Abstract
Using an in vitro selection, we have obtained oligonucleotide probes with high discriminatory power against multiple, similar nucleic acid sequences, which is often required in diagnostic applications for simultaneous testing of such sequences. We have tested this approach, referred to as iterative hybridizations, by selecting probes against six 22-nt-long sequence variants representing human papillomavirus, (HPV). We have obtained probes that efficiently discriminate between HPV types that differ by 3–7 nt. The probes were found effective to recognize HPV sequences of the type 6, 11, 16, 18 and a pair of type 31 and 33, either when immobilized on a solid support or in a reverse configuration, as well to discriminate HPV types from the clinical samples. This methodology can be extended to generate diagnostic kits that rely on nucleic acid hybridization between closely related sequences. In this approach, instead of adjusting hybridization conditions to the intended set of probe–target pairs, we ‘adjust’, through in vitro selection, the probes to the conditions we have chosen. Importantly, these conditions have to be ‘relaxed’, allowing the formation of a variety of not fully complementary complexes from which those that efficiently recognize and discriminate intended from non-intended targets can be readily selected.
INTRODUCTION
Specific interactions between macromolecules or between macromolecules and their low molecular weight ligands are crucial in all biological processes. The study of specific interactions requires the use of elaborate research tools from various field, such as molecular biology, medicine and molecular diagnostics. The specificity, describing the ability to discriminate between different ligands, is often equated with the affinity between the interacting molecules (1). A ligand of sufficiently high affinity is expected to be highly specific for its target, and the high-affinity/high-specificity paradigm was considered applicable to virtually all interacting systems (2). However, this paradigm does not apply easily to nucleic acid hybridization, which is fundamental to many techniques in molecular genetics. Although it is true that the interaction between nucleic acid strands becomes stronger with each additional base-pair and that a longer sequence precisely identifies one of many more possible nucleic acids than a shorter sequence, in practice, the ability of an oligo- or a polynucleotide to discriminate among closely related sequences through hybridization actually decreases as a function of sequence length. While there is a minimal threshold length that has to be respected to render the sequence unique within a complex genome, cross-hybridization of similar but non-identical sequences becomes more probable with longer sequences.
A number of computational tools have been developed to assist oligonucleotide probe design using different criteria and different algorithms for optimal probe selection (3–8). A large-scale microarray study showed that the majority of oligonucleotide probes did not behave according to commonly assumed hybridization models (9–11). Some authors suggested additional optimization strategies that involve empirical criteria (12–14). The possibilities of designing partly mismatched probes (15–17) or probes aiming the secondary structure of the target (8,18) are rarely considered in probe-design programs. Recently, Pozhitkov et al. (19) even recommended against the application of probe-design software tools that use thermodynamic parameters to assess probe quality in discriminating related sequences; their results implied that thermodynamic properties of oligonucleotide hybridization are far from fully understood. On the other hand, others have reported technical improvements by the application of computational methods in searching for active antisense oligonucleotides (20), or in the design of oligonucleotide microarrays (21).
To avoid false priming, when performing PCR, the annealing of primers is usually carried out at the highest possible temperature that maximizes the stability gap between complementary and mismatched duplexes. However, if sequences that have to be distinguished are similar, the difference in their binding energies is small, thereby restricting the window of adjustable experimental conditions that would allow discrimination between all potentially reacting species. Finding such conditions may become problematic in multiplex applications, when many probes and/or many targets are considered simultaneously (3,10,22,23). In such cases, there is a need to increase the room for simultaneous adjustment of these conditions. Or, as an alternative, one may try to adjust binding characteristics of the probes, given the hybridization conditions. How to create room for such adjustments that require structural manipulation of the probes? Rather than considering distinguishing between every variant of sequence length L out of 4L possible, we will only consider targeting variants that really exist, or are of diagnostic interest, and whose number is thus substantially smaller. By lowering the complexity of the investigated pool of target sequences, no more is there the need to maintain a full-sequence match. Rather than increasing the affinity to increase specificity, which has to be subsequently enforced by denaturing conditions, we propose to experimentally relax binding conditions to allow the reduction in affinity. This can be done either by manipulating the composition of the hybridization solution, by lowering the temperature or both. As a result, the number of possible structural/sequence variants of a probe(s) that can be used against a particular target sequence is substantially increased. In consequence, it allows choosing from this variety those probes that would maximize the energy gap between solicited, intended and unsolicited, non-intended complexes with their targets. In other words, resigning from fully or almost fully complementary complexes increases the number of probe–target combinations from which those that maximize the differences between all non-intended complexes can be chosen; this is of special interest in multiplex applications where a multiplicity of targets have to be simultaneously tested. Moreover, all such intended target–probe combinations can be chosen to respect minimal threshold in energy difference between all possible complexes to allow their efficient discrimination at a given experimental setting. This can be achieved by evolutionary approach, through affinity selection of probes by iterative hybridization (15).
In order to test this approach, we used human papillomavirus (HPV) as a model. The HPV family consists of >100 types, of which 39 types have been detected in the anogenital mucosa. Several of mucosal types have been associated with anogenital cancer, while other types are not carcinogenic and produce genital warts. A large variety of techniques are available for detection and subtyping of HPV types, but neither of them presents robust and complete solution to this problem (24–29). Here, we demonstrate the feasibility of this approach in designing probes against short DNA segments using target sequences representing six of the HPV subtypes that differ in only few nucleotide positions.
MATERIALS AND METHODS
Oligonucleotides
All oligonucleotides were synthesized by Integrated DNA Technologies (Coralville, IA). Target oligonucleotides corresponding to the so-called short PCR fragment, (SPF), described by Kleter et al. (25), consisted of 22-nt-long, HPV-type-specific segment, flanked by 22- and 23-nt-long PCR primers anchoring sequences as illustrated in Figure 1. These 67-nt-long oligomers were synthesized in two versions: non-modified and modified at their 5′ ends with biotin to allow for their immobilization on streptavidin-coated solid supports. The corresponding forward and reverse primers were used to amplify the synthetic targets or the corresponding HPV DNAs obtained from the clinical samples; these primers were modified at their 5′ ends by the addition of 6-carboxyflorescein (6-FAM) and the phosphate residue, respectively.
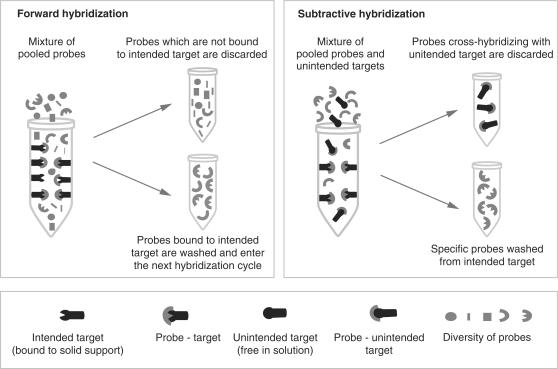
Figure 1.
Schematic presentation of iterative hybridizations, composed of two steps: forward (left) and subtractive hybridizations (right). Intended and non-intended targets, probes and complexes between them are drowned in the legend (down). Note that intended targets are attached to the solid support, while non-intended targets are free in solution.
Oligonucleotide probes were obtained by rounds of hybridizations starting with a mixture containing 22-nt-long random sequence segment embedded within constant sequence fragments to anchor PCR primers, ROM22: GCCTGTTGTGAGCCTCCTGTCGAA-(N)22-TTGAGCGTTTATTCTTGTCTCCCA, where N-corresponds to A, G, C and T (equimolar during synthesis). The following oligonucleotides were used to block the flanking primer-anchoring segments of ROM22: 5′ blocker, TTCGACAGGAGGCTCACAACAGGC and 3′ blocker, 5′P-TGGGAGACAAGAATAAACGCTCAA. The oligonucleotide GCCTGTTGTGAGCCTCCTGTCGAA, complementary to the 5′ blocker, was used as the forward primer and the 5′-phosphorylated 3′ blocker as the reverse primer, serving in PCR to amplify (i) pools of oligonucleotide mixtures [pooled probes (PPs)] obtained after each cycle of hybridization or (ii) particular probes [cloned probes (CPs)] from the plasmid clones carrying individual oligonucleotide sequences. Finally, target complements represented 22-nt-long complementary sequences of the HPV-type-specific SPF segments listed in Figure 1, all modified at 5′ end by the addition of 6-FAM.
Clinical samples
DNA was extracted from six patients containing single-type HPV. Initially, DNA was amplified with PGMY primers (30) and typed by sequencing.
Immobilization of target oligonucleotides
Streptavidin-coated tubes (Roche Diagnostics GmbH, Mannheim, Germany) and 96-well plates (Pierce Reacti-Bind Streptavidin Coated High Binding Capacity Black plates, Rockford, Il) were used for preparative and analytical purposes, respectively. After washing three times with 10 mM Tris-HCl, pH 7.5, 10 mM MgCl2 and 50 mM NaCl (TMN buffer), the tubes (or plates) were incubated with the predefined amount, between 1 and 100 pmol, of the 5′ biotinylated target oligonucleotide for at least 15 min, rinsed three times with TMN buffer and stored at 4°C until used.
Amplification and conversion of oligonucleotides into single-stranded DNA
Following hybridization, the bound oligonucleotides were dissociated from the target. These PPs were amplified by PCR: the reaction was carried out in a total volume of 50 µl containing 0.1–100 fmol of the template in the presence of 1 µM each of the primers (Figure 1), 100 μM each of dNTPs, 10 mM Tris-HCl, pH 8.3, 1.5 mM MgCl2, 50 mM KCl and 1 U of Taq polymerase (Platinum, InVitroGene). Typically, 27–30 PCR cycles were used, consisting each of 30 s at 94°C, 30 s at 53°C and 30 s at 72°C. The quantity and quality of PCR products were estimated by agarose gel-electrophoresis and/or by measuring the 6-FAM fluorescence of PCR amplicons, after eliminating the non-incorporated primers, using the Montage centrifuge filter device (Millipore, Billerica, MA). These products were rendered single stranded by incubation with 5 U of λ exonuclease (NEB, Boston, MA) that digests 5′-phosphorylated strand, for 30 min at 37°C, followed by 20 min at 65°C to inactivate the enzyme. The same procedure was used to produce single-stranded probes from PCR products from individual clones.
Hybridizations
The synthetic mixture of random oligonucleotides ROM22 (1 nmol) was used in the initial hybridization to obtain the first generation of PPs. In all subsequent hybridizations, the PPs from the preceding cycle were PCR amplified and converted to the single-stranded form. Typically, 10–50 pmol of single-stranded PP (0.05–0.25 µM) obtained in the previous cycle was mixed with two blocking oligonucleotides to obtain 0.5 µM each, in 200 μl of TMN buffer and heated to 90°C. This solution was subsequently transferred to tubes containing pre-bound biotinylated targets, then cooled down to the ambient temperature, 22–24°C, and left for at least 4 h at this temperature. The tubes were then rinsed three times with TMN buffer, and the probes that remained bound to the targets were washed off by incubation at 90°C in 200 μl of water for 2 min. There was 1 pmol of the added target per tube, except during the first hybridization when 100 pmol were added (however, the effective amount of the available target for binding was less, see below). Forward hybridizations were followed by subtractive hybridizations carried as detailed above, but in the presence of 0.5 µM (total) of the non-desired oligonucleotide targets (i.e. other than the immobilized target).
Binding experiments
Target oligonucleotides, representing SPF of different HPV types, were immobilized in separate wells of 96-well plates (under saturation with target, the resulting effective amount of the target per well was ∼17 pmol, when measured as its amount available for binding with its 6-FAM-labeled complement). PPs or CPs (0.1–0.5 µM, converted to single strands) were incubated with immobilized targets, in the presence of 1 µM each of the block oligonucleotides, in 100 μl of TMN buffer for 4 h at 22°C. The wells were rinsed three times with 100 μl of TMN buffer and the bound 6-FAM fluorescence (in relative fluorescence units, RFUs) was measured directly in Spectra MAX Gemini XS (22°C, λex = 485 nm and λem = 538 nm). The binding experiments with the 6-FAM labeled, 22-nt-long target complements were carried out using the same protocol, except that blockers were not added.
Competitive binding
The binding was measured as above, with 6-FAM-labeled oligonucleotides (PP, CP or complements) kept at a constant concentration of 10–50 pmol/well (0.1–0.5 µM), in the presence of increasing concentrations, from 0 to 10 µM, of target competitor. The latter was the non-biotinylated SPF oligonucleotide, either identical with the immobilized target (homologous competitive binding), or representing the SPF sequence of another HPV type (heterologous competitive binding). The EC50 values were estimated from the data according to the equation calculated from using the GraphPad Prism Software (Version 4).
Cloning and sequencing of individual probes
Cloning the probes from the PPs was done using TOPO TA Cloning kit (Invitrogen, CA). Typically, 20 positive clones were selected using X-Gal/IPTG-based-colorimetric reaction, following the manufacturer's protocol. The M13 forward and reverse primers were used to confirm the presence of the insert and to ‘extract’ it for subsequent direct sequence determination using LiCor apparatus (Lincoln, NE). In turn, the resulting CPs were produced by PCR using ROM22 primers and tested for binding.
Reverse format hybridization
The sequences of the CPs with the best signal-to-noise ratio were chemically synthesized Integrated DNA Technologies (IDT) with a biotin moiety at their 5′ end. Individual 5′ biotinylated probes were bound (100 pmol) to streptavidin-coated plates. The HPV SPF was generated by PCR either from the typed DNA obtained from clinical samples or from the synthetic target oligonucleotides (Figure 1), using 0.1 fmol of the template and the corresponding 6-FAM-labeled and 5′-phosphorylated forward and reverse primers (0.15 µM of each), following Kleter's procedure (25). The reaction was carried out in 50 µl in the presence of 100 µM of each of dNTPs, 10 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2, 50 mM KCl and 1 U of Taq polymerase (Platinum, InVitroGene), for 40 cycles, consisting of 30 s incubation at 94°C, 30 s at 52°C and 30 s at 72°C. The PCR products (10–30 pmol) were converted to single-stranded DNA and mixed with 200 pmol of each of the blockers (2-fold excess over the added immobilized probe). Prior to transferring into the micro-titer well, this mixture was heated to 90°C and the hybridization was performed overnight or for at least 4 h at the ambient temperature. The wells were washed three times with TMN buffer and the fluorescence was directly measured in Spectra MAX Gemini XS and at 22°C as described.
RESULTS
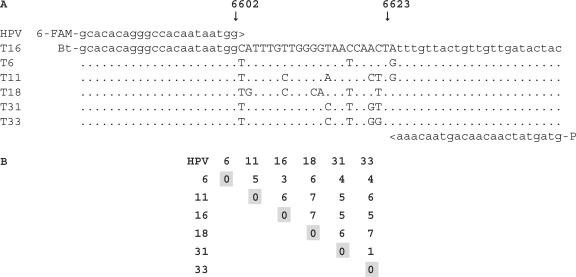
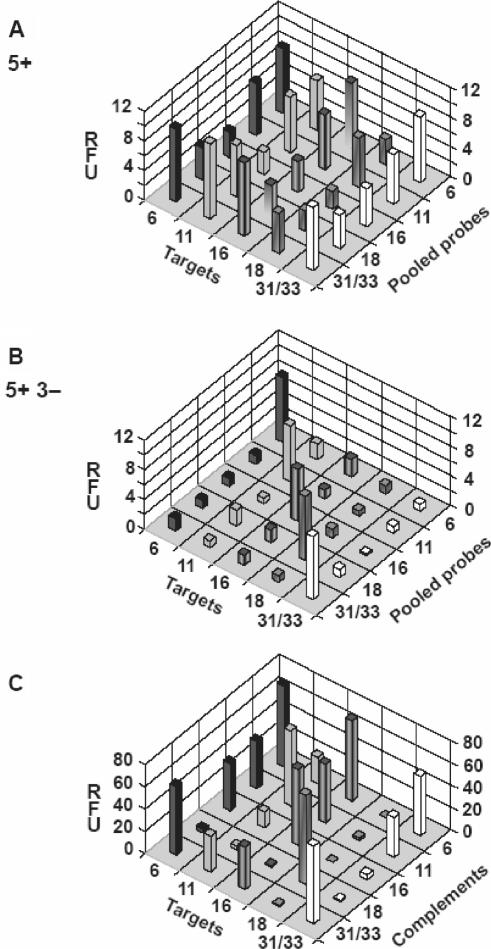
We carried out a series of iterative hybridizations to select probes recognizing six sequence variants of the ‘short PCR fragment’, SPF (25). This technique consists of two hybridization steps, as illustrated in Figure 1: forward (+) hybridization shown on the left and subtractive (−) hybridization on the right. The intended target for which we are selecting the puobes is bound to the solid support (such as streptavidin-coated tubes) in both the forward and the subtractive hybridization. The non-intended targets we want to discriminate against are present only in the subtractive steps, as a competing species, free in solution. The initial pool of random oligonucleotides is present in the hybridization solution at the first step of the selection and is subsequently replaced by the ‘reduced’ oligonucleotide pools resulting from each preceding step along the selection process. Here, SPF targets consisted of 22-nt-long amplified portion flanked by 22- and 23-nt-long primer sequences (Figure 2A). They represented different HPV types 6, 11, 16, 18, 31 and 33, differing by 3–7 nt within the amplified portion (Figure 2B) with types 31 and 33 differing only by 1 nt position that will be considered together eventually. Synthetic, biotinylated target oligonucleotides were immobilized in the streptavidin-coated tubes and were hybridized to synthetic random oligonucleotide mixture, ROM22, consisting of 22-nt random sequence flanked by two 24-nt-long primer sequences. Following the first hybridization, the unbound ROM22 oligonucleotides were washed away and the bound ones were dissociated from their targets, re-amplified by PCR and hybridized again. Each hybridization cycle enriched the resulting mixture of PPs in sequences that were efficiently binding their targets. Yet, as can be seen in Figure 3A, some of these PPs obtained after five forward hybridizations (5+), bind non-intended targets almost as well as their corresponding intended targets. As shown in Figure 3B, the specificity of the resulting PP was dramatically improved after they were submitted to three additional cycles of subtractive hybridization performed simultaneously, i.e. in the presence of mixture of non-intended targets (5+ 3−). The intensity of the specific signal (diagonal) remained the same, whereas the non-specific hybridization was substantially decreased, to the background level at several instances. Thus, the performance of PP submitted to the process of iterative hybridization that includes subtractive cycles largely surpass that of the PP obtained when this process consisted only of the forward hybridization cycles. As shown in Figure 3, PPs at the end of 5+ 3− cycles also perform much better than the 22-nt-long complements of the analyzed targets. These complements, when used as probes, readily cross-hybridize with the mismatched non-intended targets (Figure 3C).
Figure 2.
Target short PCR fragment, SPF, of distinct HPV subtypes. Twenty-two-nucleotide-long amplified sequence is flanked by sequences used to anchor the PCR primers as indicated (A) and the matrix of pairwise nucleotide differences between the SPF sequences considered (B). Bt and 6-FAM denote 5′ terminal modifications with biotin and 6-carboxyfluorescein, respectively. Dots indicate the identity with the upper sequence.
Figure 3.
Binding of probes to their intended and non-intended targets. (A) Binding of the pool of probes, PPs, obtained after five rounds of forward hybridization (5+); (B) binding of PPs after they were submitted to three additional rounds of subtractive hybridizations (5+ 3−); and (C) binding of the full 22-nt-long complements of the targets. All probes were labeled with 6-FAM at their 5′ terminus to allow quantification of the extent of hybridization, expressed in arbitrary units and corresponding to the bound measured fluorescence signal (RFU—relative fluorescence units).
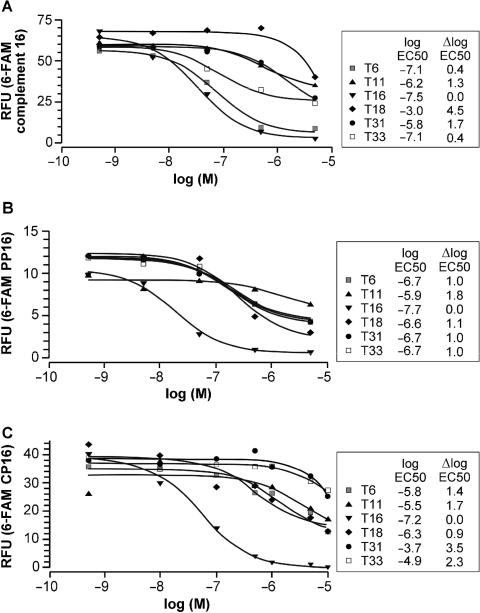
An important characteristic of a performing probe is to similarly discriminate, at given experimental conditions, all possible non-intended targets. In other words, a certain threshold of affinity difference has to be respected between the intended and all possible undesired complexes. The capacity of discrimination between different targets can be studied by competitive hybridization in which the extent of the probe–target complex is measured at varying concentrations of the competitor. If the target is immobilized and the probe is labeled, one may titrate the complex by increasing the concentration of the free targets. The effective concentration required to dissociate 50% of the original complex, EC50, provide a measure of the competitor binding. The difference between EC50 for the intended oligonucleotide target and the EC50 estimates for the non-intended oligonucleotide targets provides the measure of the discrimination capacity of the probe. It reflects the difference in the binding constants, which are expected to have similar temperature dependence here. In other words, it should not be affected by the fact that the measurements are carried out below the melting temperatures of the corresponding complexes. Figure 4 illustrates the titration experiment carried out with the immobilized HPV16 variant and different probes. In Figure 4A, the complement 16 was used as a probe. It discriminates very well against target HPV18 (T18). Yet, at the same time, it shows logEC50 difference between the intended T16 and T6 of only 0.4, indicating very poor discrimination. This can also be directly appreciated by looking at the corresponding titration curves that almost overlap in Figure 4A. In contrast, PP16 shown in Figure 4B discriminates similarly between intended T16 and other targets with logEC50 difference of 1.0 or more. Here, T18 and T6 compete with the intended T16–PP16 complex very similarly, in spite of the fact that the first differ from T16 by 7 and the second by only 3-nt positions (Figure 2B). Clearly, PP16 reveals desired characteristics of a probe that similarly discriminates multiple targets. It was chosen to be shown here since its intended target differs by only 3 nt from the closest HPV6 sequence. On the other hand, in these experiments we were unable to obtain PP31 and PP33 that would efficiently discriminate between the corresponding targets differing by only 1 nt. Instead we obtained PP31/33 recognizing both targets simultaneously.
Figure 4.
Competitive titration of the immobilized HPV-16 target (T16). T16 was hybridized with its 6-FAM-labeled complement in (A), PP16 (5+ 3−) in (B) and cloned probe CP16 (see Figure 5 for the corresponding sequence) in (C). The bound fluorescence was chased by increasing the concentrations of the non-biotinylated T16 target or each of the non-biotinylated non-intended target oligonucleotides. The effective concentration EC50 of the competitive target oligonucleotides required to reduce the binding by 50% was calculated, expressed as logEC50. ΔlogEC50 is the difference between the logEC50 values obtained for T16 and a competitive non-intended target as indicated.
Since each of the specific PP, following 5+ 3− cycles of iterative hybridization described above, consists of a mixture of different sequences, it can be expected that they comprise a smaller fraction of the sequences with even better binding characteristics. The corresponding unique sequence probes, CP for cloned probe, were obtained by cloning PPs into the plasmid vector. Individual CPs were extracted from the plasmids by PCR and tested for binding to the intended and non-intended targets. It usually took less than five clones, to obtain one with the desired, arbitrarily defined ratio of at least five to one of the specific to non-specific binding. The one with the best discrimination performance was sequenced and retained for further analysis. The sequences of the selected CPs (without flanking primer sequences) are shown in Figure 5 where they are compared to their intended targets, highlighting sites possibly engaged in the complementary interactions.
Figure 5.
Cloned probes, CPs, in the context of their intended target sequences. Differences in the SPF targets are highlighted, whereas dots indicate matches between targets and the corresponding CPs. Note that the CP sequences are flanked by priming sequences that are not shown. Alignment of given sequences was performed using BLAST engine for local alignment (36).
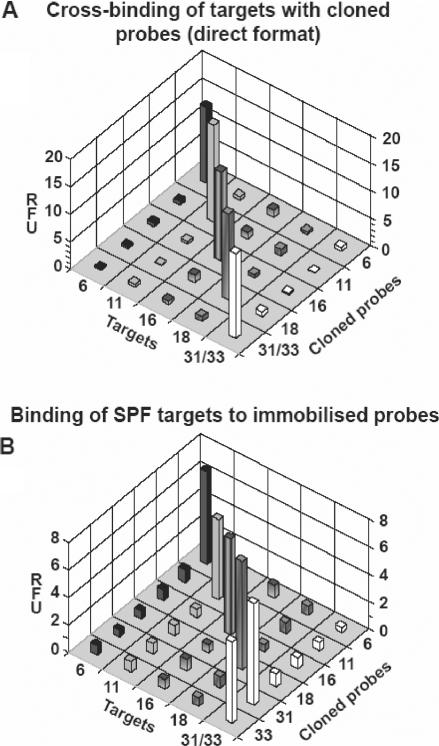
CPs performed better than PPs as far as the detection of their intended targets and discrimination against the non-intended ones is concerned (Figure 6A). In a competitive titration shown in Figure 4C, CP16 performed on average also better than its maternal PP16 (Figure 4B) as judged by ΔlogEC50 values in Figure 4B and C. Yet, presumably because of the fact that CP16 represents an individual clone, the variance in ΔlogEC50 values in Figure 4C appears greater than in Figure 4B, although some of this variance can also be accounted for by non-uniformity of the streptavidin coating between wells within the plates used in these measurements (Materials and methods section). Finally, in binding experiments including all targets (Figure 6A), CPs gave the same hybridization signal as their corresponding PPs (Figure 3B), but less background hybridization. Furthermore, the advantage of CPs over their maternal PPs is that they provide a possibility to test their eventual use as tools in diagnostic tests that require hybridization in the reverse configuration, with probes immobilized to the solid support. Indeed, all the experiments reported so far were in the ‘forward blot format’, with the immobilized targets. In the ‘reverse blot format’, the probes, with biotin moiety at their 5′ end, are themselves immobilized and therefore can provide a simultaneous test for the presence of different targets, such as distinct HPV variants in a clinical sample. This corresponds to the diagnostic situation where the target sequence amplified from a clinical sample is being tested in a panel of immobilized probes intended to identify the presence of a specific HPV subtype. As shown in Figure 6B, CPs are performed in the reverse blot format very well, although we noted a lesser binding of targets resulting in a slightly less favorable signal to the background hybridization ratio than in the direct format. This, however, did not compromise the test. Similar results were obtained when clinical samples of known HPV type were used as a source of the HPV SPF segment tested.
Figure 6.
Binding of the individual cloned probes: in (A) to the immobilized intended and non-intended HPV targets, and in (B) the same binding, but in reverse format instead, i.e. of the free PCR-amplified, tested HPV targets to the intended and non-intended immobilized cloned probes from Figure 4.
DISCUSSION
The selection in vitro (or in vitro ‘evolution’ of nucleic acid ligands) has rarely been used against nucleic acid targets (31–34) and has never been used to select probes for genotyping. One reason for this was the prevailing dogma that binding specificity between nucleic acids is best achieved through complementary interactions and that these can be easily programmed according to Watson–Crick rules (but see 31). The second reason was the experimental difficulty related to the interference of these complementary interactions in the process of selection of nucleic acid ligands against nucleic acid targets. The solution to the latter problem was proposed by Brukner et al. (15). Here, we modified the protocol described by (15), referred to as iterative hybridization to distinguish from the in vitro selection protocols involving non-nucleic acid targets. The novel application of this technology was to obtain probes for genotyping sequence variants differing by only few nucleotide positions. Because of the diagnostic importance of HPV typing and the variety of its sequence variants, we used HPV as a model system to test this approach. However, we used only six most common oncogenic and non-oncogenic HPV targets. Therefore, the probes we have obtained are not expected to discriminate against other HPV variant SPF sequences.
Iterative hybridization is an empirical approach using in vitro selection that does not rely on the rational design of hybridization probes limited by our imperfect knowledge of complex physico-chemistry of the nucleic acid interactions. Selection in vitro allows exploring a whole space of potential probes within an initial pool of random sequences and, importantly also, all possible combinations of binding configurations of these probes with their targets, given conditions of selection. As we have shown above, forward hybridization enriched PPs in the oligonucleotide ligands with binding capacities. In turn, the discriminatory binding was achieved by subtractive hybridization that purged the selected ligands also having affinity to non-intended targets. As a result, we obtained specific PPs with desired characteristics ‘programmed’ by selection. In this process, instead of adjusting hybridization conditions to the desired set of probe–target pairs, we ‘adjusted’ the probes to the conditions we have chosen. Importantly, however, these conditions have to allow a ‘relaxed’ binding, i.e. such that would readily occur even in the absence of the full saturation of all potential base pairs of the target with its complementary probe. Indeed, the key characteristics of the selected probes (Figure 5) are that they do not represent exact complements of the target sequence. It appears that when several sequences are to be simultaneously tested, different number and position of mismatches may minimize cross-reactivity of multiple similar targets without compromising the stability of each of the intended probe–target pairs. Interestingly, although we have started with the initial random sequence, which happened to be of the same length as 22-nt-long SPF targets, most of the selected probes were substantially shorter (Figure 5). Selecting shorter sequences turned to be ‘cheaper strategy’ than to accommodate non-complementary sequence gaps. CP31/33 is an exception here. It retained the initial 22-nt length forming a 9-nt 3′ end mismatched tail that apparently did not interfere by binding to non-intended targets. At the same time, some of the probes (CP6 and CP16) used an extended target sequence evolving matches within its constant flanking segment, i.e. beyond 22-nt-long SPF segment. In other words, we observe shortening of the probes and ‘extending’ of the target sequence, which may be part of the strategy of in vitro evolution. The latter perhaps because of the limited size of the SPF target itself. As for mismatches, we may speculate that increasing the temperature of the experiment could decrease their number in the selected complex.
If we have k targets to discriminate in a single test, we have to select the same number of probes. In each experiment, we use all the remaining k−1 targets as non-intended ones for subtractive hybridization. Since all iterative hybridization steps are carried out under uniform conditions, the resulting PPs are immediately ready for use in a multiplex assay where all probes should recognize their targets and discriminate against the remaining ones. Yet, they represent mixtures of probes and cloning individual sequences render them easier to propagate. Moreover, since mixture of probes also represents ‘mixture’ of binding characteristics, we expected to be able to choose from this distribution the individual probes with even better binding characteristics. This appears to be the case, at least as judged by the competitive binding experiments and the corresponding ΔlogEC50s (Figure 4).
In conclusion, we have presented here a protocol that yields oligonucleotide sequences capable of discriminating between multiple nucleic acid targets that differ by as little as 3 nt. The HPV model used here can be extended to the whole set of 39 epidemiologically important HPV types, targeting SPF or other relevant HPV segments, to create simple diagnostic devices in clinical research and in predictive testing (Brukner et al. submitted for publication). Eventually, the analysis of an extended dataset obtained from such experiments can also be used to gain new knowledge about nucleic acid–nucleic acid interactions and improve rules of the rational probe design (e.g. 31,35).
ACKNOWLEDGEMENTS
This research was supported by grant from the Canadian Institutes of Health Research (NTA-71859). Funding to pay the Open Access publication charge was provided by authors' research funds.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lomakin A, Frank-Kamenetskii MD. A theoretical analysis of specificity of nucleic acid interactions with oligonucleotides and peptide nucleic acids (PNAs) J. Mol. Biol. 1998;276:57–70. doi: 10.1006/jmbi.1997.1497. [DOI] [PubMed] [Google Scholar]
- 2.Eaton BE, Gold L, Zichi DA. Let's get specific: the relationship between specificity and affinity. Chem. Biol. 1995;2:633–638. doi: 10.1016/1074-5521(95)90023-3. [DOI] [PubMed] [Google Scholar]
- 3.Li X, He Z, Zhou J. Selection of optimal oligonucleotide probes for microarrays using multiple criteria, global alignment and parameter estimation. Nucleic Acids Res. 2005;33:6114–6123. doi: 10.1093/nar/gki914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luebke KJ, Balog RP, Garner HR. Prioritized selection of oligodeoxyribonucleotide probes for efficient hybridization to RNA transcripts. Nucleic Acids Res. 2003;31:750–758. doi: 10.1093/nar/gkg133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nielsen HB, Wernersson R, Knudsen S. Design of oligonucleotides for microarrays and perspectives for design of multi-transcriptome arrays. Nucleic Acids Res. 2003;31:3491–3496. doi: 10.1093/nar/gkg622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu C, Carta R, Zhang L. Sequence dependence of cross-hybridization on short oligo microarrays. Nucleic Acids Res. 2005;33:e84. doi: 10.1093/nar/gni082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chavali S, Mahajan A, Tabassum R, Maiti S, Bharadwaj D. Oligonucleotide properties determination and primer designing: a critical examination of predictions. Bioinformatics. 2005;21:3918–3925. doi: 10.1093/bioinformatics/bti633. [DOI] [PubMed] [Google Scholar]
- 8.Panjkovich A, Melo F. Comparison of different melting temperature calculation methods for short DNA sequences. Bioinformatics. 2005;21:711–722. doi: 10.1093/bioinformatics/bti066. [DOI] [PubMed] [Google Scholar]
- 9.Naef F, Hacker CR, Patil N, Magnasco M. Empirical characterization of the expression ratio noise structure in high-density oligonucleotide arrays. Genome Biol. 2002;3:RESEARCH0018. doi: 10.1186/gb-2002-3-4-research0018. 1-0018.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pfunder M, Frey JE. Dissociation analysis in polymerase chain reaction and 1X SSC buffer as a prerequisite for selection of 13 mer microarray probe sets with uniform hybridization behavior. Mol. Biotechnol. 2005;29:1–10. doi: 10.1385/MB:29:1:01. [DOI] [PubMed] [Google Scholar]
- 11.Harris NC, Kiang CH. Defects can increase the melting temperature of DNA-nanoparticle assemblies. J. Phys. Chem. 2006;110:16393–16396. doi: 10.1021/jp062287d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He Z, Wu L, Li X, Fields MW, Zhou J. Empirical establishment of oligonucleotide probe design criteria. Appl. Environ. Microbiol. 2005;71:3753–3760. doi: 10.1128/AEM.71.7.3753-3760.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peplies J, Glockner FO, Amann R. Optimization strategies for DNA microarray-based detection of bacteria with 16S rRNA-targeting oligonucleotide probes. Appl. Environ. Microbiol. 2003;69:1397–1407. doi: 10.1128/AEM.69.3.1397-1407.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Urakawa H, El Fantroussi S, Smidt H, Smoot JC, Tribou EH, Kelly JJ, Noble PA, Stahl DA. Optimization of single-base-pair mismatch discrimination in oligonucleotide microarrays. Appl. Environ. Microbiol. 2003;69:2848–2856. doi: 10.1128/AEM.69.5.2848-2856.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brukner I, Tremblay GA, Paquin B. Generation of amplifiable genome-specific oligonucleotide probes and libraries. Biotechniques. 2002;33:874–876, 878, 880 passim. doi: 10.2144/02334rr04. [DOI] [PubMed] [Google Scholar]
- 16.Guo Z, Smith L. Artificial mismatch hybrization. 1998. US Patent no. 5,780,233. [Google Scholar]
- 17.Lee I, Dombkowski AA, Athey BD. Guidelines for incorporating non-perfectly matched oligonucleotides into target-specific hybridization probes for a DNA microarray. Nucleic Acids Res. 2004;32:681–690. doi: 10.1093/nar/gkh196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ratushna VG, Weller JW, Gibas CJ. Secondary structure in the target as a confounding factor in synthetic oligomer microarray design. BMC Genomics. 2005;6:31. doi: 10.1186/1471-2164-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pozhitkov A, Noble PA, Domazet-Loso T, Nolte AW, Sonnenberg R, Staehler P, Beier M, Tautz D. Tests of rRNA hybridization to microarrays suggest that hybridization characteristics of oligonucleotide probes for species discrimination cannot be predicted. Nucleic Acids Res. 2006;34:e66. doi: 10.1093/nar/gkl133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Far RK, Leppert J, Frank K, Sczakiel G. Technical improvements in the computational target search for antisense oligonucleotides. Oligonucleotides. 2005;15:223–233. doi: 10.1089/oli.2005.15.223. [DOI] [PubMed] [Google Scholar]
- 21.Li W, Ying X. Mprobe 2.0: computer-aided probe design for oligonucleotide microarray. Appl. Bioinformatics. 2006;5:181–186. doi: 10.2165/00822942-200605030-00006. [DOI] [PubMed] [Google Scholar]
- 22.Gharizadeh B, Kaller M, Nyren P, Andersson A, Uhlen M, Lundeberg J, Ahmadian A. Viral and microbial genotyping by a combination of multiplex competitive hybridization and specific extension followed by hybridization to generic tag arrays. Nucleic Acids Res. 2003;31:e146. doi: 10.1093/nar/gng147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simard LR, Gingras F, Labuda D. Direct analysis of amniotic fluid cells by multiplex PCR provides rapid prenatal diagnosis for Duchenne muscular dystrophy. Nucleic Acids Res. 1991;19:2501. doi: 10.1093/nar/19.9.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klaassen CH, Prinsen CF, de Valk HA, Horrevorts AM, Jeunink MA, Thunnissen FB. DNA microarray format for detection and subtyping of human papillomavirus. J. Clin. Microbiol. 2004;42:2152–2160. doi: 10.1128/JCM.42.5.2152-2160.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kleter B, van Doorn LJ, Schrauwen L, Molijn A, Sastrowijoto S, ter Schegget J, Lindeman J, ter Harmsel B, Burger M, et al. Development and clinical evaluation of a highly sensitive PCR-reverse hybridization line probe assay for detection and identification of anogenital human papillomavirus. J. Clin. Microbiol. 1999;37:2508–2517. doi: 10.1128/jcm.37.8.2508-2517.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van den Brule AJ, Pol R, Fransen-Daalmeijer N, Schouls LM, Meijer CJ, Snijders PJ. GP5+/6+ PCR followed by reverse line blot analysis enables rapid and high-throughput identification of human papillomavirus genotypes. J. Clin. Microbiol. 2002;40:779–787. doi: 10.1128/JCM.40.3.779-787.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Ham MA, Bakkers JM, Harbers GK, Quint WG, Massuger LF, Melchers WJ. Comparison of two commercial assays for detection of human papillomavirus (HPV) in cervical scrape specimens: validation of the Roche AMPLICOR HPV test as a means to screen for HPV genotypes associated with a higher risk of cervical disorders. J. Clin. Microbiol. 2005;43:2662–2667. doi: 10.1128/JCM.43.6.2662-2667.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quint WG, Pagliusi SR, Lelie N, de Villiers EM, Wheeler CM. Results of the first World Health Organization international collaborative study of detection of human papillomavirus DNA. J. Clin. Microbiol. 2006;44:571–579. doi: 10.1128/JCM.44.2.571-579.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Hamont D, van Ham MA, Bakkers JM, Massuger LF, Melchers WJ. Evaluation of the SPF10-INNO LiPA human papillomavirus (HPV) genotyping test and the Roche linear array HPV genotyping test. J. Clin. Microbiol. 2006;44:3122–3129. doi: 10.1128/JCM.00517-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gravitt PE, Peyton CL, Alessi TQ, Wheeler CM, Coutlee F, Hildesheim A, Schiffman MH, Scott DR, Apple RJ. Improved amplification of genital human papillomaviruses. J. Clin. Microbiol. 2000;38:357–361. doi: 10.1128/jcm.38.1.357-361.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duconge F, Di Primo C, Toulme JJ. Is a closing “GA pair” a rule for stable loop-loop RNA complexes? J. Biol. Chem. 2000;275:21287–21294. doi: 10.1074/jbc.M002694200. [DOI] [PubMed] [Google Scholar]
- 32.Hartig JS, Grune I, Najafi-Shoushtari SH, Famulok M. Sequence-specific detection of MicroRNAs by signal-amplifying ribozymes. J. Am. Chem. Soc. 2004;126:722–723. doi: 10.1021/ja038822u. [DOI] [PubMed] [Google Scholar]
- 33.Kikuchi K, Umehara T, Fukuda K, Kuno A, Hasegawa T, Nishikawa S. A hepatitis C virus (HCV) internal ribosome entry site (IRES) domain III-IV-targeted aptamer inhibits translation by binding to an apical loop of domain IIId. Nucleic Acids Res. 2005;33:683–692. doi: 10.1093/nar/gki215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scarabino D, Crisari A, Lorenzini S, Williams K, Tocchini-Valentini GP. tRNA prefers to kiss. EMBO J. 1999;18:4571–4578. doi: 10.1093/emboj/18.16.4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Darfeuille F, Reigadas S, Hansen JB, Orum H, Di Primo C, Toulme JJ. Aptamers targeted to an RNA hairpin show improved specificity compared to that of complementary oligonucleotides. Biochemistry. 2006;45:12076–12082. doi: 10.1021/bi0606344. [DOI] [PubMed] [Google Scholar]
- 36.Tatusova TA, Madden TL. BLAST 2 sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol. Lett. 1999;174:247–250. doi: 10.1111/j.1574-6968.1999.tb13575.x. [DOI] [PubMed] [Google Scholar]