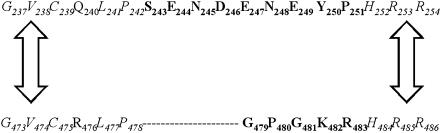Abstract
The accurate replication and transmission of genetic information is critical in the life of an organism. During its entire lifespan, the genetic information is constantly under attack from endogenous and exogenous sources of damage. To ensure that the content of its genetic information is faithfully preserved for synthesis and transmission, eukaryotic cells have developed a complex system of genomic quality control. Key players in this process are DNA polymerases, the enzymes responsible for synthesizing the DNA, because errors introduced into the genome by polymerase can result in mutations. We use DNA polymerase beta (pol β) as a model system to investigate mechanisms of preserving fidelity during nucleotide incorporation. In the study described here, we characterized the role that loop II of pol β plays in maintaining the activity and fidelity of pol β. We report here that the absence or shortening of loop II compromises the catalytic activity of pol β. Our data also show that loop variants of a specific length have a lower fidelity when compared to the wild-type polymerase. Taken together, our results indicate that loop II is important for the catalytic activity and fidelity of pol β.
INTRODUCTION
The accurate transmission of genetic information of an organism from one generation to the next is a crucial step during the lifespan of an organism. In order to ensure the absolute precision required in DNA synthesis and transmission, eukaryotic cells have derived a complex system of genome quality control to maximize accuracy and minimize mistakes resulting from DNA damage and inaccurate DNA synthesis. One of the best-known and best-described systems of DNA repair in eukaryotic cells is the base excision repair (BER) pathway. BER has been estimated to provide repairs for ∼10 000 lesions per cell per day from spontaneous oxidative and alkylation damage (1,2).
During BER, a specific DNA glycosylase recognizes its target DNA damage and excises the damaged base, leaving an abasic site (for review see 3); an apurinic/apyrimidinic endonuclease nicks the sugar–phosphate backbone and leaves a gap at the original DNA damage site (4). It is the role of DNA polymerase beta (pol β) to fill in this gap in a template-directed manner (5,6). Along with its polymerase function, pol β utilizes its 5′-deoxyribose phosphate lyase activity (7) to remove the 5′-deoxyribose phosphate moiety on the DNA substrate, and ready the product for ligation by a DNA ligase. The XRCC1- DNA ligase IIIα complex seals the nick during short-patch BER, while the same duty is thought to be performed by DNA ligase I during long-patch BER (8).
While all of the proteins involved in BER contribute an important and specialized role, it is pol β that is responsible for restoring the genetic information of the damaged DNA substrate back to its original state. Pol β is not as accurate as replicative DNA polymerases (9) because it lacks proofreading capability. However, this intrinsic lack of proofreading makes pol β an excellent candidate to study the mechanism of polymerization fidelity directly. Advantages in using pol β to study polymerization fidelity include the abundance of structural data (for example 10–14), and the small size of this protein resulting in ease of purification.
Like other template-directed polymerases, pol β selects the correct complementary base from a pool of four nucleotides. While the complementary nature of Watson–Crick base pairs lies at the heart of a template-directed polymerase's ability to select the correct incoming nucleotide opposite the templating base, the accuracy by which a DNA polymerase is able to perform its function delineates the fact that a polymerase's nucleotide discrimination is far greater than just the favorable thermodynamics of complementary base pairing (15). Therefore in order to gain a better understanding of how a polymerase achieves its fidelity, a detailed study of the structure–function relationship is imperative. Intuitively, the region of interest for fidelity would be amino acid residues near the active site region, as demonstrated by others (16,17). However, other studies have shown that regions that are located away from the active site and do not come into direct contact with either DNA substrate or incoming dNTP such as the hydrophobic hinge region, play a critical role in maintaining the fidelity of polymerization (18–26).
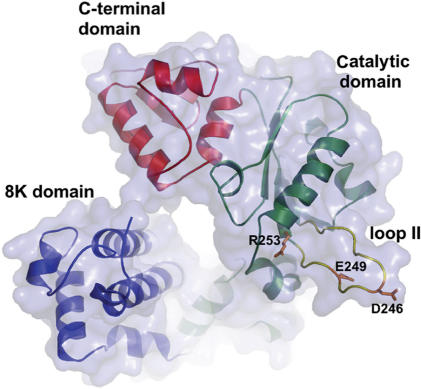
One region that is likely to be important for accurate DNA synthesis in pol β is the 14-amino acid loop II that spans residues 240–253 as shown in Figure 1. It is located on the outskirts of the palm domain, the domain in which the active site residues are located. As was first shown in the work presented by Garcia-Diaz et al. (27), structural comparison of the loop II region of two members of the Pol X family, pol β and DNA polymerase lambda (pol λ) revealed that there were some identical amino acid residues on each end of the loop as shown in Figure 2. Amino acid residues G-V-C-Q-L-P and G-V-C-R-L-P for pol β and pol λ, respectively, appear to anchor one end of loop II, whereas residues H-R-R appear to anchor the end of the loop region for both of these members of the pol X family (Figure 2).
Figure 1.
Pol β ribbon diagram of the loop region. Loop II, containing amino acid residues R253, E249 and D246, and the subdomains of pol β are labeled.
Figure 2.
Comparison of the loop II region of pol β and pol λ. Amino acid sequence surrounding and including loop II of mammalian pol β and mammalian pol λ is shown. The loop sequence of pol β is from residues 240–253, and the loop sequence of pol λ is from residues 476–485. The amino acid residues of the pol β loop that were altered in this study are in bold. The amino acids near the beginning and end of the loop that are conserved between pol β and pol λ are italicized and indicated by the two arrows in between the beginning and the end of both sequences.
Three different single amino acid alterations of loop II, R253M, D246V and E249K confer AZT resistance as depicted in Figure 1 (28). Two of these mutants, E249K and D246V, were characterized through detailed kinetic analyses (29,30). The mutant E249K was shown to have lower fidelity than wild-type pol β due to its tendency to extend mispaired bases at catalytic efficiencies greater than wild-type pol β (29). The lowered fidelity of D246V was attributed to its tendency to misincorporate T opposite template bases G and C, but the misincorporation was greatly influenced by the identity of the base 5′ to the templating base (30). Kinetic investigations of these two mutants suggest that loop II of pol β exerts a distinct influence on the primer position of the DNA substrate.
Loop II itself is solvent-exposed and highly flexible. While it is not completely disordered in known pol β crystal structures, its flexibility masks our ability to obtain a good structural understanding of this region. In order to gain a better, comprehensive understanding of its role in pol β polymerization fidelity, we constructed pol β mutants with shorter or chemically different types of loops, and characterized the resultant variants for DNA synthesis efficiencies and fidelity. Our data suggest that the length of loop II plays an important role in maintaining the activity and fidelity of polymerization.
MATERIALS AND METHODS
Chemicals and reagents
Ultrapure deoxynucleoside triphosphates (dNTPs), ATP and [γ32P]ATP were purchased from New England Biolabs, Sigma and Amersham Biosciences, respectively. All of the oligonucleotides used in these studies were synthesized by the Keck Molecular Biology Center at Yale University, and purified by denaturing polyacrylamide gel (20% acrylamide and 8 M urea) electrophoresis.
Site-directed mutagenesis of loop mutants
All the loop mutants were generated using the Stratagene site-directed mutagenesis protocol. The primers used for specific deletions and insertions are shown in Table 1. Site-directed mutagenesis was performed on wild-type rat pol β cDNA in the pET28a expression vector (28).
Table 1.
Primers for site-directed mutagenesis
| Primer | Sequence |
|---|---|
| Loopless FW | 5′ TGTTTGCCAGCTTCCC_CACAGGAGAATCGATATC 3′ |
| Loopless RV | 5′ GATATCGATTCTCCTGTG_GGGAAGCTGGCAAACA 3′ |
| 9-Ala Loop FW | 5′ GGGTGTTTGCCAGCTTCCCGCGGCGGCGGCGGCGGCGGCGGCGGCGCACAGGAGAATCGATATCAGG 3′ |
| 9-Ala Loop RV | 5′ CCTGATATCGATTCTCCTGTGCGCCGCCGCCGCCGCCGCCGCCGCCGCGGGAAGCTGGCAAACACCC 3′ |
| 5-Ala Loop FW | 5′ TTTGCCAGCTTCCCGCGGCGGCGGCGGCGCACAGGAGAATCGATATC 3′ |
| 5-Ala Loop RV | 5′ ATCGATTCTCCTGTGCGCCGCCGCCGCCGCGGGAAGCTGGCAAACA 3′ |
| 4-Ala Loop FW | 5′ TGTTTGCCAGCTTCCCGCGGCGGCGGCGCACAGGAGAATCGATATC 3′ |
| 4-Ala Loop RV | 5′ GATATCGATTCTCCTGTGCGCCGCCGCCGCGGGAAGCTGGCAAACA 3′ |
| 3-Ala Loop FW | 5′ GTTTGCCAGCTTCCCGCGGCGGCGCACAGGAGAATCG 3′ |
| 3-Ala Loop RV | 5′ CGATTCTCCTGTGCGCCGCCGCGGGAAGCTGGCAAAC 3′ |
| 2-Ala Loop FW | 5′ GGTGTTTGCCAGCTTCCCGCGGCGCACAGGAGAATCGATATC 3′ |
| 2-Ala Loop RV | 5′ GATATCGATTCTCCTGTGCGCCGCGGGAAGCTGGCAAACACC 3′ |
| 1-Ala Loop FW | 5′ GGGTGTTTGCCAGCTTCCCGCGCACAGGAGAATCGATATCAG 3′ |
| 1-Ala Loop RV | 5′ CTGATATCGATTCTCCTGTGCGCGGGAAGCTGGCAAACACCC 3′ |
| Loop1-FW | 5′ GTGTTTGCCAGCTTCCC_GAAAACGAATATCCACACA 3′ |
| Loop1-RV | 5′ TGTGTGGATATTCGTTTTC_GGGAAGCTGGCAAACAC 3′ |
| Lambda Loop FW | 5′ TGTTTGCCAGCTTCCCGGTCCGGGTAAACGTCACAGGAGAATCGATATC 3′ |
| Lambda Loop RV | 5′ GATATCGATTCTCCTGTGACGTTTACCCGGACCGGGAAGCTGGCAAACA 3′ |
| Loop2-FW | 5′ CTTCCCAGCGAGAATGAT_CACAGGAGAATCGATATC 3′ |
| Loop2-RV | 5′ GATATCGATTCTCCTGTG_ATCATTCTCGCTGGGAAG 3′ |
The underlined nucleotides in insertion primers (9-Ala, 5-Ala, 4-Ala, 3-Ala, 2-Ala, 1-Ala and Lambda-loop) indicate the nucleotides inserted into loopless sequence. The underlined space in deletion primers (Loopless, Loop1-, Loop2-)indicates where the deletion occurred in wild-type sequence.
The PCR conditions were 95°C for 30 s, 55°C for 1.5 min and 68°C for 12 min for 18 cycles. After mutagenesis, bacterial transformation and selection, the mutagenized plasmid DNA was isolated using the Qiagen Miniprep (Qiagen) kit and sequenced using the T7 forward primer for the pET constructs. All sequencing was performed at the Keck Molecular Biology Center at Yale University.
Expression and purification of proteins
Wild-type and mutant proteins were expressed from the pET28a (+) vector in the BL21DE3 strain background as described (28). The protein is expressed as a fusion protein with a hexahistidine tag at the amino terminus. Proteins were purified by nickel chelation followed by cation exchange using SP Sepharose on a fast performance liquid chromatography (FPLC) system (GE Healthcare). The purity of the proteins was determined by visualization on Coomassie blue-stained SDS-PAGE gels. Protein concentrations of wild-type pol β and loop mutants were calculated based on ɛ280 = 21 200 M−1 cm−1 and a molecular mass of 40 kDa.
DNA substrates for presteady-state and fidelity analysis
Oligonucleotides used to form the single base-pair gapped substrate (burst substrates) and five base-pair gapped substrate are shown in Table 2. The primer and template of the DNA duplex was phosphorylated at the 5′ end using T4-polynucleotide kinase (New England Biolabs), and [γ-32P]ATP (Amersham). The downstream oligonucleotide was phosphorylated at the 5′ end with non-radioactive ATP. Phosphorylated oligonucleotides were purified with Microspin P-30 or P-20 columns to eliminate the excess ATP (Bio-Rad). The labeled oligonucleotides were then annealed at the ratio of 1:1.2:1.3 for primer:template:downstream in 50 mM Tris-HCl (pH 8), 250 mM NaCl by heating at 95°C for 5 min, slow-cooling to 50°C for 30 min, and then incubating at 50°C for 20 min followed by an immediate transfer to ice. The quality of the annealed substrate was verified on 18% native polyacrylamide gel followed by autoradiography (19,23).
Table 2.
DNA duplex constructs for burst and fidelity assays
| DNA substrate | Sequence |
|---|---|
| Burst substrate 45T-22U-22D | 5′GCCTCGCAGCCGTCCAACCAAC CAACCTCGATCCAATGCCGTCC CGGAGCGTCGGCAGGTTGGTTGAGTTGGAGCTAGGTTACGGCAGG 3′ |
| Gap substrate CII45-CIIU-CIID | 5′TTGCGACTTATCAACGCCCACA AGCT GTCTT CTCAGTT CC AACGCTGAATAGTTGCGGGT GTAGTCATCGACAGAAGAGTCAAGG 3′ |
Presteady-state analysis
To determine if the rate-limiting step of the variants was similar to that of the wild-type protein, presteady-state kinetic analysis was performed. These reactions are commonly known as burst reactions and they were performed using a final concentration of 300 nM of 1-bp gapped substrate and 100 nM of wild type or variant pol β under investigation. All reactions were conducted in 50 mM Tris-HCl, pH 8.0, 2 mM dithiothreitol, 20 mM NaCl and 10% glycerol. The burst experiments were performed using a final concentration 100 μM dTTP, and initiated by rapid mixing of pol β/DNA duplex and Mg/dTTP solutions (MgCl2 was at a final concentration of 10 mM) using a Rapid Quench instrument (KinTek) (31). The reaction was quenched with 0.5 M EDTA. The product bands were resolved on a sequencing gel (Sequel NE Reagent Part A from American Bioanalytical) and visualized using a Molecular Dynamics Storm 840 Phosphorimager, and quantified using ImageQuant software.
Data analysis
The data collected for the presteady-state analysis were fit by nonlinear regression using KaleidaGraph 3.6 from Synergy Software. Data points from burst experiments were fit to the equation [product] = A [1 − exp(−kobst)] + ksst, where A is the amplitude of the burst, kobs is the observed first-order rate constant for dNTP incorporation and kss is the observed steady-state rate constant.
Base excision repair assay
Whole-cell extracts from pol β-deleted mouse embryo fibroblasts (Δβ-MEFs) were prepared as described (32). Purification, 5′-end labeling and annealing of oligonucleotides to prepare the BER substrate were as described (21) and the BER assays were performed as described (33). Each reaction was carried out in 10 μg of whole-cell extract. The final concentration of UDG-treated DNA substrate was 50 nM and that of pol β was 200 nM. The reaction products were resolved on 20% denaturing gel, exposed overnight and visualized using a Molecular Dynamics Storm 840 Phosphorimager. An unrepaired product is 16 nt in length. The 17-mer results from addition of one base by pol β. A repaired product that is ligated is 33 nt in length. Bands located between 18 and 33 nt are thought to be products of exonucleases present in the cell extract or strand-displacement synthesis from pol β. The reactions were resolved on sequencing gels, visualized using a Molecular Dynamics Storm 840 Phosphorimager, and quantified using ImageQuant software.
In vitro fidelity assay
To determine if any of the loop II variants have mutator activity, we employed the missing base primer extension assay. In this assay, we incubate 50 nM of radiolabeled substrate and 750 nM of protein (WT or mutant) in a buffer (50 mM Tris-HCl pH 8, 20 mM NaCl, 2 mM DTT and 10% glycerol) at 37°C for 2 min and then initiate the extension reaction by the addition of a dNTP cocktail missing either dATP, dTTP, dCTP or dGTP and containing 50 μM each of three dNTPs and 10 mM MgCl2 in the same buffer. The reaction was incubated at 37°C for various times. Here, 0.5 M EDTA was added at the conclusion of each of the time points to quench the reaction. The extended reaction products were resolved on a 20% Sequel NE polyacrylamide gel (American Bioanalytical). The primer and extended product bands were visualized using a Molecular Dynamics Storm 840 Phosphorimager, and quantified using ImageQuant software. The quantified product bands, those opposite template T and C for 0–10 min (in the case of Figure 6), of wild type and variants, were normalized against the background of the sequencing gel (0 min). This value was divided by the quantity of normalized primer to give a fraction of extended product. This quantity for the variants was divided by the same for wild-type, which yielded a ratio of variant product over wild-type product. The calculations were performed with Microsoft Excel.
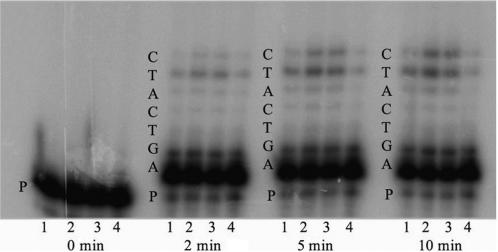
Figure 6.
Example of the in vitro five base-pair missing base primer-extension fidelity assay. Reaction products formed during the gap-filling assay when dCTP is missing from the nucleotide pool are shown. Purified enzymes were incubated with gapped DNA substrate that was radiolabeled at the 5′ end of the primer (Table 2) and a pool of three dNTPs missing dCTP for 2, 5 and 10 min at 37°C as described in the Materials and methods section. The products resulting from incorporation of nucleotides into the primer were resolved on a denaturing acrylamide gel and visualized on a Phosphorimager. Lane 1 is wild type, lane 2 is lambda-loop, lane 3 is 5-Ala loop, lane 4 is 4-Ala loop and P is the primer strand. The length of incubation time is listed underneath each group. The sequence of the template is listed on the side of each group.
In vivo Trp+ reversion assay
Loop II mutants were assayed using a genetic screen as previously described (34). The screen is based on the observation that pol β can substitute for DNA polymerase I of E. coli in DNA replication. The E.coli strain used was SC18-12 with the genotype recA718 polA12 uvrA155 trpE65 lon-11 sulA1. The SC18-12 strain containing either wild-type or mutant pol β variants that were expressed from a plasmid as described (24) were cultured overnight at 37°C in LB broth supplemented with tetracycline, ampicillin and 0.2% arabinose to induce expression of either the wild type or variant pol β. Serial dilutions of the overnight cultures were diluted in PBS of pH 7 and plated on both ET and Eglu plates. ET medium was E-salts supplemented with 0.4% glucose and 20 μg/ml of tryptophan (34). Eglu medium is ET without tryptophan. Reversion frequencies were calculated as follows: [(number of colonies/ml on Eglu)/(number of colonies/ml on ET)]. The data are presented as an increase in reversion frequency of mutants over that of wild type.
HSV-tk forward mutational assay
Selected loop II mutants and wild type were characterized in the HSV-tk forward mutational assay as previously described (18,35,36). Using the FT334 strain with the genotype recA13 upp tdk, inactivating mutations in the Herpes simplex virus type 1 thymidine kinase (HSV-tk) gene were selected in the presence of 5-fluoro-2′-deoxyuridine (FUdR). The substance is cytotoxic to cells carrying an active copy of the HSV-tk gene. The mutational frequency is calculated by dividing the total number of tk− colonies on FUdR by the total number of colonies plated as described (18,35,36).
RESULTS
The specific chemical nature of loop II does not affect activity of pol β
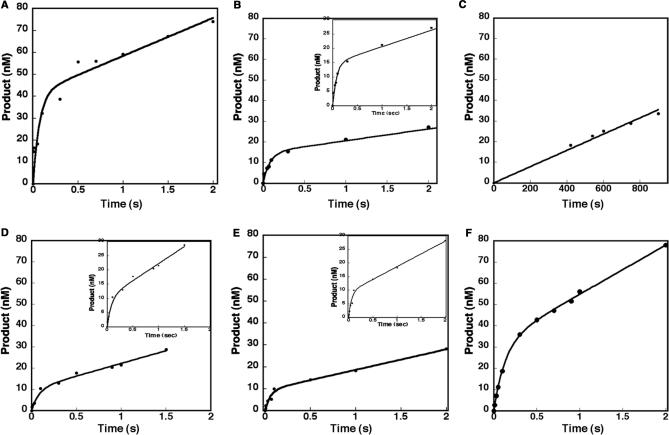
To address the question of whether the chemical nature of the amino acids of loop II are important for polymerase activity, we generated a loop mutant in which all of the loop residues were altered to alanine, called 9-Ala, as shown in Figure 3. This mutant retained the total length of the wild-type loop II, but differed considerably in its chemical nature. In order to ascertain whether the chemical nature of loop II affects the activity of pol β, we first used presteady-state kinetics to determine if 9-Ala retains the signature biphasic kinetics exhibited by wild type. After binding to DNA, pol β binds to dNTP substrate and is thought to undergo one or more conformational changes that result in activation of the enzyme for catalysis. The rate-limiting step for wild-type pol β in the pathway occurs after the polymerization step, and it is thought to be dissociation of the enzyme from the DNA (37,38). Figure 4A shows an example of a biphasic burst for wild-type pol β, in which the rate is 14.5 ± 5.9 s−1. Surprisingly, the 9-Ala loop exhibited biphasic burst kinetics, as shown in Figure 4B, with a rate of 12.2 ± 2.5 s−1, which is similar to the wild-type protein (22,39). This result shows that the chemical nature of the amino acids in loop II does not affect the rate of catalysis. We note that the burst amplitude of 9-Ala appears to be lower than that of wild type, and the reason for this is currently under investigation.
Figure 3.
Amino acid sequences of loop constructs. The name of the construct is on the left with wild-type loop sequence near the top. The residue positions above the amino acid sequences correspond to wild-type residues.
Figure 4.
Rapid burst kinetics of wild type and loop II variants. Insertion of dTTP into single nucleotide gapped DNA substrate 45T-22U-22D. Values in Table 2 were measured using the chemical quench-flow apparatus at 37°C. A preincubated cocktail of 100 nM loop mutant or WT and 300 nM gapped DNA was mixed with a cocktail of 100 μM dTTP containing 10 mM MgCl2. The reactions were quenched and monitored as described under the Materials and methods section. (A) The data for wild type were fit to the burst equation shown in the Materials and methods section. (B) An example of a loop mutant, 9-Ala, that exhibited rapid burst kinetics. Data were fit to the burst equation. The inset is an expansion of the Y-axis to show the points that define the burst. (C) Loopless does not exhibit rapid burst kinetics. Data were fit to a linear equation. (D–F) Examples of loop mutants with different residues, 5-Ala, Loop1- and Lambda-loop respectively, that exhibited rapid burst kinetics. Data were fit to the burst equation. The insets are an expansion of the Y-axis to show the points that define the burst. The kobs and steady-state constants for all loop mutants and wild type are listed in Table 3. The results are representative of at least three experiments.
Loop II is important for rapid catalysis by pol β
Next we asked whether pol β needs the flexible loop II to catalyze DNA synthesis. To answer this question we used site-directed mutagenesis to generate the Loopless mutant that is devoid of loop II as shown in Figure 3, purified the resulting protein and characterized its ability to synthesize DNA using presteady-state kinetics. In contrast to wild-type pol β and the 9-Ala variant, no products were observed for Loopless under burst reaction conditions (data not shown). However, when the reaction was incubated for longer periods of time, Loopless exhibited a slow linear phase of product formation, as shown in Figure 4C. This suggests that the rate-limiting step of this variant is different from that of wild-type pol β. The steady-state reaction rate of Loopless is 0.043 ± 0.004 s−1, which is ∼10-fold slower than the wild-type value in the linear phase (Table 3). Our results indicate that loop II is critical for pol β to exhibit a rapid burst of product formation, but that loop II is not essential for pol β to catalyze DNA synthesis.
Table 3.
Presteady-state and steady-state constants
| Loop constructs | Amino acid sequence | Presteady-state burst constant (s−1) | Steady-state constant (s−1) | |
|---|---|---|---|---|
| Wild type | QLP SENDENEYP | HR | 14.5 ± 5.93 | 0.422 |
| Loopless | QLP | HR | No burst | 0.0394 ± 0.0010 |
| 9-Ala Loop | QLPAAAAAAAAA | HR | 12.2 ± 2.55 | 0.386 |
| 5-Ala Loop | QLPAAAAA | HR | 12.0 ± 4.34 | 1.16 |
| 4-Ala Loop | QLPAAAA | HR | 12.2 ± 1.71 | 1.02 |
| 3-Ala Loop | QLPAAA | HR | 11.0 ± 1.98 | 0.343 |
| 2-Ala Loop | QLPAA | HR | No burst | 0.599 ± 0.080 |
| 1-Ala Loop | QLPA | HR | No burst | 0.407 ± 0.044 |
| Loop1- | QLPENEYP | HR | 16.8 ± 4.99 | 1.01 |
| Lambda Loop | QLP GPGKR | HR | 7.58 ± 0.589 | 0.739 |
| Loop2- | QLPSEND | HR | 14.5 ± 4.99 | 0.457 |
The minimum length of loop II required for pol β to exhibit burst kinetics is three residues
The first mutants we constructed revealed two properties of the loop. The chemical nature of the 9-Ala residues in loop II did not appear to alter the rate-limiting step of pol β, and loop II is required to maintain burst kinetics but is not essential for activity. Next, we wished to determine if there was a minimum loop II length that was critical for pol β to exhibit burst kinetics. We generated loop mutants of consecutively shorter lengths that consisted of only alanine residues, as shown in Figure 3. We started with a 5-Ala loop mutant because pol λ, a very close relative of pol β, also has five residues in loop II. We used presteady-state kinetics to determine whether these variants with shorter loops exhibited burst kinetics. The presteady-state burst constants of 5-Ala (Figure 4D), 4-Ala, and 3-Ala (not shown) are 12.0 ± 4.3, 12.2 ± 1.7, and 11.0 ± 1.9 s−1 respectively (Table 3), which are similar to wild-type pol β (22,39). Both the 5-Ala and Loop1- variants exhibited low amplitudes and this is currently under investigation. However, the 2-Ala and 1-Ala loop variants did not exhibit burst kinetics under presteady-state reaction conditions (data not shown). Like Loopless, the rate-limiting step for these two variants appears to be at or before the chemistry step. This result indicates that in order for pol β to exhibit burst kinetics, loop II must have a length of at least three residues.
Our results thus far suggested that loop II length was important for the catalytic activity of pol β and that its chemical nature does not appear to be important for catalysis. To confirm this, we generated the lambda-loop and Loop1- variants, which each have five amino acid residues in loop II. As shown in Figures 2 and 3, lambda-loop contains amino acid residues that make up the loop II region of pol λ, and Loop1- retains the last five amino acid residues of the original pol β loop II. Thus these variants contain loops that vary in their chemical nature, but are five amino acid residues in length.
Lambda-loop and Loop1- were assayed by using the same presteady-state experimental conditions as the other loop mutants. Our data indicate that both of these variants exhibit a rapid burst of product formation under presteady-state conditions (Figure 4E and F). The lambda-loop variant has a burst constant of 7.58 ± 0.58 s−1, which indicates that it catalyzes DNA synthesis at a somewhat slower rate than wild-type pol β. The Loop1- variant has a burst constant of 16.8 ± 4.9 s−1, which is similar to that of wild-type pol β (22,39). Importantly, both of these loop variants retained the biphasic burst profile which suggests that the nature of the amino acids of loop II does not appear to affect the catalytic activity of pol β.
To confirm that the length, rather than chemical nature, of loop II is the principal determinant of polymerase activity, we generated another loop mutant, Loop2-, which contains four of the original amino acids from the loop of wild-type pol β, as shown in Figure 3. When this mutant was assayed for activity under presteady-state conditions it exhibited a biphasic kinetic profile (data not shown) very similar to wild type. Loop2- has a burst constant of 12.2 ± 1.7 s−1, which is also very similar to that of wild type (22,39). In summary, our results show that loop II must consist of at least three amino acid residues in order to maintain the wild-type biphasic kinetic profile, but that deletion of the entire loop does not lead to a complete loss of activity.
Loop II mutants participate in reconstituted base excision repair
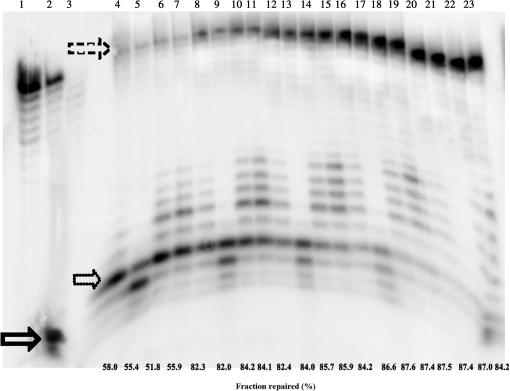
To determine if loop II mutants participate in BER, we characterized all loop II mutants in a reconstituted BER assay. A uracil-containing DNA duplex was treated with uracil DNA glycosylase (UDG). The UDG-treated DNA was then incubated with whole-cell extract from Δβ-MEFs in the presence of pol β wild-type or loop II mutants. The BER reactions were performed at five different time points, and by 20 min all the repaired n + 1 product had been ligated to form the expected elongated and sealed product. All of the loop mutants participate in BER and Figure 5 shows an example of the BER assay for wild-type pol β and the Loopless, 9-Ala and 5-Ala variants. Thus, even mutants that do not exhibit burst kinetics participate in BER. Note that variants of pol β that do not possess any polymerase activity are completely inactive in this assay (J.B.S. submitted for publication).
Figure 5.
Loop II mutants participate in BER. Lane 1 is the annealed 33-mer oligonucleotide that was the starting material; lane 2 is the 33-mer oligonucleotide that was treated with UDG; lane 3 is UDG-treated 16-mer substrate incubated with whole-cell extract from Δβ-MEF. The solid arrow points to the radiolabeled 16-mer that is the product of base removal and incision by UDG and APE I, respectively. The dotted arrow points to the 17-mer, resulting from n + 1 addition by pol β. The dashed arrow near the top points to ligated product, which is 33 nt in length. Lanes 4–7 are incubation for 1 min with Δβ-MEF. Lanes 8–11 are incubation for 2 min with Δβ-MEF. Lanes 12–15 are incubation for 5 min with Δβ-MEF. Lanes 16–19 are incubation for 10 min with Δβ-MEF. Lanes 20–23 are incubation for 20 min with Δβ-MEF. Lanes 4, 8, 12, 16 and 20 contain wild-type pol β. Lanes 5, 9, 13, 17 and 21 contain Loopless. Lanes 6, 10, 14, 18 and 22 is 9-Ala. Lanes 7, 11, 15, 19 and 23 contain 5-Ala. The fraction of 16-mer that was repaired is below each lane.
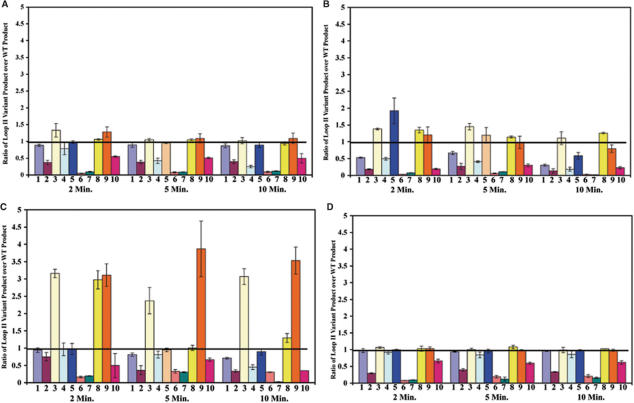
Altering the length of loop II affects the fidelity of pol β
The second major question we addressed was whether loop II of pol β is critical for accurate DNA synthesis. Each of the loop II variants was tested for their fidelity in the missing base primer extension assay (28,40). This assay qualitatively measures the ability of DNA polymerase to insert incorrect nucleotides and extend mispaired primer-termini. An example of results comparing the lambda-loop, 5-Ala and 4-Ala to that of wild type is shown in Figure 6 and the quantitative results for all loop variants are presented in Figure 7. Strikingly, each of the loop variants containing five amino acid residues, namely 5-Ala, Loop1- and lambda-loop, (Figure 7; bars 3, 8 and 9, respectively) appear to insert an incorrect nucleotide opposite template G and extend the resulting mispaired terminus. These variants consistently generate 2.5–4-fold more product than wild type in the absence of dCTP, suggesting that they could be mutator variants of pol β. The other loop variants appear to be at least as accurate at wild-type pol β in this assay.
Figure 7.
Lambda-loop, 5-Ala and Loop1- are mutators in the missing base primer extension assay in the absence of dCTP. Quantification of the products for all loop II variants was performed using Phosphoimager and ImageQuant software as described in the Materials and methods section. (A) nucleotide pool missing A; (B) nucleotide pool missing T; (C) nucleotide pool missing C; (D) nucleotide pool missing G. The X-axis lists the loop mutants. 1 is 9-Ala, 2 is Loopless, 3 is 5-Ala, 4 is 4-Ala, 5 is 3-Ala, 6 is 2-Ala, 7 is 1-Ala, 8 is Loop1-, 9 is lambda-loop and 10 is Loop2-. Loop mutants are organized in groups of 10 for each graph with the incubation time points of 2, 5 and 10 min listed underneath. Y-axis is the fold-increase of loop II variant product over wild-type product. The results are the average of three experiments. Error bars represent the standard deviation of the mean.
5-Ala, Loop1- and Lambda-loop variants exhibit a mutator phenotype
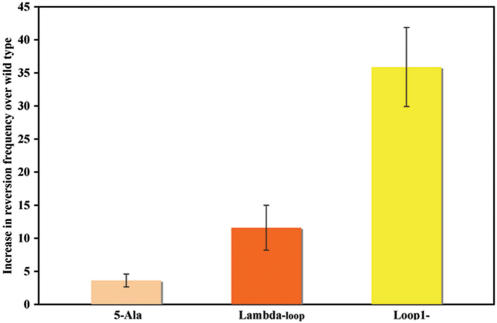
We have previously shown that rat pol β can substitute for DNA polymerase I of E.coli in the joining of Okazaki fragments during DNA replication (41). Because pol β synthesizes DNA in E.coli, it enables us to employ an E.coli genetic assay to detect mutator activity (34). In order to confirm that the five amino acid residue loop variants exhibited mutator activity, we characterized them in the Trp+ reversion assay (34,42). The SC18-12 E.coli strain, which carries the trpE65 allele, needs to be maintained in the presence of tryptophan because of an ochre mutation in the trpE gene, which ultimately prevents the synthesis of tryptophan. A Trp+ reversion in either the trpE65 gene or an anticodon loop of a tRNA can occur due to the insertion of the incorrect dNTP by an error-prone pol β variant. This permits the strain to grow in the absence of tryptophan and is a measure of the mutator activity of the pol β variant that is expressed in the strain. As shown in Figure 8, the reversion frequencies for all three of the five amino acid residue loop II mutants are greater than wild type. Loop1-, lambda-loop and 5-Ala exhibit 35-, 11- and 3-fold increases, respectively, in Trp+ reversion frequencies over wild type. Results from the Trp+ reversion assay confirm the mutator phenotypes of five residue loop II mutants that were observed in the in vitro gap-filling assay.
Figure 8.
Five residue loop II mutants exhibit mutator activity in the Trp+ reversion assay. The SC18-12 strain containing plasmid encoding either wild-type or loop II mutant were cultured, diluted and plated as described in the Materials and methods section. The bar graph illustrates the increase in reversion frequency of selected loop II mutant over that of wild type. X-axis is the loop II mutant. Y-axis is the increase in reversion frequency over that of wild type. The results are averaged from three independent experiments. Error bars represent the standard deviation.
Five residue loop II mutants have an increased mutation frequency in a forward mutation assay
The HSV-tk forward mutation assay allows us to obtain mutation frequencies of selected loop II mutants from in vitro reactions (18,35,36). This assay can detect a large range of mutations including single base substitutions, insertions, deletions and multiple mutations. All three five-residue loop variants exhibit mutator activity in this assay. Like the Trp+ reversion assay, the mutant with the highest tk mutation frequency is Loop1-, which has an almost 8-fold increase over wild type, as shown in Table 4. This was followed by 5-Ala and Lambda-loop where the mutation frequencies are 7-fold and 3-fold, respectively, greater than wild-type pol β. Taken together, the results of three different fidelity assays suggest that loop II variants of pol β consisting of five amino acid residues exhibit mutator activity.
Table 4.
HSV-tk forward mutation frequency of wild-type and five residue loop II mutants
| Polymerase | Viable cells (per ml) CmR | Mutant cells (per ml) CmR FUdRR | HSV-tk mutant frequency (10−4) | HSV-tk mutant frequency increase over WT |
|---|---|---|---|---|
| DNA pol β WT | 68 800 | 160 | 23.2 | |
| 25 800 | 100 | 38.7 | ||
| 25 200 | 100 | 39.6 | ||
| Mean (SD) | 33.8 (9.22) | |||
| 5-Ala | 28 200 | 760 | 269.5 | |
| 40 800 | 1080 | 264.7 | ||
| 60 000 | 1200 | 200 | ||
| Mean (SD) | 245 (38.8) | 7.24 | ||
| Lambda-loop | 62 040 | 820 | 132 | |
| 45 840 | 680 | 148.3 | ||
| 98 700 | 1100 | 111.4 | ||
| Mean (SD) | 130 (18.6) | 3.84 | ||
| Loop1- | 17 760 | 400 | 225 | |
| 12 420 | 340 | 273 | ||
| 19 800 | 600 | 303 | ||
| Mean (SD) | 267 (39.3) | 7.90 |
DISCUSSION
The goal of the work presented here was to determine if loop II of pol β is essential for activity and accurate DNA synthesis. This study was prompted by previous work from our laboratory in which we demonstrated that loop II was important for substrate specificity and the prevention of mispair extension (28–30).
In the study described here, we characterized a number of different variants of loop II for their ability to function in and maintain fidelity during DNA synthesis. Remarkably, we found that loops consisting of three to nine alanine residues were as or nearly as active as wild-type pol β and exhibited burst kinetics, suggesting that the overall rate-limiting step of these variants is the same as that of wild type. In addition, each of these loop variants appear to synthesize DNA with fidelity that is similar to wild-type pol β. Replacement of the nine residue loop II with either four or five residues other alanine also resulted in a polymerase that could catalyze DNA synthesis with similar activity as wild-type pol β. Loops consisting of 0–2 alanines were also able to catalyze DNA synthesis, but at much slower rates and with altered kinetics in comparison to wild-type pol β. Taken together, our results lead us to conclude that the length of the loop, rather than its chemical nature, is critical for pol β polymerase activity. Surprisingly, although loops consisting of five amino acid residues of various chemical natures were active, the resulting polymerases catalyze DNA synthesis with lower fidelity than wild-type pol β. Based upon these observations and our previous results (28–30), we conclude that loop II is important for the fidelity of pol β.
Chemical nature and size of the loop is not important for activity
Surprisingly, we found that a pol β variant with a loop consisting of nine Ala residues was just as active as wild-type pol β, and exhibited burst kinetics, suggesting that it followed a kinetic pathway quite similar to wild-type pol β. Pol β variants with loops of 5, 4 and 3 Ala residues were also found to exhibit burst kinetics. These results suggest that the chemical nature and size of loop II are not important for activity until the loop is comprised of less than three residues.
We and others (28–30,43) have proposed that loop II may somehow contribute to the formation of a gate that regulates access of dNTPs to the active site, and that the length and mobility of the loop controls the size of such a channel. The mobility of loop II is well documented through X-ray crystallographic studies (13,14,44,45). None of the structures published to date show any side-chain or main-chain interactions of any kind that would stabilize a certain loop conformation. Model building of short loop II mutants followed by molecular dynamics calculations and comparison to other members of the pol X family suggest that only the longest variants of loops could theoretically play a role in gating dNTP access not through direct steric interference but through long-range electrostatic interactions. In terminal transferase co-crystal structures (46), for example, where the loop equivalent to loop II in pol β consists of 17 residues, the geometry of the dNTP channel is not visibly affected by the loop. Pol λ, in which loop II consists of five residues, shows an increased affinity for the dNTP substrate when compared to pol β (47,48). Taken together, these results indicate that the loop and its proposed contribution to a gating function are unlikely to affect the movement of dNTP substrates in and PPi product out of the active site in a major way.
Shorter loop II mutants, however, exhibit significant kinetic effects. These are conceivably caused by strain and distortions in the main-chain geometry of residues adjacent to or within strands 4 and 5. Computer modeling and molecular dynamics of the Loopless model of pol β indicate that the position and backbone angles of Leu241, Pro242 and His252 have to be altered significantly (data not shown). The changes in alpha carbon positions required to reconnect the main chain of pol β occur only 13 Å away from Asp256 and the metal-ion-binding site in the catalytic domain. Mutations in loop residues can have wide-ranging effects, even on rigid structures as those found in beta alpha barrels (49,50). It is possible that drastic changes in the composition and length of loop II have similar effects on the stability of adjacent beta strands in the pol β active site. An increase in domain rigidity of pol β is known to affect the accuracy of DNA synthesis as observed in the case of the Met282Leu mutator mutant (22). We suggest that a loop II consisting of three amino acid residues is sufficient to maintain the backbone angles and positions of Leu241, Pro242 and His252 that are required for efficient catalysis.
We have also proposed that loop II functions to position the primer strand of the DNA by buttressing against helix I of pol β. In initiating these studies, we assumed that a smaller loop II would have diminished its ability to support helix I and the thumb domain of pol β would affect the primer position. In terms of catalytic activity, this proposal seems questionable since loops of four and three amino acid residues still exhibit burst kinetics.
A five amino acid loop affects the fidelity of pol β
We were surprised to find that variants with loops consisting of five amino acid residues, irrespective of a specific sequence context or side-chain character, synthesized DNA with lower fidelity than wild-type pol β. The simplest explanation for this result is that a loop of five residues assumes a structure that somehow impairs the accurate synthesis of DNA. It is likely that the backbone angles and positions of Leu241, Pro242 and His252 are maintained in the five residue loop variants because they are active, but that the length of these loops induces subtle alterations in the protein that result in inaccurate DNA synthesis. These subtle alterations could result in a lack of template or primer stabilization, or alteration in the dNTP-binding pocket. Characterization of the types of mutations induced by the five residue loop variants should be informative regarding their mechanisms of inaccurate DNA synthesis.
Another possible explanation for the lack fidelity of the five residue loop variants is that the position of Asp256 may be affected. The Mg2+-bound dNTP substrate is proposed to enter the active site first, and is initially positioned by the interaction of its phosphate moiety with the enzyme (51). The Mg2+ ion coordinates Asp190 and 192. Upon binding of the Mg2+–dNTP, the polymerase is thought to assume a closed conformation. The catalytic Mg2+ then binds and coordinates both Asp256 and Asp192. It also coordinates with the 3′ OH of the primer strand. After binding of the catalytic Mg2+, phosphodiester bond formation is observed in crystals (14). Asp256, which is part of a β strand, is just three amino acid residues away from loop II and could be affected by geometric strain or movements of the five residue loop variants. Thus, if a five-member loop assumes an aberrant structure or lacks proper dynamic movement, it could result in destabilization of Asp256. This could lead to slightly altered positioning of the 3′ terminus of the primer or the incoming dNTP and result in misincorporation.
Loop II and BER
In our BER assay, all of the loop II variants were able to participate in the reaction that led to the finished ligation product, including loops consisting of 0–2 amino acid residues. This suggests that pol β does not have to possess robust polymerase activity to participate in BER, and supports the idea that the dRP lyase activity of pol β is critical for repair (5). However, we have recently found that a pol β variant that possesses dRP lyase activity but no polymerase activity does not support BER in vitro (Lang et al., submitted for publication), suggesting that both the dRP lyase and polymerase activities of pol β are critical for repair. The demonstration that even the Loopless variant of pol β participates in BER also suggests that loop II is unlikely to be important for interactions with other proteins that are important for BER.
SUMMARY AND IMPLICATIONS
Our results show that a flexible loop that is located at a distance away from the active site of pol β is important for pol β activity, likely by maintaining the backbone angles and positions of Leu241, Pro242 and His252. Our results also implicate loop II as having a role in maintaining pol β fidelity. These findings support the idea that amino acid residues of pol β that are distant from its active site are critical for activity and fidelity.
ACKNOWLEDGEMENTS
We thank Shibani Dalal for help and suggestions throughout this study. We also thank Daniela Starcevic for her suggestions during the inception of this study. This work is dedicated to the memory of Richard Lin Lin This research was supported by NIH grant CA80830 (to J.B.S.). Funding to pay the Open Access publication charge was provided by the National Cancer Institute.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 2.Barnes D.E., Lindahl T. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu. Rev. Genet. 2004;38:445–476. doi: 10.1146/annurev.genet.38.072902.092448. [DOI] [PubMed] [Google Scholar]
- 3.McCullough A.K., Dodson M.L., Lloyd R.S. Initiation of base excision repair: glycosylase mechanisms and structures. Annu. Rev. Genet. 1999;68:255–285. doi: 10.1146/annurev.biochem.68.1.255. [DOI] [PubMed] [Google Scholar]
- 4.Demple B., Harrison L. Repair of oxidative damage to DNA: enzymology and biology. Annu. Rev. Biochem. 1994;63:915–948. doi: 10.1146/annurev.bi.63.070194.004411. [DOI] [PubMed] [Google Scholar]
- 5.Sobol R.W., Horton J.K., Kuhn R., Gu H., Singhal R.K., Prasad R., Rajewsky K., Wilson S.H. Requirement of mammalian DNA polymerase-beta in base-excision repair. Nature. 1996;379:183–186. doi: 10.1038/379183a0. [DOI] [PubMed] [Google Scholar]
- 6.Kubota Y., Nash R.A., Klungland A., Schar P., Barnes D.E., Lindahl T. Reconstitution of DNA base excision-repair with purified human proteins: interaction between DNA polymerase beta and the XRCC1 protein. EMBO J. 1996;15:6662–6670. [PMC free article] [PubMed] [Google Scholar]
- 7.Matsumoto Y., Kim K. Excision of deoxyribose phosphate residues by DNA polymerase β during repair. Science. 1995;269:699–702. doi: 10.1126/science.7624801. [DOI] [PubMed] [Google Scholar]
- 8.Timson D.J., Singleton M.R., Wigley D.B. DNA ligases in the repair and replication of DNA. Mutat. Res. 2000;460:301–318. doi: 10.1016/s0921-8777(00)00033-1. [DOI] [PubMed] [Google Scholar]
- 9.Kunkel T.A. The mutational specificity of DNA polymerase-beta during in vitro DNA synthesis: production of frameshift, base substitution, and deletion mutations. J. Biol. Chem. 1985;260:5787–5796. [PubMed] [Google Scholar]
- 10.Batra V.K., Beard W.A., Shock D.D., Krahn J.M., Pedersen L.C. Magnesium-induced assembly of a complete DNA polymerase catalytic complex. Structure. 2006;14:757–766. doi: 10.1016/j.str.2006.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pelletier H., Sawaya M.R., Kumar A., Wilson S.H., Kraut J. Structures of ternary complexes of rat DNA polymerase beta, a DNA template-primer, and ddCTP. Science. 1994;264:1891–1903. [PubMed] [Google Scholar]
- 12.Pelletier H., Sawaya M.R., Wolfle W., Wilson S.H., Kraut J. Crystal structures of human DNA polymerase beta complexed with DNA: implications for catalytic mechanism, processivity, and fidelity. Biochemistry. 1996;35:12742–12761. doi: 10.1021/bi952955d. [DOI] [PubMed] [Google Scholar]
- 13.Sawaya M.R., Prasad R., Wilson S.H., Kraut J., Pelletier H. Crystal structures of human DNA polymerase beta complexed with gapped and nicked DNA: evidence for an induced fit mechanism. Biochemistry. 1997;36:11205–11215. doi: 10.1021/bi9703812. [DOI] [PubMed] [Google Scholar]
- 14.Arndt J.W., Gong W., Zhong X., Showalter A.K., Liu J., Dunlap C.A., Lin Z., Paxson C., Tsai M.D., et al. Insight into the catalytic mechanism of DNA polymerase beta: structures of intermediate complexes. Biochemistry. 2001;40:5368–5375. doi: 10.1021/bi002176j. [DOI] [PubMed] [Google Scholar]
- 15.Loeb L.A., Kunkel T.A. Fidelity of DNA synthesis. Annu. Rev. Biochem. 1982;51:429–457. doi: 10.1146/annurev.bi.51.070182.002241. [DOI] [PubMed] [Google Scholar]
- 16.Osheroff W.P., Beard W.A., Wilson S.H., Kunkel T.A. Base substitution specificity of DNA polymerase β depends on interactions in the DNA minor groove. J. Biol. Chem. 1999;274:20749–20752. doi: 10.1074/jbc.274.30.20749. [DOI] [PubMed] [Google Scholar]
- 17.Beard W.A., Shock D.D., Yang X.P., DeLauder S.F., Wilson S.H. Loss of DNA polymerase β stacking interactions with templating purines, but not pyrimidines, alters catalytic efficiency and fidelity. J. Biol. Chem. 2002;277:8235–8242. doi: 10.1074/jbc.M107286200. [DOI] [PubMed] [Google Scholar]
- 18.Opresko P.L., Sweasy J.B., Eckert K.A. The mutator form of polymerase beta with amino acid substitution at tyrosine 265 in the hinge region displays an increase in both base substitution and frame shift errors. Biochemistry. 1998;37:111–119. doi: 10.1021/bi9722711. [DOI] [PubMed] [Google Scholar]
- 19.Li SX., Vaccaro J.A., Sweasy J.B. Involvement of phenylalanine 272 of DNA polymerase beta in discriminating between correct and incorrect deoxynucleoside triphosphates. Biochemistry. 1999;38:4800–4808. doi: 10.1021/bi9827058. [DOI] [PubMed] [Google Scholar]
- 20.Clairmont C.A., Narayanan L., Sun K.W., Glazer P.M., Sweasy J.B. The Tyr-265-to-Cys mutator mutant of DNA polymerase beta induces a mutator phenotype in mouse LN12 cells. Proc. Natl. Acad. Sci. USA. 1999;96:9580–9585. doi: 10.1073/pnas.96.17.9580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shah A.M., Li SX., Anderson K.S., Sweasy J.B. Y265H mutator mutant of DNA polymerase beta. Proper geometric alignment is critical for fidelity. J. Biol. Chem. 2001;276:10824–10831. doi: 10.1074/jbc.M008680200. [DOI] [PubMed] [Google Scholar]
- 22.Shah A.M., Conn D.A., Li SX., Capaldi A., Jager J., Sweasy J.B. A DNA polymerase beta mutator mutant with reduced nucleotide discrimination and increased protein stability. Biochemistry. 2001;38:11372–11381. doi: 10.1021/bi010755y. [DOI] [PubMed] [Google Scholar]
- 23.Shah A.M., Maitra M., Sweasy J.B. Variants of DNA polymerase beta extend mispaired DNA due to increased affinity for nucleotide substrate. Biochemistry. 2003;36:10709–10717. doi: 10.1021/bi034885d. [DOI] [PubMed] [Google Scholar]
- 24.Starcevic D., Dalal S., Sweasy J.B. Hinge residue Ile260 of DNA polymerase beta is important for enzyme activity and fidelity. Biochemistry. 2005;44:3775–3884. doi: 10.1021/bi047956x. [DOI] [PubMed] [Google Scholar]
- 25.Starcevic D., Dalal S., Jaeger J., Sweasy J.B. The hydrophobic hinge region of rat DNA polymerase beta is critical for substrate binding pocket geometry. J. Biol. Chem. 2005;280:28388–28393. doi: 10.1074/jbc.M502178200. [DOI] [PubMed] [Google Scholar]
- 26.Dalal S., Hile S., Eckert K.A., Sun K.W., Starcevic D., Sweasy J.B. Prostate-cancer-associated I260M variant of DNA polymerase beta is a sequence-specific mutator. Biochemistry. 2005;44:15664–15673. doi: 10.1021/bi051179z. [DOI] [PubMed] [Google Scholar]
- 27.Garcia-Diaz M., Dominguez O., Lopez-Fernandez L.A., de Lera L.T., Saniger M.L., Ruiz J.F., Parraga M., Garcia-Ortiz M.J., Kirchhoff T., et al. DNA polymerase lambda (Pol lambda), a novel eukaryotic DNA polymerase with a potential role in meiosis. J. Mol. Biol. 2004;301:851–867. doi: 10.1006/jmbi.2000.4005. [DOI] [PubMed] [Google Scholar]
- 28.Kosa J.L., Sweasy J.B. 3′-Azido-3′-deoxythymidine-resistant mutants of DNA polymerase β identified by in vivo selection. J. Biol. Chem. 1999;274:3851–3858. doi: 10.1074/jbc.274.6.3851. [DOI] [PubMed] [Google Scholar]
- 29.Kosa J.L., Sweasy J.B. The E249K mutator mutant of DNA polymerase β extends mispaired termni. J. Biol. Chem. 1999;274:35866–35872. doi: 10.1074/jbc.274.50.35866. [DOI] [PubMed] [Google Scholar]
- 30.Dalal S., Kosa J.L., Sweasy J.B. The D246V mutant of DNA polymerase β misincorporates nucleotides. J. Biol. Chem. 2004;279:577–584. doi: 10.1074/jbc.M309607200. [DOI] [PubMed] [Google Scholar]
- 31.Johnson K.A. Conformational coupling in DNA polymerase fidelity. Annu. Rev. Biochem. 1993;62:685–713. doi: 10.1146/annurev.bi.62.070193.003345. [DOI] [PubMed] [Google Scholar]
- 32.Biade S., Sobol R.W., Wilson S.H., Matsumoto Y. Impairment of proliferating cell nuclear antigen-dependent apurinic/apyrimidinic site repair on linear DNA. J. Biol. Chem. 1998;273:898–902. doi: 10.1074/jbc.273.2.898. [DOI] [PubMed] [Google Scholar]
- 33.Podlutsky A.J., Dianova I.I., Wilson S.H., Bohr V.A., Dianov G.L. DNA synthesis and dRPase activities of polymerase β are both essential for single-nucleotide patch base excision repair in mammalian cell extracts. Biochemistry. 2001;40:809–813. doi: 10.1021/bi002064s. [DOI] [PubMed] [Google Scholar]
- 34.Washington S.L., Yoon M.S., Chagovetz A.M., Li S.X., Clairmont C.A., Preston B.D., Eckert K.A., Sweasy J.B. A genetic system to identify DNA polymerase β mutator mutants. Proc. Natl. Acad. Sci. USA. 1997;94:1321–1326. doi: 10.1073/pnas.94.4.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eckert K.A., Hile S.E., Vargo P.L. Development and use of an in vitro HSV-tk forward mutation assay to study eukaryotic DNA polymerase processing of DNA alkyl lesions. Nucleic Acid Res. 1997;25:1450–1457. doi: 10.1093/nar/25.7.1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eckert K.A., Mowery A., Hile S.E. Misalignment-mediated DNA polymerase β mutations: comparison of microsatellite and frame-shift error rates using a forward mutation assay. Biochemistry. 2002;1:10490–10498. doi: 10.1021/bi025918c. [DOI] [PubMed] [Google Scholar]
- 37.Werneburg B.G., Ahn J., Zhong X., Hondal R.J., Kraynov V.S., Tsai M.D. DNA polymerase beta: pre-steady-state kinetic analysis and roles of arginine-283 in catalysis and fidelity. Biochemistry. 1995;35:7041–7150. doi: 10.1021/bi9527202. [DOI] [PubMed] [Google Scholar]
- 38.Zhong X., Patel S.S., Werneburg B.G., Tsai M.D. DNA polymerase beta: multiple conformational changes in the mechanism of catalysis. Biochemistry. 1997;39:11891–11900. doi: 10.1021/bi963181j. [DOI] [PubMed] [Google Scholar]
- 39.Vande Berg B.J., Beard W.A., Wilson S.H. DNA structure and aspartate 276 influence nucleotide binding to human DNA polymerase beta. Implication for the identity of the rate-limiting conformational change. J. Biol. Chem. 2001;276:10824–10831. doi: 10.1074/jbc.M002884200. [DOI] [PubMed] [Google Scholar]
- 40.Suzuki M., Avicola A.K., Hood L., Loeb L.A. Low fidelity mutants in the O-helix of Thermus aquaticus DNA polymerase I. J. Biol. Chem. 1997;272:11228–11235. doi: 10.1074/jbc.272.17.11228. [DOI] [PubMed] [Google Scholar]
- 41.Sweasy J.B., Loeb L.A. Mammalian DNA polymerase beta can substitute for DNA polymerase I during DNA replication in Escherichia coli. J. Biol. Chem. 1992;267:1407–1410. [PubMed] [Google Scholar]
- 42.Witkin E.M., Roegner-Maniscalco V., Sweasy J.B., McCall J.O. Recovery from ultraviolet light-induced inhibition of DNA synthesis requires umuDC gene products in recA718 mutant strains but not in recA+ strains of Escherichia coli. Proc. Natl. Acad. Sci. USA. 1987;84:6805–6809. doi: 10.1073/pnas.84.19.6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beard W.A., Wilson S.H. Structural design of a eukaryotic DNA repair polymerase: DNA polymerase β. Mutat. Res. 2000;460:231–244. doi: 10.1016/s0921-8777(00)00029-x. [DOI] [PubMed] [Google Scholar]
- 44.Davies J.F. II, Almassy R.J., Hostomska Z., Ferre R.A., Hostomsky Z. A crystal structure of the catalytic domain of DNA polymerase beta. Cell. 1994;76:1123–1133. doi: 10.1016/0092-8674(94)90388-3. [DOI] [PubMed] [Google Scholar]
- 45.Krahn J.M., Beard W.A., Miller H., Grollman A.P., Wilson S.H. Structure of DNA polymerase β with the mutagenic DNA lesion 8-oxodeoxyguanine reveals structural insights into its coding potential. Structure. 2003;11:121–127. doi: 10.1016/s0969-2126(02)00930-9. [DOI] [PubMed] [Google Scholar]
- 46.Delarue M., Boule J.B., Lescar N., Expert-Bezancon N., Jourdan N., Sukumar N., Rougeon F., Papanicolaou C. Crystal structures of a template-independent DNA polymerase: murine terminal deoxynucleotidyltransferase. EMBO J. 2002;21:427–439. doi: 10.1093/emboj/21.3.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garcia-Diaz M., Bebenek K., Krah J.M., Blanco L., Kunkel T.A., Pederson L.C. A structural solution for the DNA polymerase λ-dependent repair of DNA gaps with minimal homology. Mol. Cell. 2004;13:561–572. doi: 10.1016/s1097-2765(04)00061-9. [DOI] [PubMed] [Google Scholar]
- 48.Diaz-Garcia M., Bebenek K., Krahn J.M., Kunkel T.A., Pedersen L.C. A closed conformation for the catalytic cycle. Nat. Stru. Mol. Biol. 2005;12:97–98. doi: 10.1038/nsmb876. [DOI] [PubMed] [Google Scholar]
- 49.Luger K., Szadkowski H., Kirschner K. An 8-fold beta alpha barrel protein with redundant folding possibilities. Protein Eng. 1990;3:249–258. doi: 10.1093/protein/3.4.249. [DOI] [PubMed] [Google Scholar]
- 50.Urfer R., Kirschner K. The importance of surface loops for stabilizing an eightfold beta alpha barrel protein. Protein Sci. 1992;1:31–45. doi: 10.1002/pro.5560010105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bakhtina M., Lee S., Wang Y., Dunlap C., Lamarche B., Tsai MD. Use of viscogens, dNTPαS, and rhodium (III) as probes in stopped-flow experiments to obtain new evidence for the mechanism of catalysis by DNA polymerase. Biochemistry. 2005;44:5177–5187. doi: 10.1021/bi047664w. [DOI] [PubMed] [Google Scholar]