Abstract
The publication of a complete genome sequence is usually accompanied by annotations of its genes. In contrast to protein coding genes, genes for ribosomal RNA (rRNA) are often poorly or inconsistently annotated. This makes comparative studies based on rRNA genes difficult. We have therefore created computational predictors for the major rRNA species from all kingdoms of life and compiled them into a program called RNAmmer. The program uses hidden Markov models trained on data from the 5S ribosomal RNA database and the European ribosomal RNA database project. A pre-screening step makes the method fast with little loss of sensitivity, enabling the analysis of a complete bacterial genome in less than a minute. Results from running RNAmmer on a large set of genomes indicate that the location of rRNAs can be predicted with a very high level of accuracy. Novel, unannotated rRNAs are also predicted in many genomes. The software as well as the genome analysis results are available at the CBS web server.
INTRODUCTION
Ribosomes are the molecular machines which form the connection between nucleic acids and proteins in all living organisms. The ribosome's dependence on ribosomal RNAs (rRNAs) for its function has caused them to be conserved at both the sequence and the structure level. Because of this, rRNAs are often used in comparative studies such as phylogenetic inference. Comparative studies have become more popular as more genomes have been completely sequenced, but can potentially become complicated when some of the genes they are based on are poorly annotated or not annotated at all. Unfortunately, this is often a problem with rRNAs as genome annotation pipelines usually do not include tools specific for rRNA detection. Instead, rRNAs are often located by sequence similarity searches such as BLAST. Although such searches may give reasonable answers due to the high level of sequence conservation in the core regions of the genes, using such results for annotation purposes can be problematic. The validity of the search results depends on the program and database used. Changing one or both of these can drastically change the results. Genomic databases have grown exponentially over the past two decades and search programs have as a consequence had to undergo constant revisions in order to meet the requirements of the research community. Thus, the results of a search done today are probably very different from those produced several years ago. An added complication is that the most commonly used database search methods have poor performance for noncoding RNAs. A recent study comparing several different methods for predicting noncoding RNAs, including rRNAs, found that the most commonly used methods gave the most inaccurate results (1).
Through our work on the GenomeAtlas database (2), we have seen the results of poor annotation of rRNAs. Some genomes do not have any rRNAs annotated at all, whereas other genomes seem to have rRNAs annotated on the wrong strand. We initially tried to do systematic BLAST (3) searches, but it proved difficult to maintain consistency throughout this process. The high level of sequence conservation among the rRNAs enabled us to create hidden Markov models (HMMs) from structural alignments. Such models are more capable of capturing the sequence variation that is inherently present in the rRNA gene families than simple BLAST searches. Using HMMs also simplifies the use of common criteria for prediction assessment. A library of HMMs was constructed and the program RNAmmer was developed to make use of this library. RNAmmer is available through the CBS web site, as a web service or as a stand-alone package. It has been tested on all published genomes and gives accurate predictions of rRNAs. The program also has the added benefit of producing results that are comparable between genomes.
Our work has focused on three of the major rRNA species. The ribosome consists of two subunits, the small and the large subunit, which pair up to form the functional ribosome. The rRNAs present in prokaryotes are the 5S and 23S in the large subunit, and the 16S in the small subunit. In eukaryotes, 5S, 5.8S and 28S rRNA exist in the large subunit, and 18S rRNA in the small subunit. The 5.8S is not considered in this work. There are substantial sequence and secondary structure similarities between eukaryotic and prokaryotic rRNAs; however, the eukaryotic rRNAs commonly have longer stems and larger loops than those of the prokaryotes. The subunits are composed of both RNAs and proteins. Since their discovery in the early 1950s, it has been debated whether ribosomal function should be credited to the rRNAs or the proteins. Recent crystal studies have revealed that protein synthesis to a large extent is dependent on the rRNAs (4–7) and this has most likely been instrumental for their high level of conservation.
In prokaryotes, the 16S, 23S and 5S rRNAs are commonly transcribed together, while the 18S, 28S and 5.8S rRNAs form a transcriptional unit in eukaryotes. Eukaryotic 5S rRNA commonly appear in highly duplicated tandem repeats (8). In most organisms, there are several copies of the rRNA transcription unit, and although as much as 11% sequence divergence has been observed between units within the same genome, the difference is usually less than 1% (9). In several cases, segments are also edited out of the transcribed rRNA. These segments may be introns that after splicing leave a continuous rRNA, or they can be intervening sequences (IVS) that leave a fragmented rRNA which is still functional within the ribosome structure (10). Introns are most prevalent in eukaryotes and archaeas, while intervening sequences have been seen in eukaryotes and bacteria. Introns are predominantly found within conserved sequences close to tRNA and mRNA-binding sites (10), whereas intervening sequences are ordinarily seen in hypervariable regions (11).
MATERIALS AND METHODS
Using HMMs to find new members of a sequence family requires reliable multiple alignments. The 16S/18S and 23S/28S rRNA alignments were retrieved from the European ribosomal RNA database (ERRD) (12). In this database, annotated large and small subunit ribosomal RNA sequences from the EMBL nucleotide database with a length of at least 70% of their estimated full length have been aligned. Multiple alignments of 5S rRNAs were retrieved from the 5S Ribosomal RNA Database (13). Data from both databases were downloaded on October 27, 2005. The alignments are all structural alignments, i.e. aligned using secondary structure information gained from comparative sequence analysis. The 5S alignments were already divided into separate alignments for archaeal, bacterial and eukaryotic sequences, whereas the ERRD data were not. The alignments for 16/18S and 23/28S rRNAs were divided into the same groups as the 5S data to provide kingdom-specific predictors. The data was stored in a MySQL database for easier handling.
The ERRD data contained sequences from ‘environmental samples’. These were excluded since there was little information about them. The 5S were generally around 120 nt long, the 16/18S around 1500 nt and the 23/28S around 3000 nt long, all with no obvious outliers. The length of the eukaryotic rRNAs varied substantially, more than those of bacterial and archaeal rRNAs, but no sequences in the alignments seemed obviously wrong.
The sequences were divided into phylogenetic groups to help with further analysis. Due to sequencing bias, some phylogenetic groups dominated the data sets. Such a skew could potentially cause the predictors to be less sensitive on underrepresented phylogenetic groups. Among the bacteria, 82% of the sequences were from three phyla: Actinobacteria, Firmicutes and Proteobacteria. Around 70% of the archaeal sequences were from Euryarchaeota; among the eukaryotes, the Streptophyta comprised 15% of the data. Several of the sequences also proved to be very similar. Therefore, redundancy reduction inspired by Hobohms second algorithm (14) was performed. This algorithm starts with a sorted list of the number of neighbors each sequence has. An all-against-all comparison between the sequences is performed and neighborship is judged by the level of similarity found. Similarity was measured by Score = ∑i,jnijSij/(N-g) where i and j sum over the four nucleotides, nij counts the number of aligned nucleotide pairs (i, j), N is the length of the sequence and g is the number of gap-only positions; Sij refers to the scoring matrix EDNAFULL created by Todd Lowe. The maximum similarity level allowed was set to ensure that each phylum was represented. Similarity graphs were formed for each group, with the sequences as vertices and edges between similar sequences. The sequence with the highest connectivity and its edges were deleted from the graph, and this was repeated until no edges remained. At the end, all removed sequences were checked to see if they had any edges to vertices in the remaining set. If not, they were reinstated. This procedure was implemented as a C program.
Sequences in ERRD may contain ambiguous nucleotide symbols representing nucleotides that have not been uniquely determined. These occur more frequently in bacteria and eukaryotes than in archaea, and primarily at both ends of the alignment: in 16/18S, predominantly at the end; in 23/28S, predominantly at the beginning. In the latter case, this was mostly due the high prevalence of gaps at the end of the alignment. As we found that ambiguous nucleotides at the ends reduced the ability to predict start and stop positions accurately, we decided to remove all sequences with five or more ambiguous nucleotides in either end of the sequence. A summary of the number of sequences removed during curation of the alignments is shown in Table 1.
Table 1.
The initial number of rRNA sequences and the number of sequences excluded for different reasons.
| Kingdom | Type | Initial count | Environmental samples | Incomplete sequences | Redundancy reduction | Total in HMM |
| Archaea | 5S | 58 | 0 | 0 | 10 | 48 |
| 16S | 589 | 239 | 471 | 287 | 76 | |
| 23S | 37 | 0 | 18 | 8 | 15 | |
| Bacteria | 5S | 461 | 0 | 0 | 101 | 360 |
| 16S | 12 107 | 1429 | 10 723 | 2485 | 743 | |
| 23S | 398 | 0 | 155 | 130 | 127 | |
| Eukaryotes | 5S | 316 | 0 | 0 | 33 | 283 |
| 18S | 6585 | 24 | 5222 | 836 | 979 | |
| 28S | 157 | 0 | 91 | 8 | 58 |
Environmental samples were excluded due to lack of phylogenetic information. Sequences with too many unknown nucleotides in either end of the sequence were excluded to improve HMM accuracy. Redundancy reduction was performed to reduce bias. Note that these groups may overlap. The last column indicates the number of sequences used to build each HMM.
The software package HMMer (15) version 2.3.2 was used to create HMMs from alignments where all columns containing only gaps had been removed. It was configured for nucleotides, and to compensate for skews in the nucleotide distribution a custom null model for each alignment was used. Although redundancy reduction had been performed, the Henikoff position-based weighing scheme (16) was used to reduce any remaining biases. When using the HMMs to search genome sequences, the default alignment method was used: a match must span the entire model, and several matches may be found within one sequence.
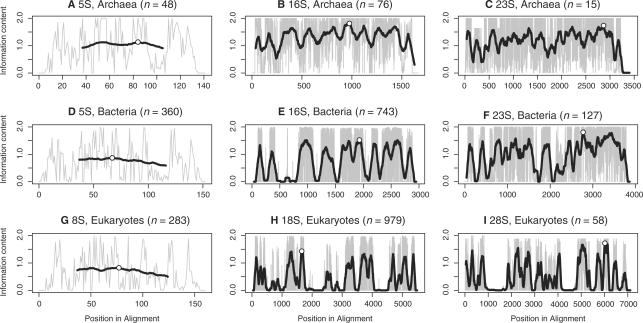
With the aim of increasing the search speed, we determined the 75 most conserved consecutive columns in each alignment, as illustrated in Figure 1, and produced ‘spotter’ HMMs based on these. Since searches with the smaller spotter models would be considerably faster, we wanted to investigate the possibility of using the spotter to pre-screen for candidates, using the full HMMs only on regions surrounding the spotter hits. Spotter and full model searches were done separately. Spotter and full model predictions were matched based on whether they had overlapping nucleotides on the same strand. A linear regression was used to express spotter score in terms of full model score. Variation was estimated as linear in full model score with non-positive regression coefficients. Least squares estimates were used in both cases. Spotter scores were assumed to be missing when negative and, hence, assumed to follow a truncated normal distribution; expected scores and square deviations were used to replace missing values in the two regressions. From this model, we computed the lowest full model score, T99 , for which there was at least a 99% likelihood of getting a corresponding spotter hit, and the likelihood, Pmin , that a full model hit with the lowest found score should have a corresponding spotter hit.
Figure 1.
The graphs show conservation in the alignments as measured by information content: C=∑ifilog2(fi/qi) where i sums over the four nucleotides, fi is the frequency of nucleotide i in the column and qi =1/4 is used as the background frequency. Ambiguous nucleotide symbols were evenly divided between the corresponding fi, gaps between all four nucleotides. The grey line represents the value for each position in the alignment, the black line is a running average over 75 nt around the current position, whereas the white dot indicates the center of the most conserved 75 nt region of the alignment.
Both the full HMMs and the spotter HMMs were run on all fully sequenced genomes found in the Genome Atlas database (listed in Supplementary Table S1). All predictions with non-negative score and E-value at most 100 were reported. Only full model hits with E-value <0.01 were accepted as reliable hits, but none with E-value between 0.01 and 100 were reported. As rRNAs within a genome tend to be very similar, usually with at least 99% identity, different full model hits within a genome corresponding to actual rRNAs should be expected to have similar scores. However, we found a substantial number of hits with far lower scores which we assume to be pseudogenes, truncated rRNAs or otherwise non-functional rRNA copies. To ensure that these did not have an adverse effect on the analyses, we excluded full model hits having a score less than 80% of the maximal score in that genome. These are listed in Supplementary Table S2.
Annotations of rRNAs were obtained from GenBank. Unfortunately, rRNAs have not been annotated in a uniform manner and it was often unclear exactly what was annotated. In some cases, both the separate rRNAs and the full operon was annotated. In all such cases, the operons were longer than 5000 nt, and all annotations longer than that were thus excluded. In our experience, this affected only operons. In other cases, different pieces of the same gene had been annotated as separate entities. Thus, some predictions matched several annotation entries; these are listed in Supplementary Table S3. A prediction was considered to match an annotation if they were on the same strand and the length of their overlap was at least half the length of the shorter of the two; it was considered to be annotated if it matched at least one annotation. The deviation between annotated and predicted start and stop positions was also examined, but predictions with multiple matching annotations were excluded from this comparison.
Additional analyses were performed for experimentally verified 16S in Anaplasma marginale St. Maries (M60313), Chlamydia muridarum Nigg (D85718), Escherichia coli K12 MG1655 (J01695), Sulfolobus tokodaii St. 7 (AB022438), Thermus thermophilus HB8 (X07998) and Nitrobacter hamburgensis X14 (L11663). Computational speed was assessed on M. capricolum ATCC 27343 (CP000123) Solibacter usitatus Ellin6076 (CP000473) and Sargasso Sea data (AACY01000001-AACY01811372). All test searches reported were performed on an SGI Altix 3000 machine using one 1.3 GHz Itanium 2 processor.
RESULTS
The predictions of the full HMM models have been compared first against annotations, then against the spotter models.
Full model predictions versus annotation
As Table 2 shows, the predictors appeared to be better at detecting bacterial rRNAs and less powerful for eukaryotic rRNAs. The highest accuracy was seen for the 16/18S rRNAs followed by the 23/28S. Two groups of rRNAs were particularly difficult to locate: the archaeal 5S and the eukaryotic 18S. The missing archaeal 5S were all from four euryarchaeotic genomes which are all anaerobic methane producers. The eukaryotic 18S that the predictors could not find were all from two genomes, Guillardia theta and Plasmodium falciparum.
Closer evaluation revealed that several annotated rRNAs that lacked a matching prediction had actually been detected, but on the opposite strand. In eukaryotes, this was only seen with Arabidopsis thaliana 5S. In bacteria, most of the reverse predictions were 5S; in archaea, they were predominantly 16S and 23S. It should be noted that for all the reverse strand predictions the predicted start and stop positions agreed well with the annotation, indicating that they have been annotated on the wrong strand. Annotated rRNAs that lacked matching predictions in either direction are listed in Supplementary Table S4.
Table 2 gives the number of predicted rRNAs that did not have a corresponding annotation: putative novel rRNAs. About 70% of them were 5S rRNAs, and only a few were archaeal. In bacteria, most of the novel rRNAs were found in Firmicutes and Gammaproteobacterias, although it should be noted that these two phyla are the two dominant groups and contain the bulk of the currently sequenced bacterial genomes. Among the eukaryotes, only A. thaliana had novel rRNAs. The scores of the new rRNA predictions did not significantly differ from those that were annotated, indicating that these are true rRNAs not yet annotated. The 5S is often omitted in the rRNA annotation; since the eukaryotic 5S is usually separated from the 18-28S sequence, they might be less visible to annotators.
Table 2.
The number of rRNAs annotated and predicted in the genomes that were examined.
| Kingdom | Type | Annotated | Same strand | Other strand | Not found | Full model predictions | Novel |
| Archaea (n = 27) | 5S | 56 (24) | 43 (21) | 1 (1) | 12 (8) | 47 (23) | 4 (3) |
| 16S | 47 (25) | 45 (25) | 2 (2) | 0 (0) | 47 (27) | 2 (2) | |
| 23S | 47 (25) | 44 (24) | 2 (2) | 1 (1) | 46 (26) | 2 (2) | |
| Bacteria (n = 321) | 5S | 1205 (285) | 1166 (285) | 30 (16) | 9 (5) | 1339 (320) | 173 (69) |
| 16S | 1172 (299) | 1146 (299) | 22 (12) | 4 (4) | 1237 (320) | 91 (34) | |
| 23S | 1197 (297) | 1154 (291) | 22 (13) | 21 (12) | 1248 (313) | 94 (36) | |
| Eukaryotes (n = 13) | 5S | 65 (7) | 46 (6) | 19 (1) | 0 (0) | 324 (9) | 278 (5) |
| 18S | 13 (4) | 6 (4) | 0 (0) | 7 (2) | 13 (6) | 7 (3) | |
| 28S | 13 (5) | 12 (4) | 0 (0) | 1 (1) | 19 (7) | 7 (3) |
The table gives the number of annotations, and splits this into those matching predictions on the same strand, on the other strand, and not found. The total number of full model predictions is given. Novel predictions are full model predictions not matching any annotation on the same strand, and include those annotated on the other strand. Numbers in parentheses indicate the number of genomes. It should be noted that the eukaryotic annotated count is somewhat uncertain due to ambiguous rRNA annotations. The genomes which were analyzed were from the GenomeAtlas database, a database over all available fully sequenced genomes.
Start and stop deviations
The differences between predicted and annotated start and stop positions are illustrated in Figure 2 and it shows that they agree well. The median of the start and stop prediction deviations were in most groups zero or very close to zero with more than half within 10 nucleotides. This was not the case for the eukaryotes.
Figure 2.
Deviation of start and stop positions between predicted and annotated RNA is presented as pairs of panels. The number of predictions among the archaea, bacteria and eukaryotes are denoted beneath the panel group heading. The zero position in each panel corresponds to the annotation start or stop position with predicted positions presented relative to these. The yellow dot indicates the median deviation and the black box the quartile range. The hinges on the side of the box extend from the side of the box to the data point that is closest to, but does not exceed, 1.5 times the interquartile range. The curves show the density of the distribution.
For eukaryotic 5S, only five genomes contained predictions with matching annotations. The predictions were uniform in length, whereas the annotations were more variable. The predictions that indicated a substantially shorter 5S than annotated were all in Schizosaccharomyces pombe: the average length of the annotations was 170 nt, whereas the corresponding predictions were all 114 nt. For eukaryotic 18S, however, predicted start and stop positions were very accurate, although many annotated 18S were missed.
For eukaryotic 28S, only two genomes had predictions with matching annotations. One of them, Encephalitozoon cuniculi, had stop positions predicted once 1112 nt and twice 4797 nt downstream of the annotation, whereas the start position was accurately predicted. In the other genome, Guillardia theta, the start positions were uniformly predicted 110 nt upstream of the annotated position, but with the stop position quite accurately predicted.
Since rRNAs tend to be very similar within a genome, predictions within each genome generally had similar lengths. This similarity within genomes as well as within groups of closely related genomes caused multiple peaks in the distributions of endpoint deviations. An example of this can be seen in the bacterial 16S predictions where some of the predicted start and stop positions were clustered downstream of the annotation and where some of the predicted start positions were clustered upstream of the annotation. Some of the major contributors to the upstream peak in the start positions were different Streptococcus pyogenes strains, Bacillus genomes and Yersinia pestis genomes. These, in addition to Streptococcus agalactiae strains and Vibrio parahaemolyticus, were also prevalent in the stop position downstream peak. There was also a downstream peak in the start positions, and the genomes causing this peak were mainly Staphylococcus aureus, Bacillus cereus and several Escherichia coli relatives.
Most of the start and stop deviations did not exceed 100 nt. However, there were a few cases of deviations exceeding 1000 nt, and these are not shown in the figure. This was the case for eukaryotic 23S and was mainly due to the three previously described stop positions predicted considerably downstream of the annotated stop position. In the two longer predictions from E. cuniculi, this was due to the HMM placing the latter 100 nt of the prediction further downstream to achieve a better score. Such inserts would most likely not appear when the spotter model is used first, since the inserted sequence would be too long. To test this, a truncated version of the sequence was run through the predictor. The stop position was then accurately predicted. This phenomenon also explains some cases among the bacterial 16S predictions where the start position was placed very far upstream of the annotation. There were 27 rRNAs that had a start position predicted to start anywhere from 13 000 to 40 000 nt upstream of the annotated start position. All but one of these were Firmicutes, mostly Streptococci and Staphylococci. Closer study of the sequences revealed that the misplaced start position predictions were again due to long sequences being inserted near the start of the rRNA, indicating that the first part of the HMM had been misplaced in the same manner as for Guillardia theta's stop predictions. To test if these were the same kind of inserts, a region ending in the same place as the predictions but starting 10 000 nt earlier was run through the full model predictor. This led to the bacterial 16S rRNAs being predicted with a deviation in start and stop positions on par with what was otherwise seen.
Comparison to experimentally verified rRNAs
Annotations were often ambiguous and considered unreliable. For discrepancies between annotations and RNAmmer predictions, it is not a priori clear which of the two is correct. However, some genomes with experimentally verified rRNAs were selected to further assess the accuracy of start and stop predictions. The genomes we examined were Anaplasma marginale Str. Maries, Chlamydia muridarum Nigg, Escherichia coli K12 MG1655, Sulfolobus tokodaii Str. 7, Thermus thermophilus HB8 and Nitrobacter hamburgensis X14. These genomes all had complete 16S sequences according to the NCBI database and had accompanying literature which said that they were experimentally determined. When checking the positions of these rRNAs with BLAST against the genome, some discrepancies were found. Due to this we used the BLAST results when comparing annotated rRNAs to predictions.
In total, there were 14 copies of the six 16S sequences, and all of them were found by our predictions. Stop predictions were more accurate than start predictions. In all but four cases, the start position was predicted to be 7 nt downstream of the annotated start position. In A. marginale and S. tokodaii, the start position was predicted to be the same as annotation, and both of the two entries from C. muridarum were predicted to start 3 nt downstream of annotated start position. In N. hamburgensis the start position was, in contrast to the other cases, predicted to start 7 nt upstream of annotated start position. The stop positions in all but three predictions ended on the same position as the annotation. In N. hamburgensis predicted stop was 9 nt downstream, whereas in S. tokoaii and A. marginale the predicted stop was 1 nt downstream of annotation. Thus, all predictions were within 10 nt of the annotated start and stop positions.
Comparison to RFAM
RFAM is a database of RNA families which incorporates secondary structure in its analyses. We have made a comparison with the 5S rRNA predictions of RFAM (17,18) for a selection of twenty prokaryotic genomes listed in Supplementary Table S5. There were a total of 55 5S annotated in these genomes. RNAmmer found 53 of them, while 54 were found in RFAM. In three of the genomes, both methods predicted a 5S to within a few nucleotides of the annotated position, but both placed it on the other strand. Both predictors identified three new 5S rRNAs within these genomes, and at approximately the same positions. Two of these new 5S rRNAs followed another annotated 5S rRNA, looking like a tandem repeat. In most cases, both methods placed the start position a few nucleotides downstream of the annotation, whereas the stop position was more evenly distributed around the annotated position. RNAmmer generally predicted rRNAs to be shorter by a nucleotide or two than RFAM, usually at start of the genes.
Spotter pre-screening
Table 3 shows that, with the exception of archaeal 5S, no full model hits were missed by the spotter model. Also, the spotter produced relatively few false positives, except for the eukaryotic 5S.
Table 3.
Evaluation of spotter and full model predictions.
| Kingdom | Type | Number of model predictions | Full model scores | T99 | Pmin | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Full | Spotter | FPS | Min | Q1 | Med | Q3 | Max | ||||
| Archaea | 5S | 47 | 35 | 7 | 2.9 | 12.7 | 20.0 | 35.3 | 50.6 | 34.9 | 0.69 |
| 16S | 47 | 47 | 0 | 1180.8 | 1891.9 | 1937.9 | 2004.0 | 2096.5 | <0 | 1.0 | |
| 23S | 46 | 46 | 1 | 2240.7 | 2714.1 | 2870.7 | 3155.3 | 3267.3 | <0 | 1.0 | |
| Bacteria | 5S | 1339 | 1339 | 123 | 39.9 | 77.7 | 89.5 | 94.6 | 109.6 | 14.0 | 1.0 |
| 16S | 1237 | 1237 | 31 | 721.9 | 1905.5 | 1989.4 | 2058.7 | 2148.5 | <0 | 1.0 | |
| 23S | 1248 | 1248 | 20 | 2502.8 | 3267.8 | 3586.5 | 3690.7 | 3876.1 | <0 | 1.0 | |
| Eukaryotes | 5S | 324 | 324 | 251 | 43.9 | 51.1 | 53.9 | 74.3 | 82.2 | <0 | 1.0 |
| 18S | 13 | 13 | 14 | 625.3 | 625.3 | 1733.1 | 1777.5 | 1777.6 | <0 | 1.0 | |
| 28S | 19 | 19 | 5 | 1434.2 | 2904.7 | 3225.0 | 3335.9 | 3380.9 | <0 | 1.0 | |
This table shows the total number of full models, the number of spotter predictions that had matching full model predictions and the number of false positive spotter model predictions. The characteristics of the full model prediction score distributions are shown. FPS denotes the number of false positive spotter predictions. T99 refers to the lowest score a full model could have while still being detected with 99% probability by a spotter model with positive score. Pmin is the probability that a spotter with positive score would find a full model with the minimum score indicated. The lowest score for a full model score can be used as a lower limit on which results could be expected to be real.
Minimum, maximum, quantile and median scores for all the full model predictions are shown in Table 3, giving some indication of the range of scores that rRNAs can be expected to have. The table also includes the threshold T99 and the likelihood Pmin which indicate that all full model predictions were expected to have corresponding spotter model predictions except some among the archaeal 5S.
Based on the relatively stable lengths of the different types of rRNAs and the corresponding full model hits and the position of the spotter hit within them, we decided on window sizes around spotter model hits to use when the spotter model is used first. These were chosen to be 300 nt for the 5S rRNA, 5000 nt for the 16/18S and 9000 nt for the 23/28S. Being roughly three times the length of the corresponding rRNAs, we consider rRNA sequences to be unlikely to extend beyond these windows.
Computational speed
Searching Mycoplasma capricolum ATCC27343, about 1 Mbp, for bacterial 16S took 14 minutes using the full HMM. Using the spotter to screen the sequence, then the full model on the spotter hits, reduced the time to 16 seconds. Search times are expected to increase proportionally to the genome size; when using the spotter model to screen the sequence, search time will also increase with increasing number of spotter hits.
Time differences between searching long and short sequences were examined by searching through the complete sequence of Solibacter usitatus Ellin6076, and through the Sargasso Sea environmental samples (19). Searching the S. usitatus genome, about 10 Mbp, took 48 seconds per Mbp. Two copies from each rRNAs family were found. The Sargasso Sea samples consisted of 811 372 entries totaling over 800 Mbp. On this set the search speed was 407 seconds per Mbp. The article (19) accompanying this set indicated 1164 small subunit rRNA genes (16/18S) or fragments of genes; we found only 332, but our predictors are not able to find fragments of rRNAs. In addition, we found 562 5S and 68 23S sequences.
DISCUSSION
Our aim has been to enable high-throughput searches for rRNA while producing accurate and consistent predictions suitable for comparative analyses. For this purpose, we have developed the RNAmmer package which relies on HMMs for both speed and accuracy. HMMs were made using HMMer (15), which from a multiple alignment produces an HMM where match states represent columns with a specific nucleotide distribution, corresponding deletion states represent the possibility of gaps, and insertion states represent columns with large numbers of gaps; transition probabilities between the states indicate how likely each of the states are. HMMs thus differ from sequence alignments in that the likelihood of insertions and deletions may vary along the sequence. When searching a sequence with an HMM, the score indicates how well the sequence segment matches the model. The information content of a position, which reflects the nucleotide distribution and the likelihood of gaps, indicates how well that position is conserved. A good match to the HMM may come either from a highly conserved region which may well be short, or from a longer region with only weak conservation. We find both these cases. Bacterial 16S are detected despite almost half of the nucleotides being assigned to insert states, as other regions are highly conserved. For archaeal 23S, however, the information content of each position is low, but the sequence is long and there are few allowed insert states.
These aspects can also explain cases of poor performance, both of the full model and of the spotter model. The low information content in the eukaryotic 5S and 18S alignments indicates that these sequences are more divergent than archaeal and bacterial 5S and 16S. In addition, 40% of the 5S and 75% of the 18S alignment give rise to insert states in the HMM. Thus, there is little for the HMM to recognize. In addition, many of the missed 18S rRNAs were from Cryptophyta, a phylum which makes up only 0.6% of the alignment data.
The archaeal 5S show the same characteristics as the eukaryotic 5S and 18S, which most likely explains the low performance for these rRNAs. The score for archaeal 5S hits were generally low, and the spotter score comes only from a 75 nt part of the sequence giving it even lower score causing it to miss 12 of the full model hits. It is notable, however, that these were the only cases missed by the spotter model: with the exception of archaeal 5S, our analyses show that the spotter should be able to detect rRNAs unless they are much further diverged than what we find in our data.
Columns at the beginning and end of the multiple alignments often have low conservation and many gaps. Such columns are generally accommodated into the HMM as insert states, but HMMer ignores them at the beginning and end of the alignment. An example is the 5S, where match states stop around 10 columns from the end of the alignments effectively causing the HMM to predict the last conserved nucleotide of the consensus sequence rather than the stop of the rRNAs. Hence, it is not uncommon for the stop position of the 5S to be predicted up to 10 nt downstream of the annotated stop position.
These effects can also explain the endpoint accuracy that was seen when we compared our results to experimentally determined 16S sequences. We tried to find sequences where the ends had been experimentally verified by RACE or PCR, but such rRNAs proved difficult to find. All the ones we selected were sequenced, but it is uncertain to what extent the authors had tried to determine the ends. These experimentally found rRNAs did show better agreement with annotation than predictions in general, although this is not sufficient to conclude that our predictions are more accurate. Our stop predictions were very accurate, but more deviation was seen in the start predictions. These results could reflect more variation in the beginning of the alignments, which as in the 5S case could effectively cause the HMM to predict the last conserved nucleotide of the consensus sequence rather than the end of the rRNAs.
In some cases, larger endpoint deviations occur. This can happen when one of the ends of the model finds a better match in a different part of the sequence. Insertion states sometimes allows the HMM to insert long gap regions and thus find a matching stop position far from the rest of the sequence. As shown for the bacterial 16S sequences that displayed this phenomenon, this is less of a problem when the spotter model is employed. The window searched around the spotter hit would most likely be too short to accommodate such an insert, and the model would match with the proper sequence.
For fragmented rRNAs, long gap regions may be correctly predicted. This was seen for Coxiella burnetii 23S where our prediction has the same start position as annotated, but where the predicted stop position is 1884 nt downstream of GenBank's stop position. However, according to Entrez Gene, this rRNA appears in four pieces and with the same stop position as ours, suggesting that in some cases ‘too long’ predictions might actually be correct. These cases should normally not be masked when using the spotter unless inserts between the fragments would make it exceed the window size.
The HMM produced by HMMer requires time of order O(NM) to search a sequence of length N using a model with M states, M being proportional to the length of the multiple alignment. However, the speed is increased by using a 75 nt long spotter model to pre-screen the sequence, which requires time of order O(N), and then running the full HMM on windows around each spotter hit which requires time of order O(KM2) for K spotter hits, and window size proportional to M. The benefit of using the spotter is clearly illustrated in the M. capricolum searches. However, the time difference between the S. usitatus and the Sargasso Sea data searches shows that the spotter might lose its mission when dealing with many shorter sequences.
There are other approaches to predicting non-coding RNA. One commonly used method is sequence alignment, e.g. BLAST (3), Paralign (20) or FASTA (21). Another is based on structure-sensitive Stochastic Context Free Grammars (SCFG) (22) which form the basis of the tRNA prediction program tRNAscan-SE (23) and of Infernal (24), which is used when creating RFAM. While the sequence alignment methods are very fast, they are not particularly suited for prediction of non-coding RNA (1). Infernal, however, has a general worst case running time of order O(MN3) , which is prohibitive. The RFAM database (17,18), which includes 5S and the 5′ domain of 16S, uses BLAST to pre-screen genome sequences, followed by Infernal; despite a more efficient approach than the general SCFG, it does not analyze the entire 16S. A search for 5S in a 1 Mbp genome using Infernal took 4 hours 45 minutes: almost 1000 times as much as the 16 seconds used by RNAmmer for the much larger 16S model. A time-saving approach to SCFGs could be to use the RaveNna (25) package which can convert an RFAM SCFG to an HMM. This drastically reduces the running time; however, its usefulness would be limited since no models for the larger rRNAs are available. Another factor is that the 5S found by RaveNna (26) which were not already in RFAM were all in organellar sequences, sequences not analyzed by RNAmmer. For further comparisons and comments on these different methods, we refer to (1).
The RNAmmer program is available as a traditional HTML-based prediction server at http://www.cbs.dtu.dk/services/RNAmmer as well as through a SOAP-based web service. It is also available for download through the same site.
SUPPLEMENTARY DATA
Supplementary Data is available at NAR online.
ACKNOWLEDGEMENTS
We are grateful for funding from EMBIO at the University of Oslo, the Research Council of Norway and the Danish Center for Scientific Computing. It was also supported by a grant from the European Union through the EMBRACE Network of Excellence, contract number LSHG-CT-2004-512092. We would also like to thank our colleagues for critical reading of the manuscript. Funding to pay the Open Access publication charge was provided by Research Council of Norway.
Conflict of interest statement. None declared.
REFERENCES
- 1.Freyhult E, Bollback J, Gardner W., Jr Exploring genomic dark matter: a critical assessment of the performance of homology search methods on noncoding RNA. Genome Res. 2007;17:117–125. doi: 10.1101/gr.5890907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pedersen A, Jensen L, Brunak S, Staerfeldt H, Ussery D. A DNA structural atlas for Escherichia coli. J. Mol. Biol. 2000;299:907–930. doi: 10.1006/jmbi.2000.3787. [DOI] [PubMed] [Google Scholar]
- 3.Altschul S, Gish W, Miller W, Myers E, Lipman D. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 4.Wimberly B, Brodersen D, Clemons W, Jr, Morgan-Warren R, Carter A, Vonrhein C, Hartsch T, Ramakrishnan V. Structure of the 30s ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 5.Schluenzen F, Tocilj A, Zarivach R, Harms J, Gluehmann M, Janell D, Bashan A, Bartels H, Agmon I, et al. Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution. Cell. 2000;102:615–623. doi: 10.1016/s0092-8674(00)00084-2. [DOI] [PubMed] [Google Scholar]
- 6.Nissen P, Hansen J, Ban N, Moore P, Steitz T. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- 7.Yusupov M, Yusupova G, Baucom A, Lieberman K, Earnest T, Cate J, Noller H. Crystal structure of the ribosome at 5.5 Å resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 8.Srivastava A, Schlessinger D. Structure and organization of ribosomal DNA. Biochimie. 1991;73:631–638. doi: 10.1016/0300-9084(91)90042-y. [DOI] [PubMed] [Google Scholar]
- 9.Acinas S, Marcelino L, Klepac-Ceraj V, Polz M. Divergence and redundancy of 16s rRNA sequences in genomes with multiple rrn operons. J Bacteriol. 2004;186:2629–2635. doi: 10.1128/JB.186.9.2629-2635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jackson S, Cannone J, Lee J, Gutell R, Woodson S. Distribution of rRNA introns in the three-dimensional structure of the ribosome. J Mol Biol. 2002;323:35–52. doi: 10.1016/s0022-2836(02)00895-1. [DOI] [PubMed] [Google Scholar]
- 11.Evguenieva-Hackenberg E. Bacterial ribosomal RNA in pieces. Mol Microbiol. 2005;57:318–325. doi: 10.1111/j.1365-2958.2005.04662.x. [DOI] [PubMed] [Google Scholar]
- 12.Wuyts J, Perriere G, Van De Peer Y. The European ribosomal RNA database. Nucleic Acids Res. 2004;32 doi: 10.1093/nar/gkh065. Database issue, D101–D103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szymanski M, Barciszewska M, Erdmann V, Barciszewski J. 5s Ribosomal RNA database. Nucleic Acids Res. 2002;30:176–178. doi: 10.1093/nar/30.1.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hobohm U, Scharf M, Schneider R, Sander C. Selection of representative protein data sets. Protein Sci. 1992;1:409–417. doi: 10.1002/pro.5560010313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eddy S. Profile hidden markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 16.Henikoff S, Henikoff J. Position-based sequence weights. J. Mol. Biol. 1994;243:574–578. doi: 10.1016/0022-2836(94)90032-9. [DOI] [PubMed] [Google Scholar]
- 17.Griffiths-Jones S, Moxon S, Marshall M, Khanna A, Eddy S, Bateman A. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 2005;33 doi: 10.1093/nar/gki081. Database Issue, D121–D124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Griffiths-Jones S, Bateman A, Marshall M, Khanna A, Eddy S. Rfam: an RNA family database. Nucleic Acids Res. 2003;31:439–441. doi: 10.1093/nar/gkg006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Venter J, Remington K, Heidelberg J, Halpern A, Rusch D, Eisen J, Wu D, Paulsen I, Nelson K, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 20.Rognes T. ParAlign: a parallel sequence alignment algorithm for rapid and sensitive database searches. Nucleic Acids Res. 2001;29:1647–1652. doi: 10.1093/nar/29.7.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pearson W, Lipman D. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Durbin R, Eddy S.R, Krogh A, Mitchison G. Biological Sequence Analysis: Probabilistic Models of Proteins and Nucleic Acids. Cambridge University Press; 2000. [Google Scholar]
- 23.Lowe T, Eddy S. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eddy S. A memory-efficient dynamic programming algorithm for optimal alignment of a sequence to an RNA secondary structure. BMC Bioinformatics. 2002;3:18. doi: 10.1186/1471-2105-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weinberg Z, Ruzzo W. Sequence-based heuristics for faster annotation of non-coding RNA families. Bioinformatics. 2006;22(1) doi: 10.1093/bioinformatics/bti743. [DOI] [PubMed] [Google Scholar]
- 26.Weinberg Z, WL R. RECOMB 04: Proceedings of the Eighth Annual International Conference on Computational Molecular Biology. ACM Press; 2004. pp. 243–251. [Google Scholar]