Abstract
Conditional expression of short hairpin RNAs (shRNAs) to knock down target genes is a powerful tool to study gene function. The most common inducible expression systems are based on tetracycline-regulated RNA polymerase III promoters. During the last years, several tetracycline-inducible U6 and H1 promoter variants have been reported in different experimental settings showing variable efficiencies. In this study, we compare the most common variants of these promoters in several mammalian cell lines. For all cell lines tested, we find that several inducible U6 and H1 promoters containing single tetracycline operator (tetO) sequences show high-transcriptional background in the non-induced state. Promoter variants containing two tetO sequences show tight suppression of transcription in the non-induced state, and high tet responsiveness and high gene knockdown efficiency upon induction in all cell lines tested. We report a variant of the H1 promoter containing two O2-type tetO sequences flanking the TATA box that shows little transcriptional background in the non-induced state and up to 90% target knockdown when the inducer molecule (dox–doxycycline) is added. This inducible system for RNAi-based gene silencing is a good candidate for use both in basic research on gene function and for potential therapeutic applications.
INTRODUCTION
The technology of small interfering RNA (siRNA)-based gene knockdown has become a common method to study gene function in mammalian cells (1,2). The introduction of short double-stranded RNA's into cells leads to sequence-specific down regulation of endogenous mRNAs that match the siRNA. This post-transcriptional gene suppression process is referred to as RNA interference or simply RNAi.
RNAi can be induced in mammalian cells either by introduction of synthetic 21–23 nt siRNA's or by plasmids and viral vectors that express the siRNA molecules (3). In the latter case, siRNA molecules can be produced intracellularly as two single-stranded complementary RNA molecules from separate promoters or, more commonly, from a single promoter as a short hairpin RNA (shRNA). The shRNA molecule is then further processed to siRNA by cellular ribonuclease complexes (4,5). RNA suppression by hairpin siRNA (shRNA) has been shown to be more efficient than other siRNA methods tested (6).
Plasmid vector based siRNA expression strategies have several advantages over other methods. First, the costs of DNA oligomers for construction are much lower than of synthetic siRNA molecules. Plasmid vectors encoding a selectable marker are expected to be more efficiently transfected into cells than naked RNA molecules. And last, vector-based siRNA expression strategies offer the advantage of inducible expression in the cases where gene knockdown is expected to have a deleterious effect on the targeted cell. Stable expression of the siRNA can easily be obtained when selectable plasmids or viral vectors are used to deliver the expression module into cells. Stable gene-knockdown studies involving genes essential for cell growth or survival require a conditional system where siRNA expression is tightly regulated.
siRNA and shRNA synthesis systems in cells are most often driven by RNA polymerase III (pol III) promoters. There are several advantages to using RNA pol III systems. siRNA transcription is high, and the fact that it is driven by cis-acting elements found exclusively in the 5′-flanking region, results in uniform RNA molecules containing defined 5′ and 3′ ends (7–9). During the last years, several inducible promoter systems have been developed to control expression of small RNAs. Most of these are based on variants of the U6 and H1 RNA pol III promoters. Several different strategies exist to make these promoters respond to external signals: the Cre-loxP system (10), the ecdysone-inducible system (11), the lac-repressor system (12) and the tet-repressor system (13).
Tetracycline-responsive variants of both the U6 and H1 RNA pol III promoters have been used in several studies to drive conditional shRNA production (14–18). In these systems, a tetracycline operator (tetO) sequence is inserted near the TATA box of the promoters. TetO sequences are high-affinity binding sites for the specific binding of the tetracycline repressor (tetR). Once bound, tetR will prevent RNA pol III from binding to the promoter and transcription is prevented. Addition of the inducer tetracycline or various analogs (e.g. dox), which have high affinity for tetR, causes the tetR to dissociate from tetO and transcription to proceed (19).
Previous studies using tetracycline-inducible small RNA expression systems have reported variable results regarding background transcription and induction potential of the systems. We and others have noted that RNA pol III promoters containing a single tetO sequence show significant leakiness, resulting in high background transcription, in the non-induced state (15,17). On the other hand, similar promoter variants have been used with considerable success in other studies (14,16,20). Therefore, we have now performed a comparison of several tetracycline-inducible U6 and H1 promoter variants for conditional shRNA expression in a number of mammalian cell lines. The results show that U6 and H1 promoters containing a single tetO sequence show variable background transcription levels and response to the inducing agent depending on the cell line used. Both U6 and H1 promoter variants containing two tetO sequences show tight regulation of shRNA expression in all cell lines used in this study. We also describe a variant of the H1 promoter containing 2 tetO sequences (H1-2O2) that is almost completely inactive in the non-induced state and gives high shRNA expression level upon induction by dox. This inducible system for RNAi-based gene silencing is a good candidate for use both in basic research on gene function and for potential therapeutic applications.
RESULTS AND DISCUSSION
Generation of tet-inducible derivatives of the U6 and H1 promoters
To compare the effectiveness of conditional shRNA expression from RNA pol III promoters, three tetracycline-responsive forms of the U6 and H1 promoters were constructed in addition to the wild-type (wt) promoters. TetO sequences were inserted to replace wt promoter sequences adjacent to the TATA box as indicated in Figure 1. Both promoters were generated with the tetO sequence in an upstream (US), downstream (DS) or upstream plus downstream (US/DS) position relative to the TATA box. Both the U6 and H1 promoters are extremely compact and correct spacings between the transcriptional start and the essential 5′ flanking sequences (TATA box and PSE- proximal sequence element) are important for proper RNA expression. Therefore, to preserve the correct spacing in these promoters, only one complete tetO sequence can be inserted in each of the abovementioned positions of these promoters (see Figure 1).
Figure 1.
Nucleotide sequence presentation of the tetracycline-inducible U6 and H1 promoter variants and anti-luc shRNA used in this study. PSE (proximal sequence element) is shown in italic letters. The TATA box is shaded in gray. O1 and O2 indicate O1-type and O2-type tetracycline operator (tetO) sequences, respectively. TetO sequences are shown in bold letters. The anti-luciferase shRNA sequence is underlined.
For the tet-inducible U6 promoter, we used three previously reported variants: U6-O1-US, contains a O1-type tetO located upstream of the TATA box in the U6 promoter (13). U6-O2-DS and U6-2O2-US/DS, contain O2-type tetO in the downstream position (14) and two O2-type tetO flanking the TATA box (15,20), respectively.
For the tet-inducible H1 promoter variants we only included the O2-type operator sequences, since Lin et al. (15) had already shown that single O2-type tetO sequences are tighter regulators than O1-type tetO. Also, a US/DS inducible H1 promoter/operator variant containing two O1-type tetO sequences reported earlier (17), showed moderate transcriptional background and required high concentrations of the inducer molecule for high levels of transcription in transient transfection studies. We therefore set out to test H1-O2 variants. H1-O2-US was generated with an O2-type tetO upstream of the TATA box in the H1 promoter. Like for the U6 promoter, we also constructed an H1 promoter containing an O2-type tetO in the DS position relative to the TATA box (H1-O2-DS) (18). Finally, we constructed H1-2O2-US/DS, which contains two O2-type tetO sequences flanking the TATA box of the H1 promoter.
In order to monitor the expression level of the promoter/operator systems in non-induced and induced states, we cloned an efficient anti-luciferase (anti-luc) shRNA construct immediately downstream of the U6 and H1 transcription start sites (Figure 1). A termination sequence consisting of six uridine nucleotides was included to define the 3′ end of the shRNA transcript.
We were also interested in examining the characteristics of the various inducible promoter variants in different mammalian cell lines. Therefore, we constructed several cell lines stably expressing tetR. These include several human neuroblastoma cell lines (SK-N-BE2, SJNB8, IMR32, Kelly and SK-N-AS), a human cervical carcinoma cell line (HeLa) and a human osteosarcoma cell line (Saos-2). In addition, we analyzed inducible shRNA expression in a human embryonic kidney cell line (HEK293T-REx) available from Invitrogen, Carlsbad, CA, USA.
Analysis of the tet-inducible U6 and H1 promoters
The tetR-expressing cell lines were transiently co-transfected with three plasmids; a plasmid constitutively expressing the firefly luciferase reporter (pGL3-control), a plasmid constitutively expressing β-galactosidase in order to correct for variations in transfection efficiencies (pCMV-β-gal) and a test plasmid expressing the anti-luc shRNA from various U6 and H1 promoter variants.
Both the wt-U6 and wt-H1 promoters showed very efficient expression of anti-luc shRNA as seen by a dramatic reduction of luciferase activity (88–99%, see Figures 2 and 3). Expression of several different scrambled shRNA constructs showed no change in reporter activity as expected (data not shown).
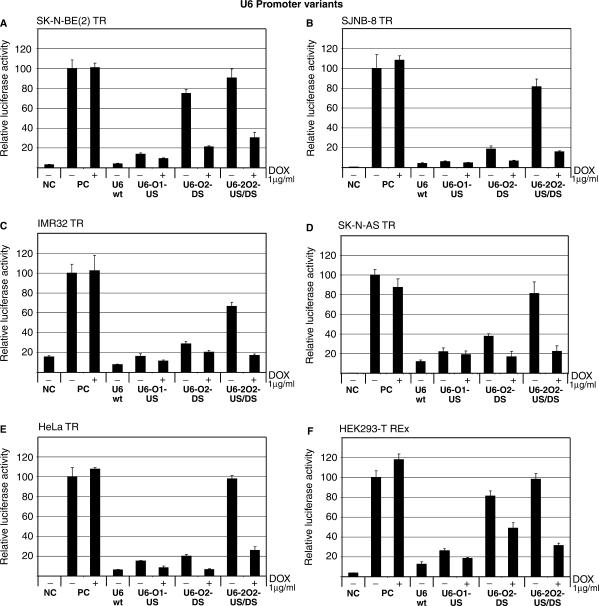
Figure 2.
Conditional shRNA expression from U6 promoter variants. TetR-expressing cells were transfected with 100 ng pCMV-β-gal, 800 ng pGL3-control (Promega) and 100 ng anti-luc shRNA expressing test plasmids under control of various U6 promoter variants. Several different tetR-expressing cell lines were used. (A) SK-N-BE (2) TR, (B) SJNB-8 TR, (C) IMR32 TR, (D) SK-N-AS TR, (E) HeLa TR, (F) HEK293-T REx (Invitrogen). NC: negative control; 100 ng pCMV-β-gal and 800 ng pGL3-basic (no luciferase expression), PC: positive control; 100 ng pCMV-β-gal and 800 ng pGL3-control (constitutive luciferase expression), + indicates addition of 1 μg/ml dox 1-day post-transfection. Cells were incubated for a total of 3 days. Error bars indicate SDs.
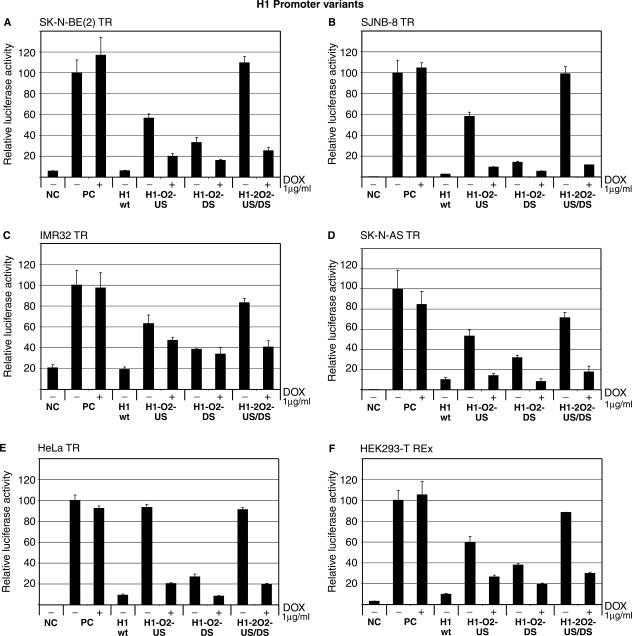
Figure 3.
Conditional shRNA expression from H1 promoter variants. TetR-expressing cells were transfected with 100 ng pCMV-β-gal, 800 ng pGL3-control (Promega) and 100 ng anti-luc shRNA expressing test plasmids under control of various H1 promoter variants. Several different tetR-expressing cell lines were used. (A) SK-N-BE (2) TR, (B) SJNB-8 TR, (C) IMR32 TR, (D) SK-N-AS TR, (E) HeLa TR, (F) HEK293-T REx (Invitrogen). NC: negative control; 100 ng pCMV-β-gal and 800 ng pGL3-basic (no luciferase expression), PC: positive control; 100 ng pCMV-β-gal and 800 ng pGL3-control (constitutive luciferase expression), + indicates addition of 1 μg/ml dox 1-day post-transfection. Cells were incubated for a total of 3 days. Error bars indicate SDs.
The addition of 1 μg/ml dox (a tetracycline derivative) to tetR-expressing cells transfected with pGL3-control and pCMV-β-gal had only minor effects on reporter expression (PC; positive control in Figures 2 and 3). This indicates that the measured differences in luciferase activity between the induced and non-induced states were due to the effect of the anti-luc shRNA. Addition of 5 or 10 μg/ml dox resulted in changes in cell morphology and growth inhibition, so in all other experiments 1 μg/ml dox was used (data not shown).
U6 promoter variants
As can be seen in Figure 2, the non-induced U6-O1-US promoter gives high background transcription of the anti-luc shRNA in all cell lines tested. This is observed as a dramatic drop in luciferase activity in the absence of dox compared to the positive controls (PC). Addition of 1 μg/ml dox had only minor effects on shRNA expression. Similar results published by other research groups support our observation that an O1-type tetO located between the PSE and TATA box of the U6 promoter is not efficiently regulated by the tetR protein in transient transfection experiments (15,16,20). Matsukura et al. (16) were able to show efficient regulation of the U6-O1-US promoter when stably transfected HCT116 cells were used. Here, induction of shRNA synthesis required high levels of dox (10 μg/ml), which might be due to high tetR levels in these cells. A difference in regulation between transient and stable transfection experiments similar to this was not observed by another research group (15).
When an O2-type tetO is placed between the TATA box and the transcriptional start of the U6 promoter (U6-O2-DS), a more variable result is observed. In the SJNB8 TR (Figure 2B), IMR32 TR (Figure 2C), SK-N-AS TR (Figure 2D), HeLa TR (Figure 2E) and Kelly TR (data not shown) cell lines, the U6-O2-DS promoter is poorly repressed by tetR, while the HEK293T-REx (Figure 2F), SK-N BE2 TR (Figure 2A) and Saos2 TR (data not shown) cell lines show a tighter regulation of the promoter. The observed difference cannot be explained by different tetR expression levels, since all these cell lines express similar levels of the tetR protein (data not shown). Previous reports also indicate a cell type difference in regulation of a similar U6-O2-DS inducible promoter. Lin et al. (15) reported low transcription repression from this promoter in the HeLa-TREx cell line (Invitrogen), while others have reported tight control of this promoter in PC-3 and 293T cells (14,20). These observations are consistent with our results.
Addition of an O2-type tetO in the US position of U6-O2-DS creates the U6-2O2-US/DS tet-inducible U6 promoter. When an anti-luc shRNA expressing plasmid under the control of this promoter is cotransfected with the reporter plasmid into tetR-expressing cell lines, high levels of luciferase were measured in the absence of dox. Addition of dox gave a dramatic reduction of luciferase activity in all cell lines tested (Figure 2). Compared to U6-wt promoter activity, the induced U6-2O2 promoter shows 69, 84, 89, 82, 77 and 78% reporter knockdown in SK-N-AS TR, SJNB-8 TR, IMR32 TR, SK-N-AS TR, HeLa TR and HEK293-T REx cell lines, respectively.
Together, these results show that the U6-2O2-US/DS promoter exhibit the best dox response in all tested cell lines compared to other tetO containing U6 promoter variants. These results are in agreement with previous published data (15), and we here show that this is valid in a wide variety of different cell lines.
H1 promoter variants
All H1 promoter variants used in this study contained the O2-type tetO sequence. When this operator was inserted in the US position (H1-O2-US), moderate to high shRNA repression (Figure 3) was seen in the absence of dox in all cell lines tested. This indicates low to moderate background transcription in the non-induced state. In contrast to the U6 promoter containing tetO in a similar position, H1-O2-US is responsive to tetR repression. The observed difference could be explained by the fact that the O2-type tetO is a tighter regulator than O1-type tetO (15). Addition of 1 μg/ml dox increased shRNA expression significantly in all cell lines.
shRNA expression from H1 promoter containing an O2-type tetO sequence in the DS position (H1-O2-DS) has previously been reported to give efficient knockdown of β-catenin in stably transfected CRC cell lines (18). Significant promoter leakage was observed in non-induced cell lines that exhibited efficient knockdown upon induction. Our results show that this promoter variant gives moderate to high transcriptional background in all tested cell lines (Figure 3). Induction of anti-luc shRNA expression by addition of dox efficiently down-regulates the luciferase reporter to levels below that observed for the similar U6 promoter variant, indicating that transcriptional activity of the H1 promoter variant is higher.
Finally, we created an H1 promoter containing two O2-type operators in the US and DS positions (H1-2O2-US/DS). This inducible promoter shows tight shRNA repression in absence of dox in all tested cell lines (Figure 3). Addition of 1 μg/ml dox induces shRNA expression resulting in low levels of luciferase after 2 days of incubation. This means that the tetR protein efficiently blocks transcription from the H1-2O2-US/DS promoter. Compared to H1-wt promoter activity, the induced H1-2O2 promoter shows 81, 91, 67, 84, 87 and 74% reporter knockdown in SK-N-AS TR, SJNB-8 TR, IMR32 TR, SK-N-AS TR, HeLa TR and HEK293-T REx cell lines, respectively. Matthess et al. (17) have reported moderate tightness and only a small difference between the induced and non-induced state of an H1 promoter containing two O1-type tetO sequences (2O1). This is in agreement with earlier observations showing that O2-type tetO are tighter regulators of RNA pol III promoters than O1-type tetO sequences (15). Very recently, Kappel et al. (21) showed that the background transcription of the 2O1-US/DS H1 promoter variant is less pronounced in stably transfected HeLa cells.
As can be seen from Figure 3, down-regulation of the luciferase reporter is most efficient in the neuroblastoma SJNB8 cell line (Figure 3B), where 91% reduction (compared to H1-wt) in luciferase activity was observed after addition of dox. Importantly, no background transcription from the promoter was observed in the non-induced state, indicating that this inducible promoter system is extremely tight.
In summary, most U6 and H1 promoter variants containing single tetO sequences are poorly regulated by tetR. In contrast, both U6-2O2-US/DS and H1-2O2-US/DS containing two O2-type tetO sequences are tight regulators of shRNA expression in all cell lines tested. For the cell lines used in this study, the H1-2O2-US/DS promoter is slightly more efficient for shRNA expression upon addition of the inducer dox than the U6-2O2-US/DS promoter The new tetO-containing RNA pol III promoters described in this study will be useful in basic research on gene function and possibly also for potential therapeutic applications.
Both single and double tetO inducible shRNA expression systems exist on the market today. In this work, we have constructed two new variants of an inducible H1 promoter (H1-O2-US and H1-2O2-US/DS) that show tight suppression of shRNA expression in the non-induced state, and high tet responsiveness and high reporter gene knock-down efficiency upon induction by dox. In addition, we have performed, for the first time, a detailed comparison of inducible shRNA expression systems in several different cell lines. Our results show that U6 and H1 promoter variants containing double tetO2 sequences are efficiently regulated by tetR in all cell lines tested, and we expect these promoter variants to be useful in other tetR-expressing cell lines as well.
MATERIALS AND METHODS
Molecular cloning
pSHAG-1 and pSHAG-Ff1 (5) contain a wt U6 promoter-driven expression cassette. pSHAG-Ff1 (U6 wt anti-luc shRNA in this study) encodes an anti-luc shRNA homologous to nucleotides 1340–1368 of the coding sequence of the firefly luciferase gene (NCBI accession number U47296) while pSHAG-1 is the negative control (NC) without shRNA sequence.
Table 1 shows the sequence of the oligonucleotides used for molecular cloning in this study.
Table 1.
Oligonucleotids used in this study
| Name | Sequence (5′–3′) |
|---|---|
| ON22 | ATAAGAATGCGGCCGCAAGGTCGGGCAGGAAGAGGGCC |
| ON31 | GATCGGATCCGGTGTTTCGTCCTTTCCACAAGATATATAACTCTATCAATGATAGAGTACTTTCAAGTTA CGGTAAGCA |
| ON34 | AGTCGGATCCAAAAAATGGATTCCAACTCAGCGAGAGCCACCCGATCAAGCTTCATCAGGTGGCTCCCG CTGAATTGGAATCCGGTGTTTCGTCCTTTCCAC |
| ON37 | ACGATCTCTATCACTGATAGGGAGATATATAAAGCCAAGAAATCG |
| ON38 | CGGGATCCAAAAAATGGATTCCAACTCAGCGAGAGCCACCCGATCAAGCTTCATCAGGTGGCTCCCGCT GAATTGGAATCCACGATCTCTATCACTGATAGGGAG |
| ON46 | GATCGAATTCGAACGCTGACGTCATCAAC |
| ON47 | GATCAGATCTGAGTGGTCTCATACAGAACTTATAAGATTCCCAAA |
| ON75 | GATCAGATCTGAGTGGTCTCATACAGAACTTATAAGTCTCTATCACTGATAGGGATTTCACGTTTATGGT GATTTCCCA |
| ON76 | GATCAGATCTCTATCACTGATAGGGAACTTATAAGTCTCTATCACTGATAGGGATTTCACGTTTATGGTGA TTTCCCA |
| ON77 | GATCCCGGATTCCAATTCAGCGGGAGCCACCTGATGAAGCTTGACGGGTGGCTCTCGCTGAGTTGGAATC CATTTTTTGGAAA |
| ON78 | AGCTTTTCCAAAAAATGGATTCCAACTCAGCGAGAGCCACCCGTCAAGCTTCATCAGGTGGCTCCCGCTGA ATTGGAATCCGG |
| ON83 | GATCGGATCCAAAAAATGGATTCCAACTCAGCGAGAGCCACCCGATCAAGCTTCATCAGGTGGCTCCCGCT GAATTGGAATCCGGTCTCTATCACTGATAGGGAGATATATAA |
| ON84 | GATCGGATCCGGTCTCTATCACTGATAGGGAGATATATAATCTCTATCACTGATAGGGAGTTTCAAGTTACG GTAAGCAT |
Plasmids containing variants of the U6 promoter were constructed by PCR. An U6 promoter containing a tetO sequence upstream of the TATA box (U6-O1-US) was made by PCR amplification of the U6 promoter from pSHAG-1 using ON22 and ON31 (encodes the operator sequence) as primers. The resulting PCR product was used as template in a second PCR with ON22 and ON34 as primers to add the anti-luc shRNA sequence downstream of the U6-O1 promoter. This PCR product was digested with NotI/BamHI and ligated into a NotI/BamH1 cut pSHAG-1 vector to produce pU6-O1-US anti-luc.
Construction of U6 promoters containing one O2-type tetO sequence downstream of the TATA box and two O2-type tetO sequences flanking the TATA box, were performed with PCR in a similar procedure using ON22/ON37 and ON22/ON84, respectively. These PCR products were further amplified using ON22/ON38 and ON22/ON83 as primers, digested and ligated into pSHAG-1 to produce pU6-O2-DS anti-luc and pU6-2O2-US/DS anti-luc.
H1 variant promoters expressing the anti-luc shRNA were made as follows: pENTRH1-O2 is a pENTR3c (Invitrogen)-based plasmid that contains an H1 promoter with an O2-type tet operator in the DS position, followed by a BglII/HindIII-removable 750 bp ‘stuffer’ fragment. The H1 promoter sequence can be removed from this plasmid by EcoRI/BglII digestion. pENTRH1-wt (containing a wt H1 promoter) was made by amplifying the H1 promoter from pENTRH1-O2-DS with ON46 (EcoRI)/ON47 (BglII) as primers, digesting the resulting PCR product with EcoRI/BglII and ligation into a EcoRI/BglII digested pENTRH1-O2-DS vector. pENTRH1-O2-US and pENTRH1-2O2-US/DS were made in a similar way using ON46/ON75 and ON46/ON76 as PCR primers, respectively. The BglII/HindIII-removable 750 bp ‘stuffer’ fragment was then replaced with an anti-luc shRNA sequence by ligation of annealed primers ON77/ON78 into these vectors to produce: pH1-wt anti-luc, pH1-O2-US anti-luc, pH1-O2-DS anti-luc and pH1-2O2-US/DS anti-luc. All plasmid constructs were verified by DNA sequencing.
Cell culture and transfection
SK-N-BE (2), Kelly, SK-N-AS, Saos-2 and HeLa cells were grown in RPMI1640 supplemented with 10% FBS. SJNB8 and IMR32 cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% FBS. HEK293-TREx cells (Invitrogen) were grown in Dulbecco's modified Eagle's medium supplemented with 10% FBS and 15 μg/ml blasticidin. All cells were maintained in a humidified 37°C incubator with 5% CO2, supplied with fresh complete medium every 3 days, and subcultured before confluence was reached.
Also, 4–5 × 105 cells were seeded into each well of a 12-well tissue culture plate and transfection was performed the following day with Lipofectamin2000 (Invitrogen) according to the manufacturer's protocol.
Generation of stable cell lines producing tetracyclin repressor (tetR)
Cell lines were maintained and transfected as described above with the plasmid pcDNA6TR (Invitrogen) carrying a gene encoding the selectable marker, blasticidin, and a gene coding for the tetracycline operon repressor protein (tetR). Stably transfected cell lines resistant to blasticidin were selected and cultured in blasticidin-containing media.
Luciferase/β-galactosidase assay
TetR-expressing cells were transfected with the luciferase reporter plasmid pGL3-control (Promega, Madison, WI, USA), the β-galactosidase expressing pCMV-β-gal (Stratagene, La Jolla, CA, USA) and a test-plasmid expressing the anti-luc shRNA from various U6 and H1 promoters. HEK293T-REx cells were not transfected with pCMV-β-gal, since these cells already express β-galactosidase from the Flp-In cassette. The NC was transfected with pGL3-Basic (Promega), which lacks a promoter for expressing the reporter gene.
An aliquot of 1 μg/ml dox was added 24 h after transfection and cells were harvested after an additional 48 h of incubation. Luciferase and β-galactosidase activities were measured in triplicate immediately using the Dual-Light® System (Applied Biosystems, Foster City, CA, USA) according to the manufacturer's instructions. Normalization of luciferase measurements from HEK293T-REx was done using total protein. All experiments were performed at least in triplicate.
ACKNOWLEDGEMENTS
This work was supported by grants from the Northern-Norwegian Health Authorities (gene therapy program to T.F.), the Erna and Olav Aakre Foundation for Cancer research (to T.F. and C.E.), the Dutch Cancer Society ‘KWF Kankerbestrijding’ (Grants UVA 2003-2849 to D.G. and R.V., and UVA 2005-3665 to D.G.) and the SKK (‘Stichting Kindergeneeskundig Kankeronderzoek’) (to R.V.). pENTRH1-O2 was a kind gift from Sylvia Sagen at University of Tromsø. Funding to pay the Open Access publication charges for this article was provided by the Northern-Norwegian Health Authorities (gene therapy program).
Conflict of interest statement. None declared.
REFERENCES
- 1.Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 2.Zhou D, He QS, Wang C, Zhang J, Wong-Staal F. RNA interference and potential applications. Curr. Top. Med. Chem. 2006;6:901–911. doi: 10.2174/156802606777303630. [DOI] [PubMed] [Google Scholar]
- 3.Amarzguioui M, Rossi JJ, Kim D. Approaches for chemically synthesized siRNA and vector-mediated RNAi. FEBS Lett. 2005;579:5974–5981. doi: 10.1016/j.febslet.2005.08.070. [DOI] [PubMed] [Google Scholar]
- 4.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 5.Paddison PJ, Caudy AA, Bernstein E, Hannon GJ, Conklin DS. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 2002;16:948–958. doi: 10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu JY, DeRuiter SL, Turner DL. RNA interference by expression of short-interfering RNAs and hairpin RNAs in mammalian cells. Proc. Natl Acad. Sci. USA. 2002;99:6047–6052. doi: 10.1073/pnas.092143499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell FE, Jr., Setzer DR. Transcription termination by RNA polymerase III: uncoupling of polymerase release from termination signal recognition. Mol. Cell. Biol. 1992;12:2260–2272. doi: 10.1128/mcb.12.5.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Myslinski E, Ame JC, Krol A, Carbon P. An unusually compact external promoter for RNA polymerase III transcription of the human H1RNA gene. Nucleic Acids Res. 2001;29:2502–2509. doi: 10.1093/nar/29.12.2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park JS, Kunkel GR. Both RNA polymerase III and RNA polymerase II accurately initiate transcription from a human U6 promoter in vitro. Biochem. Biophys. Res. Commun. 1995;214:934–940. doi: 10.1006/bbrc.1995.2376. [DOI] [PubMed] [Google Scholar]
- 10.Yu J, McMahon AP. Reproducible and inducible knockdown of gene expression in mice. Genesis. 2006;44:252–261. doi: 10.1002/dvg.20213. [DOI] [PubMed] [Google Scholar]
- 11.Gupta S, Schoer RA, Egan JE, Hannon GJ, Mittal V. Inducible, reversible, and stable RNA interference in mammalian cells. Proc. Natl Acad. Sci. USA. 2004;101:1927–1932. doi: 10.1073/pnas.0306111101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Higuchi M, Tsutsumi R, Higashi H, Hatakeyama M. Conditional gene silencing utilizing the lac repressor reveals a role of SHP-2 in cagA-positive Helicobacter pylori pathogenicity. Cancer Sci. 2004;95:442–447. doi: 10.1111/j.1349-7006.2004.tb03229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohkawa J, Taira K. Control of the functional activity of an antisense RNA by a tetracycline-responsive derivative of the human U6 snRNA promoter. Hum. Gene Ther. 2000;11:577–585. doi: 10.1089/10430340050015761. [DOI] [PubMed] [Google Scholar]
- 14.Czauderna F, Santel A, Hinz M, Fechtner M, Durieux B, Fisch G, Leenders F, Arnold W, Giese K, et al. Inducible shRNA expression for application in a prostate cancer mouse model. Nucleic Acids Res. 2003;31:e127. doi: 10.1093/nar/gng127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin X, Yang J, Chen J, Gunasekera A, Fesik SW, Shen Y. Development of a tightly regulated U6 promoter for shRNA expression. FEBS Lett. 2004;577:376–380. doi: 10.1016/j.febslet.2004.10.033. [DOI] [PubMed] [Google Scholar]
- 16.Matsukura S, Jones PA, Takai D. Establishment of conditional vectors for hairpin siRNA knockdowns. Nucleic Acids Res. 2003;31:e77. doi: 10.1093/nar/gng077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matthess Y, Kappel S, Spankuch B, Zimmer B, Kaufmann M, Strebhardt K. Conditional inhibition of cancer cell proliferation by tetracycline-responsive, H1 promoter-driven silencing of PLK1. Oncogene. 2005;24:2973–2980. doi: 10.1038/sj.onc.1208472. [DOI] [PubMed] [Google Scholar]
- 18.van de Wetering M, Oving I, Muncan V, Pon Fong MT, Brantjes H, van Leenen D, Holstege FC, Brummelkamp TR, Agami R, et al. Specific inhibition of gene expression using a stably integrated, inducible small-interfering-RNA vector. EMBO Rep. 2003;4:609–615. doi: 10.1038/sj.embor.embor865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berens C, Hillen W. Gene regulation by tetracyclines. Genet. Eng. (NY) 2004;26:255–277. doi: 10.1007/978-0-306-48573-2_13. [DOI] [PubMed] [Google Scholar]
- 20.Chen Y, Stamatoyannopoulos G, Song CZ. Down-regulation of CXCR4 by inducible small interfering RNA inhibits breast cancer cell invasion in vitro. Cancer Res. 2003;63:4801–4804. [PubMed] [Google Scholar]
- 21.Kappel S, Matthess Y, Zimmer B, Kaufmann M, Strebhardt K. Tumor inhibition by genomically integrated inducible RNAi-cassettes. Nucleic Acids Res. 2006;34:4527–4536. doi: 10.1093/nar/gkl628. [DOI] [PMC free article] [PubMed] [Google Scholar]