Abstract
BACE1 is the protease responsible for the production of amyloid-β peptides that accumulate in the brain of Alzheimer's disease (AD) patients. BACE1 expression is regulated at the transcriptional, as well as post-transcriptional level. Very high BACE1 mRNA levels have been observed in pancreas, but the protein and activity were found mainly in brain. An up-regulation of the protein has been described in some AD patients without a change in transcript levels. The features of BACE1 5′ untranslated region (5′ UTR), such as the length, GC content, evolutionary conservation and presence of upstream AUGs (uAUGs), indicate an important regulatory role of this 5′ UTR in translational control. We demonstrate that, in brain and pancreas, almost all of the native BACE1 mRNA contains the full-length 5′ UTR. RNA transfection and in vitro translation show that translation is mainly inhibited by the presence of the uAUGs. We provide a mutational analysis that highlight the second uAUG as the main inhibitory element while mutations of all four uAUGs fully de-repress translation. Furthermore, we have evidence that a sequence within the region 222-323 of the BACE1 5′ UTR has a stimulatory effect on translation that might depend on the presence of trans-acting factors.
INTRODUCTION
BACE1 (β-site APP cleaving enzyme 1) is a type 1 membrane-associated aspartyl protease of 501 amino acids (1–5). BACE1 is required for the processing of neuregulin 1 type III, an EGF-like factor that activates the EGF receptor in Schwann cells, thereby regulating the myelination of axons in the peripheral and central nervous system (6,7). In addition to neuregulin 1, other BACE1 substrates have been identified, such as the amyloid-β precursor-like proteins 1 and 2 (8,9), the ST6 β-galactosamide α-2,6-sialyltranferase 1 (10), the selectin P ligand (11), the β subunits of voltage-gated sodium channels (12) and the low-density lipoprotein receptor-related protein 1 (13).
Historically, BACE1 has been discovered as the secretase that cleaves the amyloid-β precursor protein (APP) at the β-site. In order to generate the amyloid-β peptide (Aβ), APP has to be cleaved sequentially by BACE1 (14–16) and the γ-secretase complex (17,18) at the β- and the γ-site, respectively. Aβ is the major component of the senile plaques, the histological hallmark of Alzheimer's disease (AD) and its progressive accumulation has been causally linked to neuronal impairment and neurodegeneration (19,20).
BACE1 shows a tissue-specific expression pattern (3,4,21). Post-transcriptional mechanisms must also play an important role as shown by the lack of correlation between mRNA and protein levels (2,3,22–24). Moreover, it has been shown that BACE1 protein expression was up-regulated in the brain of some sporadic AD patients without changes in the level of the corresponding mRNA (25–28), thereby suggesting that these mechanisms can play a pathogenic role.
In addition to transcription, regulation of gene expression can be achieved by mRNA processing, transport, localization and stability. Translation of mRNA can also be regulated typically by sequences within the untranslated regions (UTRs) and/or by interactions between the mRNA and trans-acting factors, such as RNA-binding proteins or micro RNAs (miRNAs) (29). These interactions are usually inhibitory and often interfere with the initiation step of translation (30).
Protein translation in eukaryotes is predominantly initiated by a cap-dependent scanning mechanism in which the small ribosomal subunits, with the associated translational initiation factors, are recruited to the 5′ cap structure and scan in a 5′–3′ orientation until the first AUG is recognized. Then, the large ribosomal subunit joins the complex to initiate the protein synthesis (31). In eukaryotes, recognition of an AUG as a translation start codon critically depends on its surrounding sequence. For mammals, the consensus sequence GCCA/GCCAUGG provides an optimal context for initiation (32), with the purine at position −3 and the G at position +4 being the most important nucleotides. The rate of initiation is strongly influenced by the primary and secondary structure of mRNAs. Two mechanisms for escaping the first-AUG rule, i.e. context-dependent leaky scanning and re-initiation, enable downstream AUG codons to be accessed in some mRNAs via scanning (33). Leaky scanning is the process in which a small ribosomal subunit can bypass an upstream AUG (uAUG) in a poor context and reach a downstream AUG codon that is in stronger context for translational initiation. Re-initiation of translation occurs when, after translation of an uORF, the small ribosomal subunit is not released from the mRNA but resumes scanning. Having re-acquired the necessary translational initiation factors, it can re-initiate translation at a downstream AUG. There are also some reports pointing towards a rare viral mechanism, known as ribosome shunting, in which the small ribosomal subunit binds the mRNA in 5′ cap-dependent manner and scans downstream until it reaches a stable RNA secondary structure, which is bypassed due to shunting to a downstream-landing site where the scanning is continued till the start AUG codon is reached (34). Another way to initiate translation implies a cap-independent mechanism that requires an internal ribosome entry site (IRES) and has been extensively described for some viral transcripts and a few cellular transcripts (35–37).
Genes for potent regulatory proteins, such as cytokines, growth factors, kinases, transcription factors, etc., often produce mRNAs in which the 5′ UTR is GC rich and/or burdened by uAUG codons. The idea that these encumbered sequences are nature's way of limiting the synthesis of proteins that would be harmful if overproduced, is supported by experimental data (31).
The 5′ UTR of BACE1 mRNA is long, evolutionally conserved, has a high GC content and four uAUGs, all features that suggest the potential of translational regulation. Indeed, two recent studies have demonstrated that BACE1 5′ UTR is inhibitory to translation (38,39). However, they differ in the interpretation of the data, with one report (38) favoring a major role of uAUGs, while the other (39) suggests that the GC-rich region of the 5′ UTR forms a translation barrier that prevents the ribosome from efficiently translating the BACE1 mRNA. In addition, a third article (40) suggests that a shunting mechanism can overcome the translational inhibition by BACE1 5′ UTR in a cell-specific manner. Therefore, the mechanism of down-regulation of translation driven by BACE1 5′ UTR remains controversial.
In this study, we analyzed the mechanism of inhibition of translation of BACE1 transcript by dissecting and mutating its 5′ UTR. We employed RNA transfection to avoid possible artifacts arising from transcriptional or splicing effects that might occur with DNA transfection. RNA transfection experiments were also compared with in vitro translation assays to highlight the possible contribution of trans-acting factors. By this approach, we demonstrate that the translational control of BACE1 expression is mainly limited by the presence of uAUGs rather than a major contribution from structural determinants.
MATERIALS AND METHODS
Cell culture
Reagents and media for cell culture were provided from Cambrex, East Rutherford, NJ, USA. HeLa cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 5% FCIII and 10% donor horse serum, 100 U/ml penicillin, 100 μg/ml streptomycin and 2 mM glutamine. The cells were cultured at 37°C in a humidified 5% CO2 atmosphere.
Plasmids
Plasmids pT3A-B1, pT3A-B1-1-218, pT3A-B1-222-323, pT3A-B1-222-455, pT3A-B1-323-455 and pT3A-B1-395-455 were generated by PCR amplification of pZac-B1x-Luc+ (38) with the following oligonucleotides (Sigma Genosys, St. Louis, MO, USA): taa agg tac cag ctg cga gcc (sense) and aat act cga gtg ggc ccc ggc ctt cgc (antisense) for B1; taa agg tac cag ctg cga gcc (sense) and aat act cga gta cat cgg cac ggc ggc ggc (antisense) for 1-218; taa agg tac cgc ggg ctc cgg atc cca gc (sense) and aat act cga ggc agg gcc ctg ggc cag c (antisense) for 222-323; taa agg tac cgc ggg ctc cgg atc cca gc (sense) and aat act cga gtg ggc ccc ggc ctt cgc (antisense) for 222-455; taa agg tac cag gcc ctg gcg tcc tga tg (sense) and aat act cga gtg ggc ccc ggc ctt cgc (antisense) for 323-455; taa agg tac cag gcg cca ggg acg gac gt (sense) and aat act cga gtg ggc ccc ggc ctt cgc (antisense) for 395-455. Amplified fragments were inserted into pT3A (41) with KpnI-XhoI digestion and verified by DNA sequencing. To generate the plasmid pT3A-B1-▵222-323, pT3A-B1 was digested with BamHI and PstI, blunted with T4 DNA polymerase and re-ligated. The same blunted backbone was used to prepare the plasmid T3A-B1-222-323AS by antisense insertion of the fragments 222-323 amplified by PCR.
pT3A-B1-mut2, pT3A-B1-mut1,2 and pT3A-B1-mut1,2,3 were obtained by PCR amplification of plasmids pBRm2L-B1x-mut1, pBRm2L-B1x-mut2 and pBRm2L-B1x-mut3 (38) with the same oligonucleotides described above.
The last uAUG was mutated into UUG by PCR performed with a sense oligo bearing the mutation (ggc cct gca ggc cct ggc gtc ctg ttg ccc) and the antisense oligo used for cloning of pT3A-B1. The amplified fragment was cleaved PstI-XhoI and inserted into plasmids pT3A-B1, pT3A-B1-mut1,2,3 and pT3A-B1-mut2 in order to generate pT3A-B1-mut4, pT3A-B1-mut1,2,3,4 (pT3A-B1-mut #4) and pT3A-B1-mut2,4 respectively. pT3A-B1-mut #4 Δ222-323 plasmid was obtained by deletion of the BamHI-PstI fragment from pT3A-B1-mut #4.
The plasmid sl200MLA.1 (42) was kindly provided by Dr Thoma. sl200MLA.1 was digested EcoRV-SphI, blunted and re-closed to obtain the empty vector sl200LA.1. For preparing sl200LA.1-B1-222-323, the vector pT3A-B1-222-323 was cleaved with KpnI, blunted with T4 DNA polymerase and digested with SphI. The fragments 222-323 obtained in this way was inserted into EcoRV-SphI-cleaved sl200MLA.1.
To obtain the plasmid pTOPO-hBACE1, RT-PCR was performed with total RNA from human brain (Clontech, Palo Alto, CA, USA, lot number 5120091). Reverse transcription was performed with Superscript II kit (Invitrogen, Carlsbad, CA, USA). PCR was performed with oligonucleotides agg agc ccg gag ccc ttg (sense) and gct cct cgg gct ctt cgt c (antisense). Amplified fragments were cloned in pTOPO vector (Invitrogen) according to the manufacturer's instructions.
In vitro transcription
Capped mRNAs were transcribed from 2 μg of linearized plasmid DNA in a 25 μl reaction containing 1 mM ATP, 1 mM CTP, 1 mM UTP, 10 mM DTT, 52 U RNAsin (Promega, Madison, WI, USA), 8.8 mM m7GpppG, 30 U T3 or T7 polymerase (Promega) and 1× transcription buffer (supplied with the RNA polymerase). After 10 min at 37°C, 2 mM GTP was added and the incubation continued for additional 60 min before treatment with 1 U RQ1 RNase-free DNase (Promega) for 15 min at 37°C. RNA was extracted with phenol–chloroform–isoamylalcohol (25:24:1), precipitated with 2.5 volumes of ethanol and 1/10 volumes of 3 M sodium acetate (pH 5.2), recovered by centrifugation, washed with 75% ethanol and dissolved in deionized water.
RNA transfection
For RNA transfection, TransMessenger Transfection Reagent (Qiagen, Valencia, CA, USA) was used according to the manufacturer's instructions. The ratio between Renilla and firefly reporters was 1:4. The transfection reagent was substituted with MEM after 3 h, and the luminescence was measured after 24 h. All in vitro-transcribed mRNAs used in transfection experiments contained 5′ end cap structure and poly(A) tail.
In vitro translation
In vitro translation was performed in HeLa cell extracts (43). The assays were performed in a volume of 10 μl with 0.07 pmol of the mRNA of interest and 0.026 pmol of the control Renilla reporter mRNA. Standard reactions contained 40% (v/v) HeLa extract, 60 μM amino acids, 20 mM creatine phosphate, 0.04 μg/μl creatine kinase, 16 mM HEPES pH 7.6, 0.8 mM ATP, 0.1 mM GTP, 50 μM spermidine, 0.6 U RNase inhibitor (Eppendorf, Hamburg, Germany), 2.5 mM magnesium acetate and 40 mM potassium acetate. The reactions were incubated at 37°C for 30 min and stopped by snap freezing in liquid nitrogen.
RNase protection
BACE1 riboprobes were prepared starting from XbaI-linearized pTOPO-hBACE1 in 20 μl reactions with 12 μM cold CTP and 50 μCi [α-32P] CTP per reaction. Transcription was driven by Sp6 polymerase at 42°C for 1 h, followed by 15 min DNA digestion by 1 U RQ1 RNase-free DNase (Promega). Hot riboprobes were extracted with phenol–chloroform–isoamylalcohol (25:24:1) and passed twice over CHROM SPIN 100 columns (BD Bioscience, Palo Alto, CA, USA). The RNase protection was performed with RPA III (Ambion, Austin, TX, USA) according to the instruction manual. Yield and specific activity were calculated for every probe. In particular, 3–10-fold molar excess of probe was added into the reaction, as recommended, to avoid problems with saturation. The hybridization was overnight at 60°C and RNase digestion was performed with 1:100 dilution of RNase A/T1 mix. Every experiment contained the undigested probe and a yeast-RNA preparation as negative control. A [γ-32P]-ATP labeled 50 bp DNA ladder (New England Biolabs, Beverly, MA, USA) was used as a size reference on the gel.
Real-time PCR analysis
Here, ∼2 μg of total RNA was used for first strand cDNAs synthesis with random primers and Superscript II reverse transcriptase (Invitrogen) according to the manufacturer's instructions. Quantitative real-time RT-PCR was performed using SYBR green and an ABI 7500 sequence detection system instrument and software (Applied Biosystems, Foster City, CA, USA). Stability assays were performed in duplicates from two independent tissue culture or in vitro translation experiments. The firefly luciferase values obtained in stability assays were normalized to Renilla luciferase mRNA levels. Analysis of endogenous BACE1 mRNA from human brain and pancreas were performed with samples from Clontech (lot number 5 120 091 and 5 090 033, respectively). Values of endogenous BACE1 mRNA levels were normalized to the mRNA levels of the ribosomal protein L19. Primers used for detection of endogenous BACE1 mRNA levels were: hqPCR1657f (tgg agg gct tct acg ttg tct t) and hqPCR1742r (cct gaa ctc atc gtg cac atg).
RESULTS
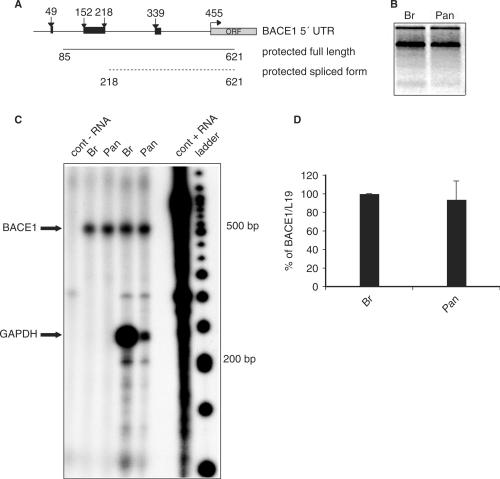
Since in a previous study (38) we suggested the presence of alternatively spliced variants of the 5′ UTR of BACE1 mRNA, we performed RNase protection assays using total RNA isolated from human brain and pancreas before starting a molecular characterization of the 5′ UTR. The riboprobe was designed to protect most of the transcript leader and 166 nt of the main open reading frame (ORF) (Figure 1A). The full-length BACE1 5′ UTR was expected to protect a fragment of 536 nt, while the putative alternatively spliced variant was expected to protect a fragment of 403 nt. Total RNA from human brain and pancreas was first loaded on an agarose gel to confirm that both RNA preparations are of comparable quality (Figure 1B). These RNAs were incubated with the BACE1 32P-labeled riboprobe alone or with a mixture containing the BACE1 riboprobe and a GAPDH riboprobe. An estimated 3–10 molar excess of the probe was used to avoid saturation and the higher signal from GADPH riboprobe in brain was considered an internal control of the procedure. In all reactions, only the band corresponding to the BACE1 full-length 5′ UTR was detected, questioning the existence of different BACE1 transcript leaders under normal physiological conditions (Figure 1C). Interestingly, the intensity of the signal protected by the BACE1 riboprobe was comparable in both tissues and quantitative RT-PCR confirmed that BACE1 mRNA is expressed in both tissues almost at the same level (Figure 1D). This finding contradicts earlier findings in which BACE1 mRNA expression in human pancreas was reported to be much higher than in human brain (3,4,39).
Figure 1.
BACE1 mRNA in human brain and pancreas contains the full-length 5′ UTR. (A) Schematic representation of the riboprobe. The riboprobe was designed to protect most of the transcript leader and 166 nt of the ORF. A full-length BACE1 5′ UTR was expected to protect a fragment of 536 nt (from 85 to 621), while the putative alternatively spliced variant was expected to protect a fragment of 403 nt (from 218 to 621). Arrows represent uAUGs, while black boxes represent uORFs. (B) Total RNAs from human brain (Br) and pancreas (Pan) have similar quality. Equal amounts of total RNAs were separate on agarose gel. (C) RNase protection assay. 10 μg of total RNA either from human brain or pancreas were incubated with 32P-labeled riboprobe for BACE1 alone, or BACE1 and GAPDH together. Protected signal for BACE1 (∼500 bp) and GAPDH (∼240 bp) are indicated with arrows. Cont − RNA = control reaction without RNA, Br = human brain, Pan = human pancreas, cont + RNA = control reaction with yeast RNA, ladder is γ-ATP labeled 50 bp DNA marker. (D) qPCR analysis. The values were normalized on L19 expression (L19 ratio brain/pancreas = 0.6).
Ruling out an alternative splicing event in the transcript leader, we moved toward the mechanism of translational regulation of BACE1 by performing a detailed deletion and mutation analysis of its 5′ UTR. Since it was previously reported that some regions of the BACE1 5′ UTR might have promoter activity (44,45), we employed a RNA transfection assay and in vitro translation analysis, techniques that allow exclusively monitoring of translation, hence avoiding putative transcriptional/post-transcriptional effects such as cryptic promoter activity and RNA splicing.
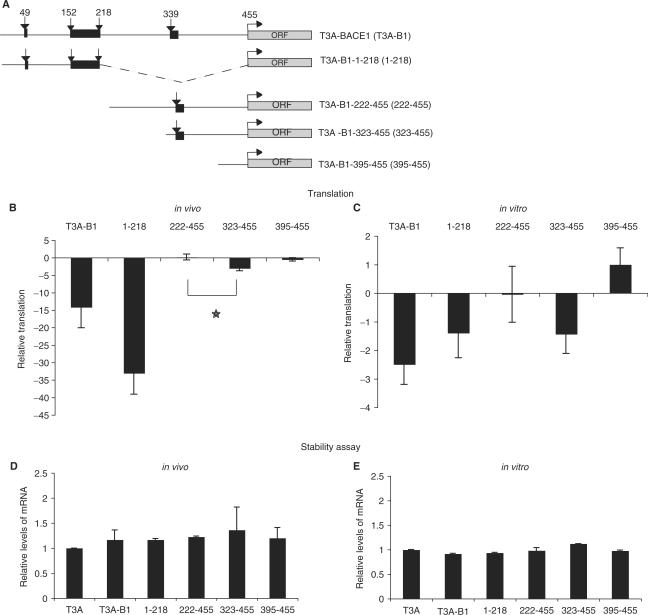
To identify regions of the BACE1 5′ UTR involved in translational regulation, we generated the deletion variants described in Figure 2A. These constructs were used to prepare in vitro-transcribed reporter mRNAs with the firefly luciferase ORF, containing a 5′ m7GpppG cap structure and a poly(A) tail of 98 A-residues. Firefly mRNAs together with a Renilla luciferase control reporter mRNA were used either in RNA-transfection experiments in HeLa cells or in an in vitro translation assay based on HeLa cell extracts. Deletion mutants showed a similar trend in both systems, although the effect on translation was more pronounced in vivo (compare Figure 2B and C). The 5′-half of the sequence (1-218) bearing three out of four uAUGs manifests a clear translational repression that is even stronger in vivo (compare T3A-B1 and 1-218 in Figure 2B and C). This enhanced repression might be ascribed to the fact that this construct lacks the putative stop codon of the uORF that originates from the second uAUG. This creates an out-of-frame overlap of that uORF with the main ORF, thereby enhancing the possibility that the ribosomal scanning misses the start codon of the reporter gene. Surprisingly, the 3′-half of the sequence (222-455) did not show any repression (compare T3A-B1 and 222-455 in Figure 2B and C). Since it has been previously reported that the 3′-half of the BACE1 5′ UTR might have strong inhibitory effect via highly structured regions (39), we performed a more detailed deletion analysis of this region by generating constructs 323-455 and 395-455 (Figure 2A). Interestingly, the reporter mRNA-bearing region 323-455 showed again an inhibitory effect that was more evident in vitro when compared with the full-length reporter, while the 395-455 reporters did not show any significant inhibitory effect as expected (Figure 2B and C, compare T3A-B1 with 323-455 and 395-455). These effects on translational efficiency could not be explained by differences in mRNA stability, since qPCR analysis shows that the transcripts are equally stable under given conditions (Figure 2D and E). The in vivo analysis of deletion mutants reveals that the BACE1 5′ UTR represses translation by sequence elements that are mainly present in the first half of the sequence. Interestingly, in addition to those regulatory elements, the deletion analysis revealed a possible presence of a translational activator: the comparison of translation efficiency of reporter mRNAs bearing the region 222-455 and 323-455 suggests a possible positive effect on translation by the intermediate region 222-323.
Figure 2.
Different regions of BACE1 5′ UTR affect translation in a positive or negative manner. (A) Schematic representation of mRNAs bearing different BACE1 5′ UTR deletion mutants and firefly luciferase as reporter gene. Black boxes represent uORFs, and arrows uAUGs. (B) RNA transfections of different deletion mutants in HeLa cell line together with Renilla control reporter mRNA. The results are presented in fold variation. Error bars denote the standard deviation from the mean of at least three independent experiments. The star indicates a P < 0.05. (C) HeLa cell extracts were co-programmed with firefly reporter mRNAs and Renilla reporter mRNA, used to account for differences in translatability of HeLa extracts. (D and E) Reporter mRNAs stability is not affected either in vivo or in vitro. Total RNA, isolated from RNA-transfected cells (B) and in vitro translation reactions (C) at the beginning and end of experiment (upon 24 h for RNA transfections or 30 min for in vitro translations, respectively) was transcribed into cDNA and amplified by qPCR with specific primers for the reporter genes. Rate of decay was calculated for each reporter mRNA and the values were normalized to the control.
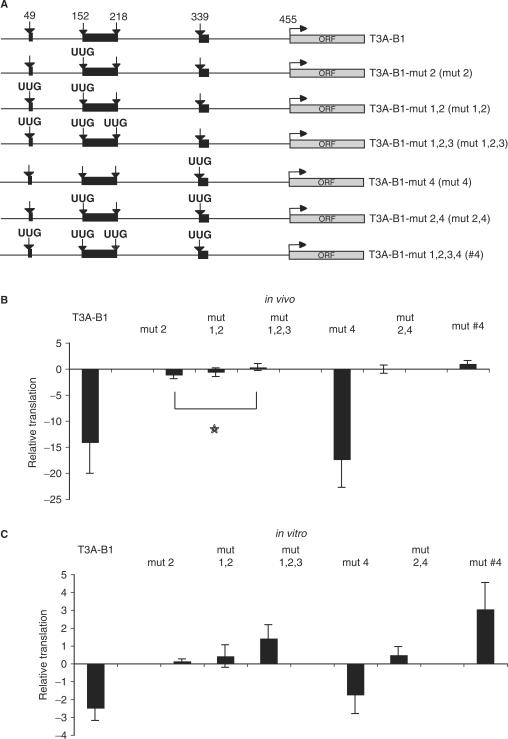
To address the function of uAUGs in translational regulation by BACE1 5′ UTR in more detail, we performed a mutational analysis (Figure 3A). According to Kozak's model (31), the first uAUG in BACE1 5′ UTR is in an optimal context for translational initiation, since it contains a purine at position −3, and a G at position +4; however, an U at position +5 can reduce this optimal context (46–48). The third and fourth uAUGs contain pyrimidines at both positions −3 and +4 and therefore are not likely to be in an optimal context for efficient translation initiation. The context of the second uAUG has a G at position −3, and a pyrimidine at position +4, thus, it is in a good, but not optimal context for translation initiation.
Figure 3.
uAUGs are responsible for low translational efficiency driven by BACE1 5′ UTR. (A) Schematic representation of reporter mRNAs bearing mutations of uAUGs. Arrows represent uAUGs or UUGs when mutated; black boxes symbolize uORFs. (B) Transfection of reporter mRNAs into HeLa cell line. Renilla mRNA was used to normalize for transfection efficiency. Results are presented in fold variation, and the error bars present the mean of at least three independent experiments. Star indicates a P < 0.05 (C) HeLa cell extracts were programmed with reporter mRNAs as in (B). The data are normalized and presented in the same manner as in (B).
Based on this prediction and on the results from the deletion analysis, we started the evaluation of the uAUGs by mutating the second uAUG into UUG (Figure 3A). Translation of this reporter in vivo and in vitro gave surprising results since this mutation (mut 2) diminished translational repression from ∼14 to ∼2-fold in vivo, and it was enough to completely bypass translational repression in vitro (Figure 3B and C, compare T3A-B1 and mut 2), suggesting that the second uAUG indeed is efficiently recognized. Additional mutation of the first and the third uAUG increased further translational efficiency, but at least in vivo, the increase was very modest when compared with the effect of the mutation of the second uAUG alone (Figure 3B and C, compare T3A-B1 with mut 2, mut 1,2 and mut 1,2,3). The role of the fourth uAUG was intriguing, since the deletion mutant that starts in its close proximity (323-455) showed a lower translational efficiency than the reporter mRNAs bearing the putative positive regulatory element (i.e. 222-455), suggesting that this uAUG may contribute to the translational repression. However, mutation of this uAUG alone did not have any significant impact on translational efficiency in vivo and in vitro (Figure 3B and C, compare T3A-B1 and mut 4). Nonetheless, mutation of the fourth uAUG in addition to the second uAUG increased additionally translational efficiency compared with the effect of the mutated second uAUG alone (Figure 3B and C, compare mut 2 with mut 2,4).
When we mutated all four uAUGs, the translational efficiency was 2–4-fold higher than control, in vivo and in vitro (Figure 3B and C, see mut #4), in spite of the fact that the BACE1 5′ UTR has a very high GC content and a strong predicted secondary structure (39).
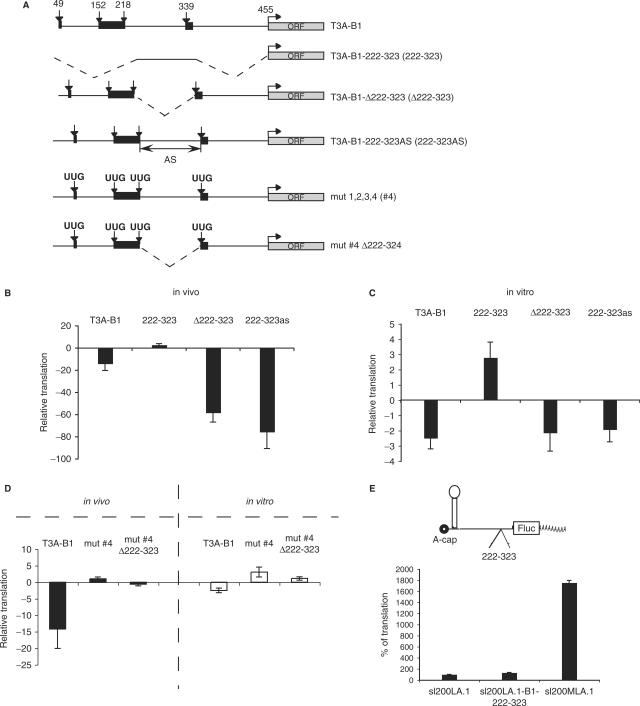
We hypothesized that the good translatability of the BACE1 5′ UTR without uAUGs could be due to the presence of the positive regulatory element in the region 222-323. This region could play a role in re-initiation efficiency by acting as a spacer between the end of the translation of the uORF and the beginning of the main ORF and/or because of the sequence that surround the stop codon of the uORF. Therefore, we evaluated the role of the region 222-323 in translational regulation in the context of both the wild-type BACE1 5′ UTR, in order to appreciate its effect on re-initiation, and the 5′ UTR without uAUGs. We generated a number of additional mutants (Figure 4A), which were used in both experimental systems, in vivo and in vitro. The translational efficiency driven by the region 222-323 alone was ∼3-fold higher when compared to the control mRNA (Figure 4B and C, see 222-323). This effect was also analyzed in the context of the full-length BACE1 5′ UTR. In RNA-transfection experiments, deletion of the 222-323 region led to an additional decrease in translational efficiency (∼4-fold when compared with the wild-type sequence) down to ∼60-fold below control (Figure 4B, see Δ222-323 and compare with T3A-B1). Replacement in antisense orientation led to further decrease of translation efficiency (Figure 4B, see 222-323as), showing that also the primary sequence plays an important role. Interestingly, deletion or antisense orientation of the region 222-323 in the context of full-length BACE1 5′ UTR did not influence translation of the reporter gene in experiments performed in vitro (Figure 4C).
Figure 4.
Effect of the 222-323 region of BACE1 5′ UTR to translation. (A) Schematic representation of used reporter mRNAs. Arrows represent uAUGs or UUGs when mutated; black boxes symbolize uORFs; dashed lines delimit deleted sequences; AS––antisense orientation. (B) The region 222-323 affects strongly translation of wild-type BACE1 5′ UTR in vivo. Transfection of reporter mRNAs into HeLa cell line. Renilla mRNA was used to normalize for transfection efficiency. Results are presented in fold variation, and the error bars present the mean of at least three independent experiments. (C) The region 222-323 does not affect translation of wild-type BACE1 5′ UTR in vitro. Renilla mRNA was used to normalize differences in translatability of different HeLa extract aliquots. Results are presented in fold variation, and the error bars present the mean of at least three independent experiments. (D) The stimulatory effect of the region 222-323 is uAUG independent in vivo and in vitro. Black bars represent RNA transfection and white bars are in vitro translation experiments. Renilla mRNA was used to normalize for transfection efficiency and translatability of different HeLa extract aliquots. Results are presented in fold variation, and the error bars present the mean of at least three independent experiments. (E) The region 222-323 does not act as IRES in vitro, in HeLa cell extract. Schematic representation of the IRES-containing reporter mRNAs is shown above the graph. HeLa cell extracts were programmed with ApppG-capped c-myc (sl200MLA.1) or BACE1-222-323 reporter mRNAs bearing a stabile hairpin upstream of a spacer sequence and the IRES (sl200A.1-B1-222-323). A reporter mRNA lacking an IRES element upstream of the firefly luciferase ORF (sl200LA.1) was used as a control. Results are presented as percentage of translation. Error bars denote the standard deviation from mean of at least three independent experiments.
To see whether the positive effect of the region 222-323 in vivo is uAUG dependent, we deleted this region from the construct in which all of the four uAUGs were mutated (Figure 4A, mut #4 Δ222-323). Deletion of this region revealed a decrease in translational efficiency, (3-fold in vivo and 2-fold in vitro), indicating that the positive effect of the region 222-323 on translation is, at least partially, uAUG independent (Figure 4D, compare mut #4 with mut #4 Δ222-323, both in vivo and in vitro), i.e. not only due to the positive effect that this sequence may exert on re-initiation efficiency. Although the decrease in translation might look similar in the two conditions (4-fold when uAUGs are present and 3-fold when absent), it would be difficult to compare those values due to the different translation efficiency of the corresponding controls (T3A-B1 in Figure 4B and mut #4 in Figure 4D). Therefore, further investigation is needed to assess the precise extent of uAUG-dependent and -independent effects. One possible explanation for the uAUG-independent positive effect of this region could be that it can act as an IRES. However, the IRES activity within this region was undetectable (Figure 4E). Furthermore, we wanted to test if there is a specific interaction between RNA-binding protein(s) and the region 222-323 in an electro-mobility shift assay. Under the standard experimental conditions we applied, we could not detect specific interactions, possibly due to the high GC content of BACE1 5′ UTR (data not shown). Further testing is required to evaluate the presence of any trans-acting factor able to bind this region.
DISCUSSION
We have previously demonstrated that BACE1 translation is inhibited by uAUGs in its 5′ UTR (38). Other studies suggested different mechanisms involving inhibition by highly structured regions (39) and the possibility to bypass inhibitory regions in a cell-specific way by ribosomal shunting (40). To shed more light on the molecular mechanism of translational regulation of BACE1, we performed a detailed analysis of its 5′ UTR.
We have previously reported an alternative splicing within BACE1 5′ UTR (38). Since spliced forms of the 5′ UTR of mRNAs have been reported to control protein translation (31), we re-evaluated our findings. However, the data obtained in this study question the existence of BACE1 5′ UTRs different from the full-length form under normal physiological conditions.
In the previous report, we used the MVA-T7pol vaccinia virus expression system, a strategy that allows the transcription of gene of interest to occur solely in the cytosol (49). In spite of this, the robust transcription and translation in cytosol driven by the virus can override the physiological mechanisms of translational regulation. For this reason, we preferred to use RNA transfection and in vitro translation assay in this study, i.e. techniques that mimic better the physiological conditions and should, in principle, allow us to monitor exclusively translational regulation and avoiding the interference of transcriptional or additional post-transcriptional events.
The BACE1 5′ UTR has a GC content of 77% and it is predicted to form very strong secondary structures (39). Secondary structures can play an important role in translational regulation. A hairpin structure with a free energy <−30 kcal/mol situated close to the cap structure can impede scanning by interfering with the binding of the small ribosomal subunit complex to the mRNA (50), while a hairpin with a free energy <−50 kcal/mol can inhibit scanning of the small ribosomal subunit (50–52). In the case of BACE1, our results clearly imply that the high GC content of BACE1 5′ UTR, predicted to represent a constitutive translational barrier, does not hinder the scanning process of the small ribosomal subunit complex as previously suggested (39). Instead, our data suggest that uAUGs fully account for the observed inhibition of BACE1 translation. It is noteworthy that we mutated uAUGs into UUGs, i.e. we changed a purine with a pyrimidine. Even though this kind of substitution might produce subtle structural changes in the transcript, it is unlikely that such mutations (as in case of the second uAUG) would abolish a very strong secondary structure that does not allow the small ribosomal subunit to go through. Indeed, bioinformatic tools (such as MFOLD) did not predict any significant structural change upon these substitutions.
The progressive rescue of BACE1 translation by cumulative uAUG mutations also questions a possible shunting mechanism in which the small ribosomal complex could bypass inhibitory segments of the 5′ UTR. In our analysis, instead, the contribution of all uAUGs to translational efficiency, with the strongest impact of the second uAUG, is more consistent with the scanning model of translational initiation. The possibility that shunting might affect BACE1 levels in pathological conditions is certainly intriguing, but it would require further investigation.
The presence of uAUGs is usually inhibitory to translation of the main ORF. uORFs can regulate mRNA stability and translational efficiency in different ways (53). Our stability assay on deletion-containing mRNA reporters suggests that the 5′ UTR of BACE1 does not affect translation efficiency by changes in stability of the mRNA. More notably, the mutational analysis of uAUGs shows that all four uAUGs affect translation, but to a different extent, being the second uAUG efficiently recognized, while the others probably skipped by leaky scanning. Interestingly, to make the construct 1-218, we deleted the last nucleotide in the stop codon of the second uORF, thereby we created an out of frame, overlapping uORF. Efficient recognition of the second uAUG (or even the third in-frame uAUG) and translation of one of these newly created overlapping uORF(s) could explain the observed low translational efficiency of this reporter mRNA in vivo. The rescue due to mutation of the second uAUG revealed that it is efficiently recognized, and the low-translational efficiency of construct 1-218, indeed, could be ascribed to the translation of the newly created overlapping uORF. Taking together these results, we propose that BACE1 translation is mainly limited by the low re-initiation frequency after translation of the second uORF, while leaky scanning of the other uAUGs seems rather efficient and does not give a major contribution to the translational inhibition of BACE1. These data on the key role of the second uAUG are consistent with a recent report on the same subject (45), while the different results obtained by other groups (39,40) might be explained by taking into account that the high GC content of the BACE1 5′ UTR hides a cryptic promoter activity (38) that, albeit weak, might influence results in certain conditions or in specific cell types.
For the process of re-initiation, the length and the nucleotide sequence between the termination codon of the translated uORF and the downstream initiation site can be critical (54,55). Re-initiation can be influenced by the nucleotide sequence around the termination codon, up to 12 nt downstream. The decrease in translational efficiency observed upon deletion of the 222-323 region might indicate the importance of this region as a spacer between the translated uORF and the downstream AUG. On the other hand, the construct with the 222-323 region in antisense orientation showed an even stronger translational repression suggesting that the primary sequence of this region play a more important role than just as spacer for the process of re-initiation, i.e. it could function as positive element by keeping high the re-initiation efficiency, acting at the level of the termination event of the uORF. Together with the stimulation of translation obtained with the 222-323 construct, these results highlight this region as an element that might act positively in cis on the transcript and/or as a binding site for positive trans-acting factors. Further work will address this issue in light of the higher translation efficiency that has been detected in primary neuronal cultures when compared with other cell types, and considering the increase in translation efficiency that can be obtained in astrocytes upon activation with cytokines (38). Furthermore, even if our data are not in favor of a specific model to explain the striking difference in BACE1 translation between human brain and pancreas, they keep open the two most notable possibilities, i.e. the expression in pancreas of high levels of a trans-acting factor (either a protein or a miRNA) that inhibits translation of BACE1, and the existence in brain of a regulator/mechanism that allows a partial skipping of the uAUG block occurring in the other tissues. The presence of multiple variants of the BACE1 transcript, ascribed to alternative polyadenylation signals, may add further complexity to this scenario.
ACKNOWLEDGEMENTS
M.M. was guest in Hentze's laboratory for a year and thanks the other members of the lab for useful discussions and help. The work was carried out within the framework of the Italian Ministry of Research Center of Excellence in Physiopathology of Cell Differentiation. Financial support was from the EU contract LSHM-CT-2003-503330 (APOPIS) to D.Z., and the Italian Ministry of Research (PRIN project 2006054051 to F.G. and FIRB project RBLA03AF28_001 to D.Z.). Funding to pay the Open Access publication charges for this article was provided by the Italian Institute of Technology.
Conflict of interest statement. None declared.
REFERENCES
- 1.Hussain I., Powell D., Howlett D.R., Tew D.G., Meek T.D., Chapman C., Gloger I.S., Murphy K.E., Southan C.D., et al. Identification of a novel aspartic protease (Asp 2) as beta-secretase. Mol. Cell Neurosci. 1999;14:419–427. doi: 10.1006/mcne.1999.0811. [DOI] [PubMed] [Google Scholar]
- 2.Sinha S., Anderson J.P., Barbour R., Basi G.S., Caccavello R., Davis D., Doan M., Dovey H.F., Frigon N., et al. Purification and cloning of amyloid precursor protein beta-secretase from human brain. Nature. 1999;402:537–540. doi: 10.1038/990114. [DOI] [PubMed] [Google Scholar]
- 3.Vassar R., Bennett B.D., Babu-Khan S., Kahn S., Mendiaz E.A., Denis P., Teplow D.B., Ross S., Amarante P., et al. Beta-secretase cleavage of Alzheimer's amyloid precursor protein by the transmembrane aspartic protease BACE. Science. 1999;286:735–741. doi: 10.1126/science.286.5440.735. [DOI] [PubMed] [Google Scholar]
- 4.Yan R., Bienkowski M.J., Shuck M.E., Miao H., Tory M.C., Pauley A.M., Brashier J.R., Stratman N.C., Mathews W.R., et al. Membrane-anchored aspartyl protease with Alzheimer's disease beta-secretase activity. Nature. 1999;402:533–537. doi: 10.1038/990107. [DOI] [PubMed] [Google Scholar]
- 5.Lin X., Koelsch G., Wu S., Downs D., Dashti A., Tang J. Human aspartic protease memapsin 2 cleaves the beta-secretase site of beta-amyloid precursor protein. Proc. Natl Acad. Sci. USA. 2000;97:1456–1460. doi: 10.1073/pnas.97.4.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu X., Hicks C.W., He W., Wong P., Macklin W.B., Trapp B.D., Yan R. Bace1 modulates myelination in the central and peripheral nervous system. Nat. Neurosci. 2006;9:1520–1525. doi: 10.1038/nn1797. [DOI] [PubMed] [Google Scholar]
- 7.Willem M., Garratt A.N., Novak B., Citron M., Kaufmann S., Rittger A., DeStrooper B., Saftig P., Birchmeier C., et al. Control of peripheral nerve myelination by the beta-secretase BACE1. Science. 2006;314:664–666. doi: 10.1126/science.1132341. [DOI] [PubMed] [Google Scholar]
- 8.Li Q., Sudhof T.C. Cleavage of amyloid-beta precursor protein and amyloid-beta precursor-like protein by BACE 1. J. Biol. Chem. 2004;279:10542–10550. doi: 10.1074/jbc.M310001200. [DOI] [PubMed] [Google Scholar]
- 9.Pastorino L., Ikin A.F., Lamprianou S., Vacaresse N., Revelli J.P., Platt K., Paganetti P., Mathews P.M., Harroch S., et al. BACE (beta-secretase) modulates the processing of APLP2 in vivo. Mol. Cell Neurosci. 2004;25:642–649. doi: 10.1016/j.mcn.2003.12.013. [DOI] [PubMed] [Google Scholar]
- 10.Kitazume S., Nakagawa K., Oka R., Tachida Y., Ogawa K., Luo Y., Citron M., Shitara H., Taya C., et al. In vivo cleavage of alpha2,6-sialyltransferase by Alzheimer beta-secretase. J. Biol. Chem. 2005;280:8589–8595. doi: 10.1074/jbc.M409417200. [DOI] [PubMed] [Google Scholar]
- 11.Lichtenthaler S.F., Dominguez D.I., Westmeyer G.G., Reiss K., Haass C., Saftig P., De Strooper B., Seed B. The cell adhesion protein P-selectin glycoprotein ligand-1 is a substrate for the aspartyl protease BACE1. J. Biol. Chem. 2003;278:48713–48719. doi: 10.1074/jbc.M303861200. [DOI] [PubMed] [Google Scholar]
- 12.Wong H.K., Sakurai T., Oyama F., Kaneko K., Wada K., Miyazaki H., Kurosawa M., De Strooper B., Saftig P., et al. Beta subunits of voltage-gated sodium channels are novel substrates of beta-site amyloid precursor protein-cleaving enzyme (BACE1) and gamma-secretase. J. Biol. Chem. 2005;280:23009–23017. doi: 10.1074/jbc.M414648200. [DOI] [PubMed] [Google Scholar]
- 13.von Arnim C.A., Kinoshita A., Peltan I.D., Tangredi M.M., Herl L., Lee B.M., Spoelgen R., Hshieh T.T., Ranganathan S., et al. The low density lipoprotein receptor-related protein (LRP) is a novel beta-secretase (BACE1) substrate. J. Biol. Chem. 2005;280:17777–17785. doi: 10.1074/jbc.M414248200. [DOI] [PubMed] [Google Scholar]
- 14.Cai H., Wang Y., McCarthy D., Wen H., Borchelt D.R., Price D.L., Wong P.C. BACE1 is the major beta-secretase for generation of Abeta peptides by neurons. Nat. Neurosci. 2001;4:233–234. doi: 10.1038/85064. [DOI] [PubMed] [Google Scholar]
- 15.Luo Y., Bolon B., Kahn S., Bennett B.D., Babu-Khan S., Denis P., Fan W., Kha H., Zhang J., et al. Mice deficient in BACE1, the Alzheimer's beta-secretase, have normal phenotype and abolished beta-amyloid generation. Nat. Neurosci. 2001;4:231–232. doi: 10.1038/85059. [DOI] [PubMed] [Google Scholar]
- 16.Roberds S.L., Anderson J., Basi G., Bienkowski M.J., Branstetter D.G., Chen K.S., Freedman S.B., Frigon N.L., Games D., et al. BACE knockout mice are healthy despite lacking the primary beta-secretase activity in brain: implications for Alzheimer's disease therapeutics. Hum. Mol. Genet. 2001;10:1317–1324. doi: 10.1093/hmg/10.12.1317. [DOI] [PubMed] [Google Scholar]
- 17.De Strooper B. Aph-1, Pen-2, and nicastrin with presenilin generate an active gamma-secretase complex. Neuron. 2003;38:9–12. doi: 10.1016/s0896-6273(03)00205-8. [DOI] [PubMed] [Google Scholar]
- 18.Haass C. Take five—BACE and the gamma-secretase quartet conduct Alzheimer's amyloid beta-peptide generation. EMBO J. 2004;23:483–488. doi: 10.1038/sj.emboj.7600061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hardy J., Selkoe D.J. The amyloid hypothesis of Alzheimer's disease: progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 20.Selkoe D.J. Cell biology of protein misfolding: the examples of Alzheimer's and Parkinson's diseases. Nat. Cell Biol. 2004;6:1054–1061. doi: 10.1038/ncb1104-1054. [DOI] [PubMed] [Google Scholar]
- 21.Marcinkiewicz M., Seidah N.G. Coordinated expression of beta-amyloid precursor protein and the putative beta-secretase BACE and alpha-secretase ADAM10 in mouse and human brain. J. Neurochem. 2000;75:2133–2143. doi: 10.1046/j.1471-4159.2000.0752133.x. [DOI] [PubMed] [Google Scholar]
- 22.Huse J.T., Byant D., Yang Y., Pijak D.S., D'Souza I., Lah J.J., Lee V.M., Doms R.W., Cook D.G. Endoproteolysis of beta-secretase (beta-site amyloid precursor protein-cleaving enzyme) within its catalytic domain. A potential mechanism for regulation. J. Biol. Chem. 2003;278:17141–17149. doi: 10.1074/jbc.M213303200. [DOI] [PubMed] [Google Scholar]
- 23.Laird F.M., Cai H., Savonenko A.V., Farah M.H., He K., Melnikova T., Wen H., Chiang H.C., Xu G., et al. BACE1, a major determinant of selective vulnerability of the brain to amyloid-beta amyloidogenesis, is essential for cognitive, emotional, and synaptic functions. J. Neurosci. 2005;25:11693–11709. doi: 10.1523/JNEUROSCI.2766-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Velliquette R.A., O’Connor T., Vassar R. Energy inhibition elevates beta-secretase levels and activity and is potentially amyloidogenic in APP transgenic mice: possible early events in Alzheimer's disease pathogenesis. J. Neurosci. 2005;25:10874–10883. doi: 10.1523/JNEUROSCI.2350-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fukumoto H., Cheung B.S., Hyman B.T., Irizarry M.C. Beta-secretase protein and activity are increased in the neocortex in Alzheimer disease. Arch. Neurol. 2002;59:1381–1389. doi: 10.1001/archneur.59.9.1381. [DOI] [PubMed] [Google Scholar]
- 26.Holsinger R.M.D., McLean C.A., Beyreuther K., Masters C.L., Evin G. Increased expression of the amyloid precursor beta-secretase in Alzheimer's disease. Ann. Neurol. 2002;51:783–786. doi: 10.1002/ana.10208. [DOI] [PubMed] [Google Scholar]
- 27.Tyler S.J., Dawbarn D., Wilcock G.K., Allen S.J. Alpha- and beta-secretase: profound changes in Alzheimer's disease. Biochem. Biophys. Res. Commun. 2002;299:373–376. doi: 10.1016/s0006-291x(02)02635-9. [DOI] [PubMed] [Google Scholar]
- 28.Yang L.B., Lindholm K., Yan R., Citron M., Xia W., Yang X.L., Beach T., Sue L., Wong P., et al. Elevated beta-secretase expression and enzymatic activity detected in sporadic Alzheimer disease. Nat. Med. 2003;9:3–4. doi: 10.1038/nm0103-3. [DOI] [PubMed] [Google Scholar]
- 29.Wilkie G.S., Dickson K.S., Gray N.K. Regulation of mRNA translation by 5′- and 3′-UTR-binding factors. Trends Biochem. Sci. 2003;28:182–188. doi: 10.1016/S0968-0004(03)00051-3. [DOI] [PubMed] [Google Scholar]
- 30.Gebauer F., Hentze M.W. Molecular mechanisms of translational control. Nat. Rev. Mol. Cell Biol. 2004;5:827–835. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kozak M. Pushing the limits of the scanning mechanism for initiation of translation. Gene. 2002;299:1–34. doi: 10.1016/S0378-1119(02)01056-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene. 1999;234:187–208. doi: 10.1016/s0378-1119(99)00210-3. [DOI] [PubMed] [Google Scholar]
- 33.Kozak M. Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene. 2005;361:13–37. doi: 10.1016/j.gene.2005.06.037. [DOI] [PubMed] [Google Scholar]
- 34.Ryabova L.A., Pooggin M.M., Hohn T. Viral strategies of translation initiation: ribosomal shunt and reinitiation. Prog. Nucleic Acid Res. Mol. Biol. 2002;72:1–39. doi: 10.1016/S0079-6603(02)72066-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hellen C.U., Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 2001;15:1593–1612. doi: 10.1101/gad.891101. [DOI] [PubMed] [Google Scholar]
- 36.Stoneley M., Willis AE. Cellular internal ribosome entry segments: structures, trans-acting factors and regulation of gene expression. Oncogene. 2004;23:3200–3207. doi: 10.1038/sj.onc.1207551. [DOI] [PubMed] [Google Scholar]
- 37.Baird S.D., Turcotte M., Korneluk R.G., Holcik M. Searching for IRES. RNA. 2006;12:1755–1785. doi: 10.1261/rna.157806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Pietri Tonelli D., Mihailovich M., Di Cesare A., Codazzi F., Grohovaz F., Zacchetti D. Translational regulation of BACE-1 expression in neuronal and non-neuronal cells. Nucleic Acids Res. 2004;32:1808–1817. doi: 10.1093/nar/gkh348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lammich S., Schobel S., Zimmer A.K., Lichtenthaler S.F., Haass C. Expression of the Alzheimer protease BACE1 is suppressed via its 5′-untranslated region. EMBO Rep. 2004;5:620–625. doi: 10.1038/sj.embor.7400166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rogers G.W., Jr, Edelman G.M., Mauro V.P. Differential utilization of upstream AUGs in the beta-secretase mRNA suggests that a shunting mechanism regulates translation. Proc. Natl. Acad. Sci. USA. 2004;101:2794–2799. doi: 10.1073/pnas.0308576101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iizuka N., Najita L., Franzusoff A., Sarnow P. Cap-dependent and cap-independent translation by internal initiation of mRNAs in cell extracts prepared from Saccharomyces cerevisiae. Mol. Cell. Biol. 1994;14:7322–7330. doi: 10.1128/mcb.14.11.7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thoma C., Bergamini G., Galy B., Hundsdoerfer P., Hentze M.W. Enhancement of IRES-mediated translation of the c-myc and BiP mRNAs by the poly(A) tail is independent of intact eIF4G and PABP. Mol. Cell. 2004;15:925–935. doi: 10.1016/j.molcel.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 43.Bergamini G., Preiss T., Hentze M.W. Picornavirus IRESes and the poly(A) tail jointly promote cap-independent translation in a mammalian cell-free system. RNA. 2000;6:1781–1790. doi: 10.1017/s1355838200001679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ge Y.W., Maloney B., Sambamurti K., Lahiri D.K. Functional characterization of the 5′ flanking region of the BACE gene: identification of a 91 bp fragment involved in basal level of BACE promoter expression. FASEB J. 2004;18:1037–1039. doi: 10.1096/fj.03-1379fje. [DOI] [PubMed] [Google Scholar]
- 45.Zhou W., Song W. Leaky scanning and reinitiation regulate BACE1 gene expression. Mol. Cell Biol. 2006;26:3353–3364. doi: 10.1128/MCB.26.9.3353-3364.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kozak M. Recognition of AUG and alternative initiator codons is augmented by G in position +4 but is not generally affected by the nucleotides in positions +5 and +6. EMBO J. 1997;16:2482–2492. doi: 10.1093/emboj/16.9.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sloan J., Kinghorn J.R., Unkles S.E. The two subunits of human molybdopterin synthase: evidence for a bicistronic messenger RNA with overlapping reading frames. Nucleic Acids Res. 1999;27:854–858. doi: 10.1093/nar/27.3.854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stallmeyer B., Drugeon G., Reiss J., Haenni A.L., Mendel R.R. Human molybdopterin synthase gene: identification of a bicistronic transcript with overlapping reading frames. Am. J. Hum. Genet. 1999;64:698–705. doi: 10.1086/302295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sutter G., Ohlmann M., Erfle V. Non-replicating vaccinia vector efficiently expresses bacteriophage T7 RNA polymerase. FEBS Lett. 1995;371:9–12. doi: 10.1016/0014-5793(95)00843-x. [DOI] [PubMed] [Google Scholar]
- 50.Kozak M. Circumstances and mechanisms of inhibition of translation by secondary structure in eucaryotic mRNAs. Mol. Cell. Biol. 1989;9:5134–5142. doi: 10.1128/mcb.9.11.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 52.Gray N.K., Hentze M.W. Regulation of protein synthesis by mRNA structure. Mol. Biol. Rep. 1994;19:195–200. doi: 10.1007/BF00986961. [DOI] [PubMed] [Google Scholar]
- 53.Morris D.R., Geballe A.P. Upstream open reading frames as regulators of mRNA translation. Mol. Cell. Biol. 2000;20:8635–8642. doi: 10.1128/mcb.20.23.8635-8642.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kozak M. Effects of intercistronic length on the efficiency of reinitiation by eucaryotic ribosomes. Mol. Cell. Biol. 1987;7:3438–3445. doi: 10.1128/mcb.7.10.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grant C.M., Hinnebusch A.G. Effect of sequence context at stop codons on efficiency of reinitiation in GCN4 translational control. Mol. Cell. Biol. 1994;14:606–618. doi: 10.1128/mcb.14.1.606. [DOI] [PMC free article] [PubMed] [Google Scholar]