spectral change on the CD patterns. In addition, the induced CD spectrum shows discernible difference right after the first 10 mM K+ titration and no appreciable difference for the further K+ titration. Note that the melting temperature of H78 in Na+ solution increases by ∼17°C upon interaction with BMVC at a molar ratio 10 (data not shown). Considering that the G4 structure of H78 surrounded by local environments is more complex and its structure can be stabilized by BMVC, the rapid spectral change upon K+ titration is very unlikely due to the switch between different types of the G4 structures. The difference in the induced CD spectrum upon the first 10 mM K+ titration may be due to the interaction of K+ with the complexes of BMVC with the minor binding sites in the H78.
Figure 9A shows the time-dependent CD spectra of H54 mixed with BMVC at a molar ratio 10 in Na+ solution and further mixed with 100 mM K+. A gradual change on CD spectra as a function of incubation time is found in both cases. However, a drastic change is found right after adding 100 mM K+. Figure 9B shows the plots of the CD intensity change at 265 nm of H24, H54 and H78 in 150 mM Na+ solution after adding 110 mM K+ as a function of time. It is found that the spectral change of H24 is very fast and almost negligible after adding K+ for 10 min. On the other hand, a drastically spectral change followed by a slow change is observed for both H54 and H78. Figure 9C shows the CD spectra of BMVC–H54 at a molar ratio 10 in Na+ solution upon K+ titration. Again a discernible CD change is detected right after the first 10 mM K+ titration followed by a slight change upon further K+ titration. It appears that the rapid spectral change is very likely due to the fast ion exchange.
Figure 9.
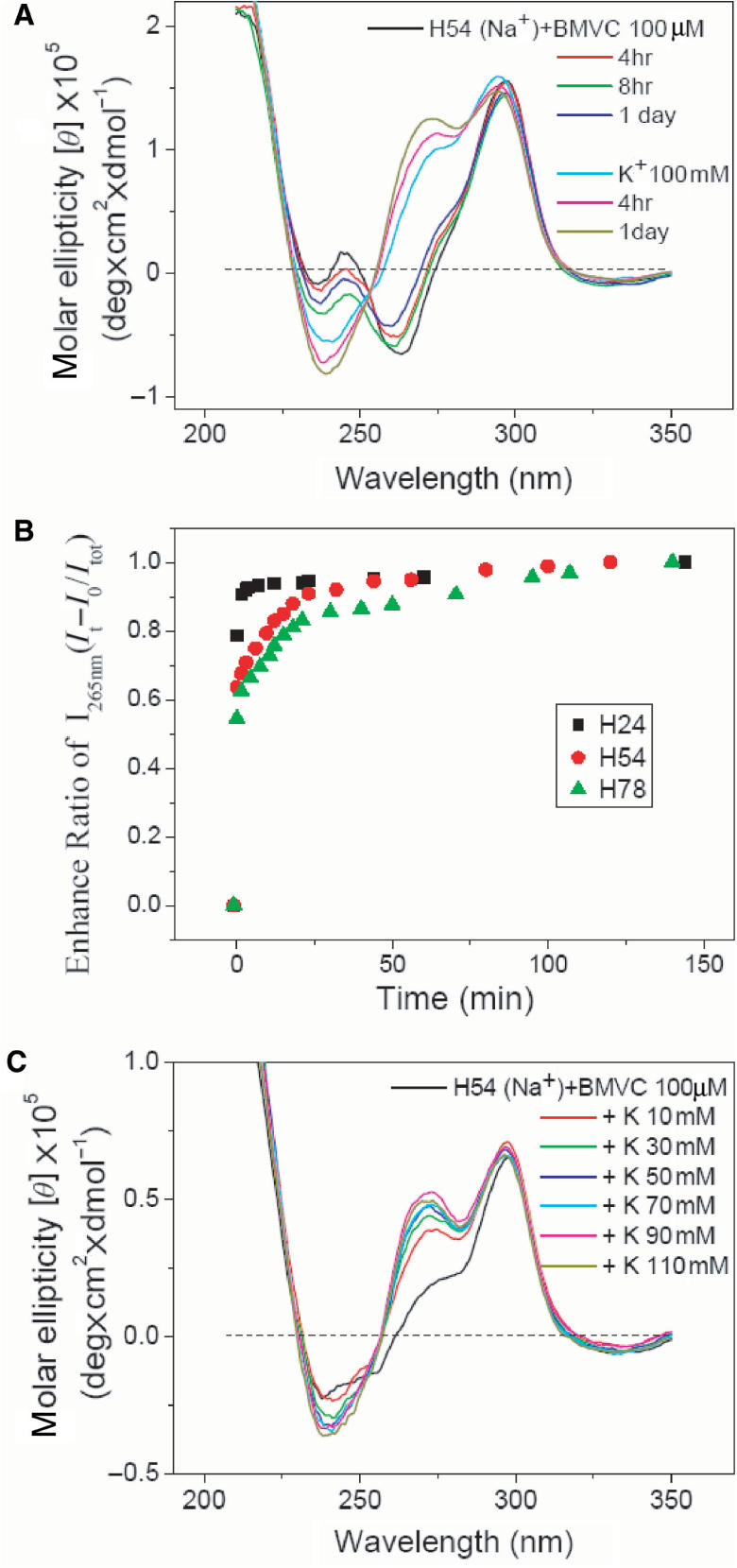
(A) Time-dependent CD spectra of H54 mixed with BMVC–H54 at a molar ratio 10 in Na+ solution and further mixed with 100 mM K+. (B) The plots of the CD intensity change at 265 nm of H24, H54 and H78 in 150 mM Na+ solution as a function of time after adding 110 mM K+. (C) CD spectra of BMVC–H54 at a molar ratio 10 in Na+ solution upon K+ titration.
Gel electrophoresis assays
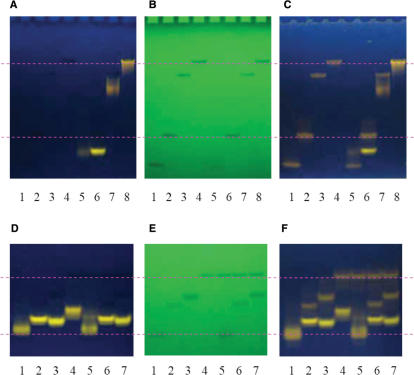
Gel analysis may allow us to evaluate the possible existence of different structural forms. Figure 10A and B shows the pre-stained gels and UV shadowing of the gels of 20 μM each DNA of H21, H24, H54 and H78 (lanes 1–4) and their complexes with 20 μM BMVC pre-stained for 10 min (lanes 5–8) in the presence of Na+, respectively. A major component is found for each lane with very similar position level between the free DNA and its pre-stained DNA in the UV shadowing, implying that the major component is the free DNA in the pre-stained gels. However, the level of the fluorescence band due to BMVC–DNA complexes in the pre-stained gels differs from the level of the band in the UV shadowing. In addition, Figure 10C shows the same gels after being post-stained by 10 μM BMVC for 10 s. The band in the UV shadowing resulting from the free DNA is clearly revealed by BMVC fluorescence in the post-stained gels. Of particular interest is that the BMVC bound H24, H54 and H78 complexes migrate faster than their free forms. In contrast, BMVC-bound H21 complexes migrate slower than their free forms. Different migrations between BMVC–H24 and BMVC–H21 complexes to their free forms indicate that BMVC bound to the end of the G-quartet with a diagonal loop is stronger than that with two TTA loops. It further supports the involvement of the 5′-TTA sequence in the BMVC binding. In addition, our data show that the high-order structural form does not exist.
Figure 10.
(A) The pre-stained gels and (B) UV shadowing of H21, H24, H54, and H78 (lanes 1–4) and their complexes with BMVC pre-stained for 10 min (lanes 5–8) in the presence of Na+. (C) The post-stained gels after 10 μM BMVC post-staining with the same gels for 10 s. (D) The pre-stained gels and (E) UV shadowing of the BMVC incubated with DNA of H22, H24, H24-B, and H26 (lanes 1–4) and the mixture of H26 with each H22, H24-B, and H24 (lanes 5–7) for 10 min in the presence of K+. (F) The post-stained gels after 10 μM BMVC post-staining with the same gels for 10 s.
We have further examined the effect of tail sequence to the G4 structure formed by a four-repeat human telomeric sequence using the gel competition assay. Figure 10D and E shows the pre-stained gels and UV shadowing of the BMVC incubated with each DNA of H22, H24, H24-B and H26 (lanes 1–4) and the mixture of H26 DNA to each DNA of H22, H24 and H24-B (lanes 5–7) for 10 min in K+ solution, respectively. Figure 10F shows the same gels after being post-stained by 10 μM BMVC for 10 s. Similar gel competition patterns in Na+ solution are also observed. Our data show that the binding strength of BMVC to different DNA follows the order of H24 ≥ H24-B > H22 ≥ H26. It implies that the two T bases in the tail play an important role in BMVC binding. However, it is of interest why the binding strength of BMVC to H26 is weaker than H24 and H24-B if they have similar G4 structures.
DISCUSSION
The binding sites of H24 to BMVC
The induced CD spectra and a binding ratio of ∼2 suggest that the two ends of the G-quartet are the main binding sites of H24 to BMVC. Moreover, we have found that the binding affinity of BMVC to the end of the G-quartet depends upon the associated loops and tails. The binding strength is given as (5′-TTA tail + diagonal loop) ∼ (5′-TTA tail + lateral loops) > (diagonal loop) ∼ (two lateral loops) > (one lateral loop). It appears that the end of the G-quartet with a diagonal loop and a 5′-TTA tail is the major binding site of H24 to BMVC at the molar ratio 1:1. Our finding is consistent with the study of acridine derivative (30). Besides the two ends of the G-quartet, BMVC could externally interact with the G4 structure of H24 at a high molar concentration. However, BMVC does not externally stack to the end surface of the G-quartet of H12. It is likely that groove interaction is a major binding mode of BMVC to the dimeric G4 structure of H12. Determining the major binding mode is critical for the design of structure-specific DNA-binding agent as a drug agent.
The major G4 structure of H24 in K+ solution
Determination of the folding structure of H24 in K+ solution is a challenge. A number of studies suggested that the parallel propeller structure is unlikely the major form of H24 in K+ solution (1,3,6,31). Others suggested that the antiparallel G4 structures are either dominated or coexisted in both Na+ and K+ solutions (4,5). Recently, three groups have determined the folding topology of four-repeated human telomeric sequences. Yang et al. (1) found that the mixed-I-type form (Scheme ID) is the major structure to a modified sequence, d(AAAG3(T2AG3)3AA) (M26), by NMR analysis. Since the CD and NMR patterns of H26 are very similar to that of M26, they strongly suggested that the mixed-I-type form (Scheme ID) is also the major structure of the wild-type H26. Sugiyama et al. (3) substituted the guanines in the H22 with 8-bromoguanine and found that the H22 exists as a mixture of mixed-I (Scheme ID) form and chair form (Scheme IC) using CD spectroscopy. Patel et al. (31) studied the modified d[TTGGG(T2AG)3]A sequence and concluded that the major structure is the mixed-I-type form (Scheme ID, ∼95%) by NMR analysis. They all suggested that the mixed-I-type form is a major structure for the four-repeated human telomeric sequences in K+ solution. It should be noted that these G-rich sequences are modified from the wild telomeric sequences.
Since the CD spectrum of H24 in K+ solution is very different from that in Na+ solution, we first examine if they have different G4 structures. Note that the basket form is dominated in the G4 structure of H24 in Na+ solution (13), while multiple conformational isomers are suggested to the G4 structures of H24 in K+ solution (1,6,15). It is possible that the basket form may coexist with other G4 structures for the H24 in K+ solution (4). Nevertheless, very similar binding characters and induced CD spectra of BMVC upon binding to H24 in Na+ or K+ solutions suggest that the G4 structures of H24 should have very similar binding sites in these solutions. Since the tails and loops in the G4 structure are important to stabilize the external stacking of BMVC to the end of the G-quartet (17), the three TTA side loops in the propeller structure (Scheme IB) are not able to hold BMVC to the end of the G-quartet by simply external stacking. Similar binding characters of BMVC to H24 in Na+ and in K+ solutions could eliminate the parallel propeller G4 structure (Scheme IB) as a major component of H24 in K+ solution (4).
Sugiyama et al. (6) suggested that the chair form and the mixed-I form are the major G4 structures of H24 in K+ solution. A highly relevant example, the thrombin- binding aptamer sequence d(G2T2G2TGTG2T2G2) (TBA), which has been shown to form a chair-type structure in the presence of Na+ or K+ (17,32), gives a very similar CD pattern to that of H24 and M23 in Na+ solution, but is different from that of H24 in K+ solution. At present, we are not able to distinguish the basket form from the chair form. It is possible that both basket and chair forms coexist for the H24 in K+ solution (5). On the other hand, the Tetrahymena telomeric DNA d(T2G4)4 (Tet24), which has been shown to form a mixed-I-type structure in Na+ solution (33), gives a similar CD spectra to those of H24 and H26 in K+ solution. Moreover, Yang et al. (1) strongly suggested that the mixed-I-type form is the major structure of H26, implying that this form is also the major structure of H24 in K+ solution. It is of interest to examine if the H24 and H26 have similar G4 structures.
Similar CD patterns of H24 and H26 in K+ solution suggest that they could have similar mixed-I-type structures. If so, one would expect to observe similar binding behaviors to BMVC. However, the gel competition assays show that the binding strength of BMVC to H26 is weaker than H24 and H24-B. The binding preference of BMVC to the (5′-TTA tail + a lateral loop) and (3′-TT tail + a lateral loop) in H26 is more likely over the (5′-TTA tail + a lateral loop) and (a lateral loop) in H24 at two ends of the G-quartets. The gel competition results suggest that the H24 and H26 have different G4 structures. If we assume that the antiparallel G4 structure is the major form to H24 and H26, it is possible that the 5′-TTA and the 3′-TT sequences could affect the binding preference of BMVC to H26 with respect to H24 since the two tails are located at the same end of the G-quartet. Therefore, we have incubated BMVC with H26 for 24 h and then mixed with H24 for 10 min. It is surprising that the pre-stained gels show no appreciable fluorescent band for the H26–BMVC complexes, but a bright fluorescent band for the H24–BMVC complexes (data not shown). It further supports that H24 and H26 have different G4 structures. Since the binding strength of BMVC to H24 is stronger than H26, the mixed-I-type form is more likely dominated in H26, but not in H24. It appears that similar CD spectra of G-rich sequences could have different G4 structures.
Recently, Neidle et al. (26) argued that the mixed-I-type form of the sequences of d[AAAG3(T2AG3)3AA] (1) and d[TTG3(T2AG)3]A (31) based on NMR analysis may not be valid to the G4 structure of H22, since both sequences have been slightly altered at the termini from telomeric regularity. Note that the flanking sequence plays a critical role for the folding of the G4 structure in K+ solution (1,34,35). Furthermore, Patel et al. (31) showed that the extra flanking residues are stacked one on each end of the core of G-tetrads, and help to stabilize this particular topology. However, such a fold has not been observed with the H22, which cannot form such base pairs. They concluded that the precise nature of all the species of H22 in K+ solution deserves further study by fine structure methods (26).
Since the induced CD patterns of BMVC in H24-T15 and Oxy12 are similar in Na+ solution, it is suggested this signal is mainly due to the interaction of BMVC with a TTT and a TTTT diagonal loop. With reference to similar CD patterns of H24 and H24-T15, we consider that the basket-type form is a major component to both of them. If so, substitution of a base in the diagonal loop of H24 does not change the folded structure. On the other hand, if we assume that different CD spectra of H24 and H24-T9 in Na+ solution are due to different G4 structures, it is not clear why substitution of one base in the lateral loop of H24 changes the folded structure. Alternatively, is it possible that different CD spectra could have similar type of the G4 structure?
Of particular interest is that several antiparallel duplex DNAs of various sequences, such as [d(C4G4)]2, also show a CD pattern with a positive band at 265 nm and a negative band at 240 nm, which is very similar to the CD spectra of the parallel G4 structure (36). Moreover, similar CD patterns have been found for a single strand d(GA)10 and homoduplexes of d(GA3)5 and d(GA2)7 under certain conditions (37,38). These results open a possibility that distinct CD spectra of H24 in Na+ or K+ solutions and different CD spectra of H24 and H24-T9 in Na+ solution may be due to different loop base stacking and various intramolecular hydrogen bonding effects. If it is so, the basket and the chair forms are more likely the main components of H24 in K+ solution.
Alternatively, we anticipate that the mixed-II form (Scheme IE) may be a candidate to coexist with other G4 structures for the H24 in K+ solution. Note that the mixed-II-type form characterized by a diagonal loop at one end and a lateral loop with a 5′-TTA at the other end of the G-quartet could have two major binding sites, which are relatively comparable to the binding sites of the basket form of H24 in Na+ solution. Nevertheless, our results favor the coexistence of the basket form and the chair form. Further experiments are necessary to verify the G4 structures of H24 in K+ solution.
Structural conversion upon K+ titration
The next question is to evaluate if the rapid spectral conversion of H24 from the Na+ form to the K+ form upon K+ titration is due to structural conversion of the basket-type form (Scheme IA) to the mixed-type form (Scheme IE). Considering the presence of multiple G4 structures of H24 in K+ solution, the CD spectrum is a collection of contributions from all the conformational isomers, such as the possible G4 structures described in the previous section. Neidle et al. (26) suggested that more attention is required to assign the CD spectra for the G4 structures, since multiple components cannot readily be identified simply by CD spectra. In addition, although the mixed-type form was suggested by the structural analysis of NMR fine lines (1,6), no direct correlation between the fine structure of NMR and the CD pattern was established. Particularly, DNA concentration is about two order magnitudes higher for the NMR studies than the CD measurements. Aggregation is possibly formed at high concentration. Furthermore, structural conversion from the Na+ form to the K+ form upon K+ titration may not be the same as a direct formation of the G4 structure of H24 in K+ solution. For example, the mixed type structure may be directly formed in the presence of K+. However, the conversion from the basket form (Scheme IA) to the mixed-type form (Scheme IE) upon K+ titration takes a lot of work to change the syn/anti configuration of guanine residues. Moreover, the binding preference of BMVC to the both ends of the G-quartet could stabilize the G4 structure of H24 in Na+ solution by increasing the melting temperature >10°C. The switch of the 5′ end GGG strand from the basket-type form to the mixed-II-type form involves first unfolding and then refolding the G-quartet; it is very unlikely that the rapid spectral conversion is due to structural conversion between different types of G4 structures.
A number of G4 structural conversions have been documented (15,39–41). Phan and Patel (15) found that the dimeric antiparallel and parallel G4 structures of H12-B coexist and interconvert in K+ solution. However, direct interconversion between these two G4 structures is improbable. It involves unfolding and folding processes. They further found that the antiparallel form is more favorable at low temperatures (<50°C). Sugimoto et al. (40) reported structural transition from antiparallel to parallel G4 structures of Oxy12 induced by Ca2+. Their kinetic data showed that the structural transition includes at least three steps: cation binding, isomerization and oligomerization. On the other hand, Feigon et al. (41) suggested that the addition of K+ to a basket G4 structure of Oxy12 in the presence of Na+ induces modest changes on local structures but does not change the type of quadruplex structures. For a unimolecular G4 structure of a TBA, the binding of the second K+ between the loops and the adjacent quartets of the chair-type G4 structure could primarily alter the loop structure but not the type of G4 structure (42). It appears that cation binding could perturb the loop structure easily and isomerizes the type of G4 structure.
Our results show that the CD spectra of H24 in Na+ solution could be perturbed by BMVC binding. The spectral change from the ∼265 nm negative band associated with a ∼245 nm positive band to the ∼268 nm positive band associated with a ∼240 nm negative band at high molar ratio of BMVC is not due to the switch between different types of G4 structures. This is because BMVC favors to bind the ends of the G-quartet and thermally stabilizes the G4 structure of H24 in Na+ solution by >10°C. Thus, we consider that the spectral change is due to changes on local conformational structures, while the overall fold is the same upon BMVC binding. The stacking of the BMVC to the end of the G-quartet could change the loop base interaction with the G-quartet and intramolecular hydrogen bonding.
Feigon et al. (32) monitored spectral conversion of [d(G3T4G3)]2 from the Na+ form to the K+ form upon K+ titration. This spectral conversion was attributed to the displacement of Na+ by K+ from G-quartet ion coordination sites. In addition, Yang et al. (1) suggested that ion exchange occurs before structural conversion. Note that cations bound by a G4 structure of Oxy12 are exchanging with ions quite fast, the bound lifetime of Na+ is ∼180 μs in a solution containing both Na+ and K+ (32,43). It is likely that the displacement of Na+ by K+ inside the G-quartet initiates the spectral conversion of H24 upon K+ titration. In addition, Chaires et al. (3) suggested that the K+ form is hydrodynamically more compact than the Na+ form and the conformational change resulting from the substitution of K+ for Na+ may involve an alternation in loop adenine residue interaction with the G-quartet. With reference to the close similarity of the CD spectra of the parallel G4 structure and the homoduplexes of d(GA2)7 and d(GA3)5 (36), we suggest that the spectral conversion of H24 upon K+ titration is likely due to the local conformational change resulting from different loop base interaction and various intramolecular hydrogen bonding effects. Thus, the loop base interaction with the G-quartet and the loop–loop interaction deserve further detailed study.
Recently, Davis et al. (44) reported that unimolecular G-quadruplex could mediate Na+/K+ cation exchange. Sen and Gilbert have proposed that the G4 structures might participate in ion-driven regulatory mechanisms in vivo (45). Further elucidating the mechanism of the spectral conversion of H24 upon K+ titration may be more important in its biological function.
Spectral conversion of long telomeric sequences
Considering longer telomeric sequences H54 and H78, similar CD patterns both in Na+ or K+ solutions indicate that the G4 structures are also formed in these sequences. In addition, the job plots suggest that two units of the G4 structure occur in H78. It is likely that the CD spectra of telomeric sequences are mainly determined by the G4 structure. At present, we are not able to elucidate the local environment of the G4 structure surrounded by the residues of various single-stranded T2AG3 repeats. Since the induced CD pattern of BMVC depends upon the binding site, one could give a brief comparison on the induced CD pattern. The induced CD pattern of BMVC–H78 at molar ratio ∼8 is also similar to that in BMVC–H24 at molar ratio ∼2. Although there are two units of G4 structure in H78, it appears that the BMVC binding to the ends of the G-quartet is perturbed by the local environments of the G4 structure in H78. Together with the melting temperature at ∼51°C for H78 and ∼68°C for BMVC–H78 at a molar ratio 10, the spectral results suggest that the two units of the G4 structure in H78 are likely independent with each other. It further implies that the study of the G4 structure of H24 is valid for the long human telomeric sequences.
Furthermore, we have also detected similar spectral conversion of H54 and H78 from Na+ form to K+ form upon K+ titration. Considering more complex unfolding and refolding of the G4 structure surrounded by additional T2AG3 repeats for a long telomeric sequence, the rapid spectral conversion is very unlikely due to the switch between different types of G4 structures upon K+ titration. In addition, the G4 structure can be thermally stabilized by BMVC. It is more probable that the rapid spectral conversion of H54 and H78 is also induced by different base stacking and intramolecular hydrogen bonding. Moreover, the CD spectra of H54 and H78 in Na+ solution slightly change as a function of incubation time after adding BMVC or K+. This spectral change is much smaller than the spectral change right after K+ titration. It appears that the rapid spectral conversion is very likely due to fast ion exchange resulting in local conformational changes.
In summary, we have determined the major binding mode of BMVC to the G4 structure of H24. The binding properties of BMVC have allowed us to eliminate the parallel propeller (Scheme IB) in K+ solution. Our data suggest that the basket G4 structure (Scheme IA) is likely a major component of the H24 in K+ solution. The chair form (Scheme IC) and the mixed-II form (Scheme IE) may coexist as the main components. In addition, our results support that the flanking sequence is critical for the folding of the G4 structure in K+ solution. Furthermore, the binding ratio together with the CD spectra indicates that H78 could form two units of G4 structure in both Na+ and K+ solutions. Similar spectral conversion observed both in H24–BMVC and H78–BMVC complexes upon K+ titration suggests that the rapid spectral conversion of H24 is very unlikely due to structural conversion between different types of the G4 structures. With reference to the CD spectra of d(GAA)7 and d(GAAA)5 (37,38), we suggest that the spectral change of H24 is attributed to fast ion exchange resulting in different loop base interaction and various hydrogen bonding effects upon K+ titration. Moreover, we have illustrated that the study of the G4 structure of H24 can be applied to a long human telomeric sequence. In addition, the G4 structure deserves more attention because the G4 structure is not only a promising target as an antitumor agent, but also it exists in many G-rich sequences involved in gene regulation.
SUPPLEMENTARY DATA
Supplementary Data is available at NAR online.
ACKNOWLEDGEMENTS
This work was supported by Academia Sinica (5202401023) and the National Science Council of the Republic of China (Grant NSC-94-2113-M001-047). We thank the invaluable discussion from referees, particularly providing the references of the CD spectra of homoduplexes. Funding to pay the Open Access publication charge was provided by Academia Sinica.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ambrus A, Chen D, Dai J, Bialis T, Jones RA, Yang D. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006;34:2723–2735. doi: 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rujan IN, Meleney JC, Bolton PH. Vertebrate telomere repeat DNAs favor external loop propeller quadruplex structures in the presence of high concentrations of potassium. Nucleic Acids Res. 2005;33:2022–2031. doi: 10.1093/nar/gki345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li J, Correia JJ, Wang L, Trent JO, Chaires JB. Not so crystal clear: the structure of the human telomere G-quadruplex in solution differs from that present in a crystal. Nucleic Acids Res. 2005;33:4649–4659. doi: 10.1093/nar/gki782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Redon S, Bombard S, Elizondo-Riojas MA, Chottard JC. Platinum cross-linking of adenines and guanines on the quadruplex structure of the AG3(T2AG3)3 and (T2AG3)4 human telomere structure in Na+ and K+ solutions. Nucleic Acids Res. 2003;31:1605–1613. doi: 10.1093/nar/gkg259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He Y, Neumann RD, Panyutin IG. Intramolecular quadruplex conformation of human telomeric DNA assessed with 125I-radioprobing. Nucleic Acids Res. 2004;32:5339–5367. doi: 10.1093/nar/gkh875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu Y, Noguchi Y, Sugiyama H. The new models of the human telomere d[AGGG(TTaGGG)3] in K+ solution. Bioorg. Med. Chem. 2006;14:5584–5591. doi: 10.1016/j.bmc.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 7.Vorlíčková M, Chládková J, Kejnovská I, Fialová M, Kypr J. Guanine tetraplex topology of human telomere DNA is governed by the number of (TTAGGG) repeats. Nucleic Acids Res. 2005;33:5851–5860. doi: 10.1093/nar/gki898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bodnar AG, Ouellette M, Frolkis M, Holt SE, Chiu CP, Morin GB, Harley CB, Shay JW, Lichtsteiner S, et al. Extension of life-span by instruction of telomerase into normal human cells. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 9.Zahler AM, Williamson JR, Cech TR, Prescott DM. Inhibition of telomerase by G-quartet DNA structure. Nature. 1991;350:718–720. doi: 10.1038/350718a0. [DOI] [PubMed] [Google Scholar]
- 10.Mergny JL, Hélène C. G-quadruplex DNA: a target for drug design. Nat. Med. 1998;4:1366–1367. doi: 10.1038/3949. [DOI] [PubMed] [Google Scholar]
- 11.Hurley LH. DNA and its associated processes as targets for cancer therapy. Nat. Rev. Cancer. 2002;2:188–200. doi: 10.1038/nrc749. [DOI] [PubMed] [Google Scholar]
- 12.Neidle S, Parkinson G. Telomere maintenance as a target for anticancer drug discovery. Nat. Rev. Drug Discov. 2002;1:383–393. doi: 10.1038/nrd793. [DOI] [PubMed] [Google Scholar]
- 13.Wang Y, Patel DJ. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure. 1993;1:262–283. doi: 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- 14.Parkinson GN, Lee MPH, Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 15.Phan AT, Patel DJ. Two-repeat human telomeric d(TAGGGTTAGGGT) sequence forms interconverting parallel and antiparallel G-quadruplexes in solution: Distinct topologies, thermodynamic properties, and folding/unfolding kinetics. J. Am. Chem. Soc. 2003;125:15021–15027. doi: 10.1021/ja037616j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang CC, Wu JY, Chang T-C. A carbazole derivative synthesis for stabilizing the quadruplex structure. J. Chin. Chem. Soc. 2003;50:185–188. [Google Scholar]
- 17.Chang CC, Wu JY, Chien CW, Wu WS, Liu H, Kang CC, Yu LJ, Chang T-C. A fluorescent carbazole derivative: high sensitivity for quadruplex DNA. Anal. Chem. 2003;75:6177–6183. doi: 10.1021/ac034789i. [DOI] [PubMed] [Google Scholar]
- 18.Chang CC, Kuo I-C, Ling I-F, Chen CT, Chen HC, Lou PJ, Lin JJ, Chang T-C. Detection of quadruplex DNA structures in human telomeres by a fluorescent carbazole derivative. Anal. Chem. 2004;76:4490–4494. doi: 10.1021/ac049510s. [DOI] [PubMed] [Google Scholar]
- 19.Chang CC, Chu JF, Kao FJ, Chiu YC, Lou PJ, Chen HC, Chang T-C. Verification of antiparallel G-quadruplex structure in human telomeres by using two-photon excitation fluorescence lifetime imaging microscopy of the 3,6-bis(1-methyl-4-vinylpyridinium)carbazole diiodide molecule. Anal. Chem. 2006;78:2810–2815. doi: 10.1021/ac052218f. [DOI] [PubMed] [Google Scholar]
- 20.Richard EG. Handbook of Biochemistry and Molecular Biology. Cleveland, Ohio: CRC Press; 1975. p. 589. [Google Scholar]
- 21.Balagurumoorthy P, Brahmachari SK, Mohanty D, Bansal M, Sasisekharan V. Hairpin and parallel quartet structures for telomeric sequences. Nucleic Acids Res. 1992;20:4061–4067. doi: 10.1093/nar/20.15.4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giraldo R, Suzuki M, Chapman L, Rhodes D. Promotion of parallel DNA quadruplexes by a yeast telomere binding protein: a circular dichroism study. Proc. Natl Acad. Sci. USA. 1994;91:7658–7662. doi: 10.1073/pnas.91.16.7658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu JY, Chang CC, Yan CS, Chen KY, Kuo I-C, Mou CY, Chang T-C. Structural isomers and binding sites of guanine-rich quadruplexes investigated by induced circular dichroism of thionin: loops and tails. J. Biomol. Struct. Dyn. 2003;21:135–140. doi: 10.1080/07391102.2003.10506911. [DOI] [PubMed] [Google Scholar]
- 24.Hazel P, Huppert J, Balasubramanian S, Neidle S. Loop-length-dependent folding of G-quadruplexes. J. Am. Chem. Soc. 2004;126:16405–16415. doi: 10.1021/ja045154j. [DOI] [PubMed] [Google Scholar]
- 25.Qi J, Shafer RH. Covalent ligation studies on the human telomere quadruplex. Nucleic Acids Res. 2005;33:3185–3192. doi: 10.1093/nar/gki632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chaires JB. Analysis and interpretation of ligand-DNA binding isotherms. Meth. Enzymol. 2001;340:3–22. doi: 10.1016/s0076-6879(01)40415-0. [DOI] [PubMed] [Google Scholar]
- 28.Kang C, Zhang X, Ratliff R, Moyzis R, Rich A. Crystal structure of four-stranded oxytricha telomeric DNA. Nature. 1992;356:126–131. doi: 10.1038/356126a0. [DOI] [PubMed] [Google Scholar]
- 29.Schultze P, Smith FW, Feigon J. Refined solution structure of the dimeric quadruplex formed. Nature. 1994;356:164. [Google Scholar]
- 30.Read M, Harrison RJ, Romabnoli B, Tanious FA, Gowan SH, Reszka AP, Wilson WD, Lelland LR, Neidle S. Proc. Natl Acad. Sci. USA. 2001;98:4844. doi: 10.1073/pnas.081560598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luu KN, Phan AT, Kuryavyi V, Lacroix L, Patel DJ. Structure of the human telomere in K+ solution: an intramolecular (3 + 1) G-quadruplex scaffold. J. Am. Chem. Soc. 2006;128:9963–9970. doi: 10.1021/ja062791w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hud NV, Smith FW, Anet F.AL, Feigon J. The selectivity for K+ versus Na+ in DNA quadruplexes is dominated by relative free energies of hydration: a thermodynamic studies by 1H NMR. Biochemistry. 1996;35:15383–15390. doi: 10.1021/bi9620565. [DOI] [PubMed] [Google Scholar]
- 33.Wang Y, Patel DJ. Solution structure of the tetrahymena telomeric repeat d(T2G4)4 G-tetraplex. Structure. 1994;2:1141–1156. doi: 10.1016/s0969-2126(94)00117-0. [DOI] [PubMed] [Google Scholar]
- 34.Ambrus A, Chen D, Dai JX, Jones RA, Yang DZ. Solution structure of the biologically relevant G-quadruplex element in the human c-MYC promoter: implications for G-quadruplex stabilization. Biochemistry. 2005;44:2048–2058. doi: 10.1021/bi048242p. [DOI] [PubMed] [Google Scholar]
- 35.Phan AT, Modi YS, Patel DJ. Propeller-type parallel stranded G-quadruplexes in the human c-myc promotor. J. Am. Chem. Soc. 2004;31:2097–2107. doi: 10.1021/ja048805k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kypr J, Fialová M, Chládková J, Tůmová M, Vorlíčková M. Conserved guanine-guanine stacking in tetraplex and duplex DNA. Eur. Biophys. J. 2001;30:555–558. doi: 10.1007/s002490100174. [DOI] [PubMed] [Google Scholar]
- 37.Kypr J, Vorlíčková M. Dimethylsulfoxide-stabilized conformer of guanine-adenine repeat strand of DNA. Biopolymers. 2001;62:81–84. doi: 10.1002/bip.1000. [DOI] [PubMed] [Google Scholar]
- 38.Kypr J, Kejnovská I, Vorlíčková M. DNA homoduplexes containing no pyrimidine nucleotide. Eur. Biophys. J. 2003;32:154–158. doi: 10.1007/s00249-003-0287-x. [DOI] [PubMed] [Google Scholar]
- 39.Wu G, Wong A. Solid-state 23Na NMR determination of the number and coordination of sodium cations bound to Oxytricha nova telomere repeat d(G4T4G4) Biochem. Biophys. Res. Commun. 2004;323:1139–1144. doi: 10.1016/j.bbrc.2004.08.210. [DOI] [PubMed] [Google Scholar]
- 40.Miyoshi D, Nakao A, Sugimoto N. Structural transition from antiparallel to parallel G-quadruplex of d(G4T4G4) induced by Ca2+ Nucleic Acids Res. 2003;31:1156–1163. doi: 10.1093/nar/gkg211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schultze P, Hud NV, Smith FW, Feigon J. The effect of sodium, potassium and ammonium ions on the conformation of the dimeric quadruplex formed by the Oxytricha nova telomere repeat oligonucleotide d(G4T4G4) Nucleic Acids Res. 1999;27:3018–3028. doi: 10.1093/nar/27.15.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marathias VM, Bolton PH. Determinants of DNA quadruplex structural type: sequence and potassium binding. Biochemistry. 1999;38:4355–4364. doi: 10.1021/bi982604+. [DOI] [PubMed] [Google Scholar]
- 43.Deng H, Braunlin WH. Kinetics of sodium ion binding to DNA quadruplexes. J. Mol. Biol. 1996;255:476–483. doi: 10.1006/jmbi.1996.0039. [DOI] [PubMed] [Google Scholar]
- 44.Kaucher MS, Harrell WA, Davis JT. A unimolecular G-quadruplex that functions as a synthetic transmembrane Na+ transporter. J. Am. Chem. Soc. 2006;128:38–39. doi: 10.1021/ja056888e. [DOI] [PubMed] [Google Scholar]
- 45.Sen D, Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988;334:364–366. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.