Abstract
Deadenylation is the first and probably also rate-limiting step of controlled mRNA decay in eukaryotes and therefore central for the overall rate of gene expression. In yeast, the process is maintained by the mega-Dalton Ccr4-Not complex, of which both the Ccr4p and Pop2p subunits are 3′–5′ exonucleases potentially responsible for the deadenylation reaction. Here, we present the crystal structure of the Pop2p subunit from Schizosaccharomyces pombe determined to 1.4 Å resolution and show that the enzyme is a competent ribonuclease with a tunable specificity towards poly-A. In contrast to S. cerevisiae Pop2p, the S. pombe enzyme contains a fully conserved DEDDh active site, and the high resolution allows for a detailed analysis of its configuration, including divalent metal ion binding. Functional data further indicates that the identity of the ions in the active site can modulate both activity and specificity of the enzyme, and finally structural superposition of single nucleotides and poly-A oligonucleotides provide insight into the catalytic cycle of the protein.
INTRODUCTION
Controlled removal of mRNA is a crucial regulatory point of gene expression in all cells. In eukaryotes, cytoplasmic mRNA degradation usually begins with shortening of the 3′ poly-A tail (deadenylation), which is followed by removal of the 5′ cap structure (decapping) and 5′–3′ exonucleolytic degradation of the mRNA body [reviewed in (1)]. In an alternative pathway, processive 3′–5′ degradation by the cytoplasmic exosome can directly follow deadenylation. In both scenarios, however, deadenylation is the first and probably also rate-limiting step and therefore a central control point of mRNA turnover and overall gene expression in eukaryotes (2).
The major deadenylase function resides in the mega-Dalton Ccr4-Not complex and in the yeast, Saccharomyces cerevisiae, enzymatic activity has been localized to two proteins with exonuclease signatures, Ccr4p and Pop2p (also called Caf1p) (3), both potentially responsible for the process. However, several lines of evidence from both in vitro and in vivo experiments indicate that Ccr4p may be the main nuclease responsible for deadenylation (4,5). Nevertheless, Pop2p orthologues from many organisms show strong homology to DEDD-type exonucleases and both the mammalian and S. cerevisiae protein can function as an independent deadenylase in vitro (6–9). In addition, disruption of the caf1 gene encoding Pop2p leads to longer mRNA half-lives and incomplete deadenylation in S. cerevisiae (3,7). Nevertheless, the deadenylase function of Pop2p does not appear to be absolutely required for poly-A removal in yeast in vivo (9). These conflicting observations mean that the functional importance of the dual nuclease activity in the deadenylase complex is still not understood.
In mammals, and most likely other higher eukaryotes, the Ccr4-Not complex is part of a biphasic deadenylase system that also includes the exonuclease Pan2 and its partner, Pan3 (10). In these organisms, deadenylation is initiated by the Pan2-Pan3 deadenylation complex, which shortens the poly-A tail to ∼110 nt before the Ccr4-Not complex takes over and completes the process. An additional eukaryotic deadenylase enzyme, poly-A-specific RiboNuclease (PARN), has also been identified (11). This nuclease may be required for more specialized processes in some eukaryotes, such as AU-rich-element (ARE)-mediated mRNA decay, which involves the rapid decay of mRNAs containing AREs in their 3′ UTR (12). However, PARN is not part of the major deadenylation system (10) and is not conserved in all eukaryotes (1).
Pop2p, Pan2 and PARN all belong to the DEDDh subgroup of the DEDD family of nucleases (7,13), while Ccr4p is related to DNase I and Exonuclease III (14). DEDD nucleases have three aspartates (D) and a glutamate (E) in their active site and the DEDDh subgroup additionally requires a nearby histidine for activity (DEDDh). The four acidic residues assist in coordinating two divalent metal ions, which are involved in both substrate binding and interaction with a water molecule essential for the proposed general, hydrolytic reaction mechanism (15,16).
Crystal structures of two of the DEDDh-type deadenylases have previously been reported; the exonuclease domain of the S. cerevisiae Pop2p (8) and the N-terminal part including the exonuclease domain of the human PARN protein (17). In addition, the structure of PARN co-crystallized with a poly-A oligomer has also been described (17). However, none of these structures contain the two metal ions required in the active site, and in S. cerevisiae Pop2p, the core DEDDh consensus motif is replaced by an unusual SEDQt sequence. Although the nuclease domain of S. cerevisiae Pop2p was shown to possess RNase activity on polynucleotide substrates in vitro (8), this activity was not specific to poly-A and therefore left open the question of whether Pop2p really acts as an active subunit during deadenylation in yeast. In the light of this, we decided to investigate the structure and function of the Pop2p orthologue from fission yeast, Schizosaccharomyces pombe, which in contrast to the S. cerevisiae protein contains a fully conserved DEDDh active site and also naturally lacks the repetitive N-terminal sequence that was removed for crystallization of the S. cerevisiae protein. In this article, we present the crystal structure of the full-length S. pombe Pop2p protein determined to 1.4 Å resolution by X-ray crystallography. The high-resolution data allows for a precise description of the active site of the enzyme, including the two metal ions. We show that the protein can function as a competent 3′–5′ riboexonuclease in vitro and further that the specificity and activity of the enzyme depend on the identity of the available divalent metal ions. A comparison of the active site with those of homologous enzymes allows us to propose a model for substrate binding and cleavage, and homology analysis leads to the identification of Pop2p and deadenylase-specific regions.
MATERIALS AND METHODS
Cloning and expression
In the initial PCR, Pop2pombeLICtev1-22: 5′-GACGACGACAAGATGGAGAATCTTTATTTTCAGGGC ATGAATTCGAATTTTTCTTATC-3′ and Pop2pombeLIC999-979,1M: 5′-GAGGAGAAGCCCGGTTTAAACTACACGAGGAGGAAA-3′ were used as forward and reverse primers to amplify the full-length caf1 (Systematic name: SPCC18.06c) open reading frame from S. pombe contig c1676, clone c18 obtained from GeneDB (http://www.genedb.org) (18). Sequences used for the ligation-independent cloning procedure are underlined. The forward primer introduces a Tev-protease cleavage site in the construct (bold) allowing for removal of the fusion tag during protein purification, and the reverse primer contains a single nucleotide mismatch (italics) that optimizes the stop codon from UAG to UAA. The PCR product was inserted into the pET-30 Ek/LIC vector (Novagen) according to the manufacturer's instructions and the cloning was verified by sequencing. The pET-30 Ek/LIC vector adds a polyhistidine tag in the N-terminus of the construct, which is used in the first step of protein purification. The D50A active site mutant was constructed by PCR using the pET-30 Ek/LIC-Pop2p vector as a template and the reverse complementary mutagenesis primers 5′-CCAGTTGTTTCTATGGCTACAGAATTTCCAGG-3′ (sense, bold = mutation) and 5′-CCTGGAAATTCTGTAGCCATAGAAACAACTGG-3′ (anti-sense). After the PCR, the template vector was removed by digestion with the methylation-sensitive restriction endonuclease, DpnI, before transforming E. coli cells with the unligated PCR product. The single A to C point mutation, which changes the codon GAU (D50) into GCU (A50), was confirmed by sequencing. For both wt and mutant proteins, positive clones were transformed into E. coli Rosetta (DE3) or Rosetta (DE3) pLysS cells (Novagen) and grown overnight at 37°C in 2 × TY media containing 30 μg/ml kanamycin and 34 μg/ml chloramphenicol to an OD600 of 0.6–0.8, then cold-shocked on ice for ∼30 min before induction of fusion protein expression with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Expression continued at 20°C overnight in a shaking incubator. Cells were collected by centrifugation and either lysed immediately or stored at −20°C.
Protein purification
Cells were resuspended in lysis buffer [50 mM Tris pH 8.0, 200 mM KCl, 20% glycerol, 5 mM MgCl2, 1 mM phenylmethylsulphonyl fluoride (PMSF) and 3 mM β-mercaptoethanol (BME)] and ruptured using an EmulsiFlex-C5 high-pressure cell homogenizer (Avestin). The cell lysate was cleared by centrifugation and 20 mM imidazole was added to the supernatant before loading onto a pre-packed 5 ml Ni2+-agarose column (GE Healthcare) pre-equilibrated with lysis buffer and 20 mM imidazole. After thorough washing with 50 mM Tris pH 8.0, 200 mM KCl, 5 mM MgCl2, 3 mM BME and 20 mM imidazole, the protein was eluted by stepping to 250 mM imidazole. The protein concentration was estimated by the Bradford protein assay (Bio-Rad) and the polyhistidine tag removed from the fusion protein by proteolytic cleavage with recombinant Tev protease (100:1 (w/w) fusion protein to Tev protease) for 1 h at room temperature. Following digestion, the sample was dialysed against 50 mM Tris pH 8.0, 100 mM KCl, 5 mM MgCl2 and 3 mM BME overnight at 4°C to lower the imidazole and salt concentrations. Subsequently, the sample was passed over the Ni2+-column again (pre-equilibrated in dialysis buffer) to remove the fusion tag, His-tagged Tev-protease and any non-cleaved fusion protein from the preparation. The flow-through and wash (dialysis buffer) containing the cleaved fusion protein was filtrated through a 0.2 µm Minisart plus filter (Sartorius) to remove any aggregates, then loaded onto a MonoQ 4.6/100 PE anion exchange column (GE Healthcare) pre-equilibrated in buffer A (50 mM Tris pH 8.0, 100 mM KCl, 5 mM MgCl2 and 5 mM BME). The protein was eluted with a gradient into buffer B (50 mM Tris pH 8.0, 1 M KCl, 5 mM MgCl2 and 5 mM BME), and fractions containing Pop2p were pooled and precipitated in 70% saturated ammonium sulphate overnight on ice. The precipitate was collected by brief centrifugation (13.200 r.p.m. in a tabletop centrifuge at 4°C) and dissolved in a small volume buffer A. The sample was centrifuged again (13.200 r.p.m. in a tabletop centrifuge at 4°C for 15 min) before loading onto a Superdex 200 10/300 GL gel-filtration column (GE Healthcare) pre-equilibrated in buffer A. Peak fractions containing monodisperse Pop2p protein were pooled and concentrated using a Centricon YM-10 filtration device (Millipore) to a protein concentration of 7–10 mg/ml as determined by the Bradford assay. Finally, the sample was ultracentrifuged at 70 000 × g for 15 min at 4°C to remove any aggregates from the concentration step before being used for crystallization.
In vitro deadenylation assay
Both wt and D50A mutant proteins used for the in vitro assays were purified as described above with the exception that the final gel filtration buffer contained no magnesium chloride (50 mM Tris pH 8.0, 100 mM KCl and 5 mM BME). RNA substrates Generic-polyA (5′-CAGCUCCGCAUCCCUUUCCCAAAAAAAAAA-3′) and Stemloop-polyA (5′-GGGCGGGCGCAAGCCCGCCCAAAAAAAAAA-3′) were synthesized with a 5′ fluorescein label and gel purified (Invitrogen). All in vitro reactions contained a 100-fold excess of Pop2p (1 μM) over RNA substrate (10 nM) and were carried out in 10 mM Tris-Cl, pH 8.0, 10 mM DTT, 50 mM KCl, 5 U/μl RNase inhibitor (New England Biolabs), and 5 mM MgCl2, 5 mM MnCl2 and/or 1 mM ZnCl2 as indicated. The reaction with the physiological mix of ions contained 7.1 mM MgCl2, 75 μM MnCl2 and 220 μM ZnCl2 (19). Reactions were allowed to proceed for the indicated amount of time at 30°C, before they were stopped with 10 μg proteinase K (Sigma-Aldrich) for 2 min at 37°C followed by the addition of RNA loading buffer (8 M urea, 5 mM Tris-HCl, pH 7.5, 0.5% (w/v) bromophenol blue, 0.5% (w/v) xylene cyanol FF and 20 mM EDTA, pH 8.0). The RNAs were separated by denaturing electrophoresis on pre-run 20% urea-acrylamide gels for 50 min at 300 V (Bio-Rad PROTEAN III system), and visualized using a Typhoon Trio imager (GE Healthcare). The RNA ladder was constructed by limited alkaline hydrolysis of the Generic-polyA RNA by incubation in 50 mM NaCO3, pH 8.9 for 10 min at 96°C followed by neutralization with 300 mM Na(CH3COO), pH 4.5.
Crystallization and crystal handling
The wt S. pombe Pop2p was crystallized by the sitting drop vapour diffusion technique by mixing 1–4 µl concentrated protein solution with 1–2 µl of reservoir solution (18–24% PEG 8000, 200 mM Mg(CH3COO)2, 100–200 mM Mes, pH 6.5, 0–10% glycerol, 5 mM BME and 1 mM NaN3) in protein:precipitant volume ratios of 1:1 to 2:1. Single crystals appeared within days at 4 or 19°C and grew to full size (100 μm) in ∼1–3 weeks. For cryo-protection, native crystals were transferred to a reservoir solution containing 25% PEG 8000 and 5% glycerol followed by 30% PEG 8000 and 10% glycerol before being flash-frozen in liquid nitrogen. Crystals used for phase determination were transferred into reservoir solution and 500 mM caesium chloride or sodium iodide and frozen directly as the salt itself provided enough additional cryo-protection.
Data collection and structure determination
Native data collection was carried out at the X11 beam line at DESY (Hamburg, Germany) at 0.8126 Å, whereas data collection on derivative crystals was done at the BL1 beam line at BESSY (Berlin, Germany) at 1.4586 Å (Table 1). Crystals were pseudo-tetragonal but belonged to space group P212121 with one molecule in the asymmetric unit. Native crystals diffracted to ∼1.4 Å with cell dimensions a = 52.370 Å, b = 54.089 Å, c = 90.959 Å and an unusually low solvent content of 28%. In contrast, the CsCl-soaked crystals only diffracted to 2.4 Å and had larger cell dimensions, a = 53.927 Å, b = 54.817 Å, c = 95.764 Å and a solvent content of 35%. Similarly, the NaI-soaked crystals diffracted to 2.2 Å and had a = 53.985 Å, b = 54.869 Å, c = 96.397 Å. Diffraction data was processed with Denzo and Scalepack (20).
Table 1.
Data Collection and Structure refinement statistics
| Native | Cs | I | |
|---|---|---|---|
| Data collection | |||
| Beam line | DESY X11 | BESSY BL1 | BESSY BL1 |
| Soak | 500 mM CsCl | 500 mM NaI | |
| Anomalous signal (f″) (e−) | 7.23 | 6.25 | |
| Wavelength (Å) | 0.834 | 1.4586 | 1.4586 |
| Space group | P212121 | P212121 | P212121 |
| Cell dimensions a/b/c (Å) | 52.370/54.089/90.959 | 53.927/54.817/95.764 | 53.985/54.869/96.397 |
| Resolution (Å) | 1.4 | 2.4 | 2.2 |
| Reflections | 51 215 | 14 874 | 15 043 |
| Completeness (outer shell) (%) | 99.6 (99.7) | 96.2 (78.0) | 99.7 (98.2) |
| Data redundancy (outer shell) | 4.7 (4.2) | 5.5 (2.1) | 6.6 (4.3) |
| Mean I/σI (outer shell) | 26.7 (2.0) | 17.6 (4.9) | 21.9 (8.4) |
| Rsym (outer shell) (%) | 5.2 (63.6) | 8.6 (18.0) | 8.1 (15.3) |
| Refinement | |||
| Resolution range used (Å) | 46.474–1.404 | – | – |
| R (%) | 13.9 | – | – |
| Rfree (%) | 20.4 | – | – |
| Number of atoms/solvent molecules/ions | 2387/327/2 | – | – |
| RMS deviation on bond lengths (Å) | 0.013 | – | – |
| RMS deviation on bond angles (°) | 2.3 | – | – |
| Ramachandran plot statistics (%) | |||
| Most favourable regions | 93.0 | – | – |
| Additionally allowed regions | 6.6 | – | – |
| Generously allowed regions | 0.4 | – | – |
| Disallowed regions | 0.0 | – | – |
The strong anomalous signal from iodine at 1.4586 Å gave clear peaks in the anomalous Patterson map calculated from these data, and was subsequently used to locate a single heavy atom site using the direct methods program, RANTAN (ccp4) (21), which was confirmed by initial phasing in MLPHARE/DM (ccp4). Phasing was expanded in SHARP (22) to a total of 12 iodine sites in a SIRAS phasing scheme using the Cs-derivative data as native due to the non-isomorphism between native and derivative data. Solvent flattening was carried out at 32, 35 and 38% solvent using SOLOMON as built into the SHARP interface. Of these, the 38% solvent map was judged visually to be the clearest, and therefore used for the subsequent model building. The strong phasing signal from iodine provided an exceptionally clear initial SIRAS density map at 2.2 Å in which ARP/wARP (23) was able to auto-trace ∼85% of the final model correctly. The ARP/wARP model was manually checked against the experimental SIRAS map and rebuilt in O (24) whereever necessary. Refinement of the model was done in iterative cycles of maximum-likelihood minimization in CNS (25) followed by manual rebuilding in O. Due to the non-isomorphism, it was necessary to use molecular replacement to transfer the model into the 1.4 Å high-resolution native data set before further refinement. The model was then manually inspected and rebuilt in O before the refinement was finalized in SHELX-97, including both anisotropic temperature factors and modelling of multiple conformations (26).
Structural alignment
Proteins with structural homology to S. pombe Pop2p were identified by a DALI search [http://www.ebi.ac.uk/dali (27)], whereas structural alignment of individual structures was done in O by least-squares minimized superposition of the conserved active site residues. Comparison of the S. pombe Pop2p structure and the S. cerevisiae Rrp6p structure was done manually and evaluated by DALI comparison. Manually determined insertions only deviated with few amino acids from DALI determined ones.
RESULTS
S. pombe Pop2p is a compact, one-domain protein
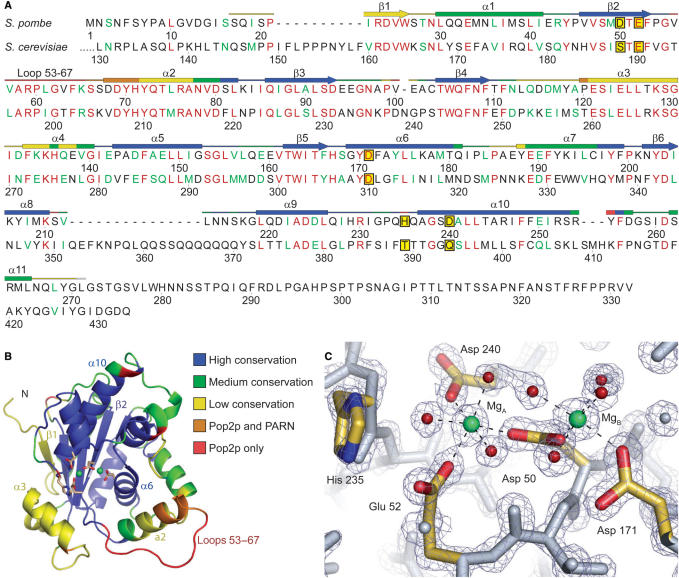
Intact S. pombe Pop2p was expressed in E. coli, purified to homogeneity and crystallized. Single crystals belonging to the space group P212121 diffracted to ∼1.4 Å. The structure was determined by single isomorphous replacement with anomalous scattering (SIRAS) phasing using data obtained from an iodide-soaked crystal diffracting to 2.2 Å and an isomorphous CsCl2 derivative data set as a reference (Table 1). The initial model was subsequently refined against the non-isomorphous 1.4 Å native data following molecular replacement, and to our knowledge represents the highest resolution structure of any exonuclease to date. The crystallographic asymmetric unit contains a single protein molecule and the final model covers residues 17–271 of the 332 amino acids in the protein, 327 solvent molecules and 2 magnesium ions (Figure 1A). The polypeptide chain is clearly defined in all parts of the electron density map except the region 61–67, which could be traced, but has relatively poor electron density.
Figure 1.
Overview of the S. pombe Pop2p structure. (A) Alignment of Pop2p from S. pombe and S. cerevisiae. Fully conserved residues are shown in red and conserved functionality in green. The secondary structure of S. pombe Pop2p is indicated above the sequence and colour-coded according to similarity to other homologous structures identified by a DALI search (see text) with the extremes being structural elements solely found in Pop2p (red) and regions found in most or all of the homologous structures (blue). Other colours represent intermediate occurrence as indicated in (B). The five active site residues are shown framed on a yellow background. Sequence diagram was produced with SecSeq (39). (B) Cartoon representation of S. pombe Pop2p. The structure is colour coded as indicated and described in (A). The side chains of the active site residues are shown as sticks and coloured by element, whereas ions in the active site are shown as green spheres. All structure figures were produced with PyMOL (40). (C) Close-up of the active site. Active site residues are coloured by element and associated magnesium ions are shown in green, waters involved in coordination of the active site ions are coloured red, and the octahedral coordination of the active site magnesium ions is indicated by dashed lines. The refined 2mFo-DFc electron-density map is shown contoured at 2.5σ.
Overall, the molecule appears as a single compact and globular domain with an irregular surface in the active site region (Figure 1B), essentially like the previously published structure of the Pop2p exonuclease domain from S. cerevisiae (8). A protrusion formed by the N-terminus contributes to the irregular surface, but the most striking feature is the deep central cavity holding the active site residues and ions. The cavity extends from the β-sheet in the inner part of the protein and is surrounded by α-helices and loops. The protein is more compact around the active site than its S. cerevisiae counterpart due to shifts of the helices surrounding the active site cavity, mainly α2 and α3. There is a strong crystal contact in the active site region, which apparently prevents binding of substrate analogues in this crystal form. It is also possible that this affects the conformation of some amino acid side chains in α3 and the N-terminal half of the loop 53–67.
S. pombe Pop2p contains a competent active site
Despite differences on the surface of the exonuclease core, the active site region is highly conserved among the DEDD nucleases. The active site of Pop2p is located in the innermost part of the central cavity on the edge of the β-sheet and consists of residues from the C-terminal part of β2 as well as α10 and α6 (Figure 1A and B). The histidine, which is important for catalysis in the DEDDh-subgroup, is situated in a loop extending from the N-terminal end of α10. Though Pop2p from S. cerevisiae was found to be active in vitro, its unconventional SEDQt active site raised doubts as to whether these proteins indeed retain the classical reaction mechanism of the DEDD nucleases (8). Schizosaccharomyces pombe Pop2p, on the other hand, contains all of the conserved residues in the active site, and the presence of the divalent metal ions and the high-resolution data has allowed for a very detailed analysis of the active site configuration in the present structure (Figure 1C).
The active site of S. pombe Pop2p consists of the two metal ions, which are magnesium in this structure due to its presence in the crystallization buffer (MgA and MgB), in addition to the conserved acidic residues, D50, E52, D171, D240 and H235. Of these, D50 takes a central position by contacting both metal ions. MgA is also coordinated by E52 and D240, while D171 contacts MgB. In addition, seven water molecules, three associated with MgA and four with MgB, complete the dual octahedral coordination spheres of the two ions. One of the water molecules associated with MgA also forms a hydrogen bond with H235, thereby bridging these. The overwhelming similarity between the active site of S. pombe Pop2p and other known DEDDh nucleases (17,28,29) thus strongly suggests that Pop2p is a catalytically competent protein that uses the conserved hydrolytic reaction mechanism of the DEDD nucleases.
The active site ions define the activity and specificity of S. pombe Pop2p
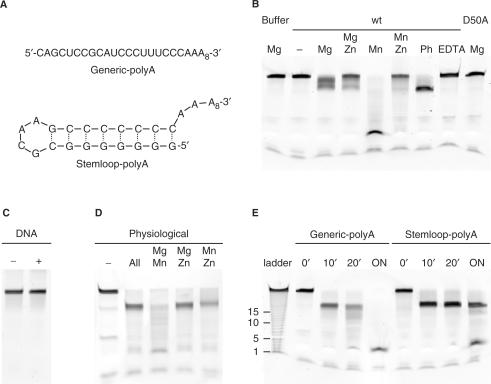
In order to further investigate the possibility that Pop2p from S. pombe might be a competent 3′–5′ exonuclease, we set up an in vitro deadenylation assay using defined RNA substrates (Figure 2A). One substrate, Generic-polyA, contained a 20-nt generic RNA sequence with little or no secondary structure followed by a 10-nt poly-A tail whereas the other substrate, Stemloop-polyA, contained a strong 8-bp stemloop followed by a 10-nt poly-A tail. In order to be able to define the ions present in the active site, protein used for the experiment was first changed into a buffer devoid of any divalent metal ions. Though this may not completely remove ions from the active site of the enzyme, it provides the best possible starting point for defining the composition of the ions. As shown in Figure 2B, there is no degradation of the Generic-polyA RNA substrate by wt Pop2p in the absence of divalent metal ions in the buffer (lane marked ‘wt/−’), nor with 5 mM Mg2+ in the buffer in the absence of protein (‘Buffer/Mg’). On the other hand, incubation of the RNA substrate in the presence of wt Pop2p and 5 mM Mg2+ resulted in limited, but consistent 3′–5′ exonuclease activity (‘wt/Mg’), while incubation with 5 mM Mn2+ results in almost complete degradation of the Generic-polyA RNA in 60 min (‘wt/Mn’). To check that the observed nuclease activity is indeed due to the specific activity of the purified Pop2p, we constructed a D50A mutant that removes the carboxyl group from the central bipartite aspartate responsible for coordinating the ions in the active site. The mutant enzyme has no activity in the presence of 5 mM Mg2+ (‘D50A/Mg’), and likewise, activity is also completely lost when the ions are removed from the wt enzyme by incubation with 10 mM with EDTA (‘wt/EDTA’). To confirm that Pop2p is specific to RNA, we synthesized the Generic-polyA substrate as an oligodeoxynucleotide substrate (‘DNA’), which is also not degraded illustrating that Pop2p is specifically a 3′–5′ riboexonuclease (Figure 2C).
Figure 2.
In vitro deadenylation assays. (A) Sequence and secondary structure of the RNA substrates used [as predicted my MFold (41)]. (B) Overview of the activity of wt Pop2p and the D50A mutant. In each experiment, 10 nM Generic-polyA RNA was incubated with either 1 μM Pop2p or Pop2p D50A for 60 min at 30°C, except the first lane which is a buffer control containing buffer with 5 mM Mg2+ but no protein. The remaining experiments contained 5 mM Mg2+ or Mn2+, and 1 mM Zn2+ as indicated. The lane marked ‘Ph’ contained the physiological mix of ions (7.1 mM Mg2+, 75 μM Mn2+, and 220 μM Zn2+), and the lane ‘EDTA’, 10 mM EDTA. (C) Incubation of an oligodeoxynucleotide substrate (Generic-polyA DNA) in the absence ‘−’ or presence ‘+’ of wt Pop2p for 120 min at 30°C in a buffer containing 5 mM Mg2+. (D) Overnight incubation of the Generic-polyA RNA substrate in the presence of wt Pop2p with no added ions (‘−’), all three ions at their physiological concentrations (‘All’), or two ions at these concentrations, as indicated. (E) Time-course experiments, where 1 μM Pop2p was incubated with 10 nM Stemloop-polyA or 10 nM Generic-polyA as indicated in a buffer containing 5 mM Mn2+ and aliquots removed at 0, 10 and 20 min, and following overnight incubation (‘ON’). ‘Ladder’ is an RNA ladder produced by limited alkaline hydrolysis of the Generic-polyA substrate with approximate nucleotide length indication.
To further analyse the observed differences in activity as a result of the ions present in the active site, Pop2p was incubated for 60 min with the Generic-polyA substrate in the presence of 5 mM Mg2+ ± 1 mM Zn2+ (Figure 2B, lanes ‘wt/Mg’ and ‘wt/Mg Zn’), 5 mM Mn2+ ± 1 mM Zn2+ (Figure 2B, lanes ‘wt/Mn’ and ‘wt/Mn Zn’), or a mix containing all three ions in their approximate, physiological concentrations, namely 7.1 mM Mg2+, 75 uM Mn2+, and 220 μM Zn2+ (19) (Figure 2B, lane ‘wt/Ph’). Under the physiological conditions, the RNA is quickly degraded to ∼20-nt, corresponding to removal of the 10-nt poly-A tail (lane ‘wt/Ph’), while the enzyme appears to be inhibited by the high zinc concentration in the experiments with only two ions and 1 mM Zn2+ (lanes ‘wt/Mg Zn’ and ‘wt/Mn Zn’)
Assuming that only two ions can bind in the active site at any one time, we next performed drop-out experiments to clarify which of the ions in the physiological mix are responsible for the observed specificity (Figure 2D). To make sure that the enzyme does not simply pause at the end of the poly-A tail, experiments were allowed to run to completion by overnight incubation at 30°C. In the absence of divalent metal ions, we observe a limited residual activity from the ion-depleted wt Pop2p after the long incubation (lane ‘−’), while the combination of all three ions consistently makes the enzyme stop after degradation of ∼10-nt (lane ‘All’). This illustrates that S. pombe Pop2p is able to specifically degrade the poly-A tail under the right conditions. In the drop-out experiments using the same concentrations of ions but leaving one ion out in each case, the combination of Mg2+ and Mn2+ appears to create a fast and non-specific enzyme (lane ‘Mg Mn’), while both the combinations Mg2+–Zn2+ and Mn2+–Zn2+ give the same specificity as observed for all three ions. However, the 20-nt band is more smeared in the Mn2+–Zn2+ case, probably reflecting full degradation of part of the substrate molecules in this case, in accordance with the above results for Mn2+ alone showing a fast but non-specific enzyme. In the final experiment, we used Mn2+ alone in the active site to investigate whether S. pombe Pop2p is able to degrade stable secondary structure under the least discriminative conditions (Figure 2E). When there is no secondary structure (‘Generic-polyA’), Pop2p almost completely degrades the RNA in 20 min, while it stops immediately before the hairpin with the Stemloop-polyA substrate, showing that Pop2p most likely cannot degrade strong secondary structure.
The Pop2p structure is compatible with the general two metal ion hydrolytic reaction mechanism
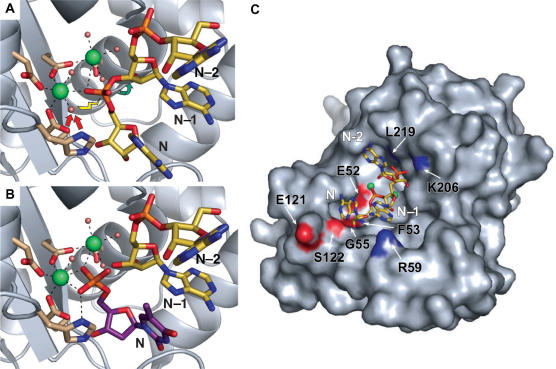
To directly visualize RNA binding by Pop2p, crystals were soaked in either single monophosphate nucleotides (product) or non-hydrolysable RNA oligonucleotides (substrate) and analyzed by X-ray diffraction. Unfortunately, due to crystal contacts between adjacent molecules in the active site region, none of these experiments were successful. However, the high degree of structural conservation among DEDD nucleases combined with the accurately defined active site in the present structure has allowed us to propose a precise model for RNA binding by Pop2p in the lack of experimental data. Human PARN (17) and the ε-subunit of the E. coli DNA polymerase III (exonuclease subunit) (29) are the closest structural neighbours to Pop2p, and as structures of these proteins bound to either poly-A RNA or single nucleotides are available, we have a rigorous framework for modelling interactions between Pop2p and RNA.
The PARN poly-A RNA substrate fits well and with no clashes into the Pop2p active site when the proteins are superposed by least-square-based alignment of the conserved active site residues, and provides a snapshot of the presumed conformation of the protein in the pre-hydrolysis state (Figure 3A). An oxygen atom from the 3′-most phosphordiester is placed between the two metal ions that probably orient and restrain the phosphate for the following nucleophilic attack. In agreement with the proposed two metal ion reaction schemes (16,29,30), H235 is poised to act as a general base by activating one of the water molecules associated with MgA for an attack on the phosphate in an SN2 nucleophilic substitution reaction. This leads to breakage of the scissile bond between the phosphate group and the O3′ of the penultimate pentose sugar and subsequent release of a monophosphate nucleotide from the 3′ end of the RNA.
Figure 3.
Pop2p–RNA interactions. (A) The pre-hydrolysis state as illustrated by superposition of poly-A from the structure of human PARN (PDB entry 2A1R, yellow sticks) onto the S. pombe Pop2p structure. The 3′ nucleotide (N), the penultimate nucleotide (N–1) and the third last nucleotide (N–2) are shown. Arrows indicate the hydrolytic reaction mechanism in which a water molecule associated with one of the active site ions is activated (red arrows) for attack on the most 3′ phosphate of the RNA (yellow lightening) leading to breakage (green arrow) of the bond between the phosphate group and the N–1 nucleotide. (B) The post-hydrolysis state illustrated by superposition of dTMP from the ε-subunit of E. coli DNA pol III (PDB entry 1J54, purple sticks) showing the position of the 3′ nucleotide (N) immediately after cleavage. The N–1 and N–2 nucleotides (PDB entry 2A1R, yellow sticks) from the PARN poly-A illustrate the position of the new 3′ end of the substrate. (C) Possible Pop2p–RNA contacts. Poly-A from PARN (PDB entry 2A1R, yellow sticks) superposed on the active site region of the S. pombe Pop2p, shown as a surface representation with atoms of residues possibly involved in the RNA recognition coloured by element.
Alignment of the two single nucleotides found in the structure of the ε-subunit of E. coli DNA polymerase III soaked in dTMP at pH 5.8 also shows a good fit, and illustrates a possible post-cleavage situation (Figure 3B) (29). In this structure, two partially occupied dTMPs were observed in the active site. The minor conformation (∼15%) mimics the 3′ nucleotide of an uncleaved RNA, whereas the major conformation has a phosphate oxygen atom that seems to contact H235 as well as MgA, much like the activated water in the pre-hydrolysis state. This appears to correspond to the position of the 3′ nucleotide in the post-cleavage state of the enzyme.
The modelling of nucleotides into the active site of the S. pombe Pop2 structure provides snapshots of the reaction cycle of the enzyme and strongly suggests that the enzyme uses a classical DEDDh-type reaction pathway in the hydrolysis of RNA. One caveat is that the octahedral coordination of the ions (Figure 1C) most likely gets interrupted at some point during the reaction cycle due to sterical constraints in the RNA substrate. This may be one of the reasons why the presence of the more promiscuous zinc ion in the active site gives higher specificity, as it would allow more flexibility with respect to substrate orientation and possibly permit direct recognition of the base in a nearby region. In support of this, it was recently found that the DEDDh exonuclease, Rrp6p is able to bind two octahedral manganese ions in the active site simultaneously, but only binds nucleotides with a combination of Mn2+ and Zn2+ (31). A similar ion substitution experiment was carried out for S. pombe Pop2p and showed that the distance between the two active site ions decreases with ∼1 Å as a result of the presence of both magnesium and zinc as opposed to magnesium alone (data not shown). This result further substantiates the idea that the nature of the ions influences the finer architecture of the active site and may therefore be important for the catalytic function of the protein in vivo.
Pop2p may interact with RNA in a wider region
Superposition of the poly-A RNA substrate from the human PARN structure onto the S. pombe Pop2p structure shows a good fit also outside of the core active site (Figure 3C). This allows potential Pop2p–RNA interactions in a wider region to be postulated, though the obvious limitations of such a model must be borne in mind.
The model predicts several potential contacts between Pop2p and the RNA backbone. At the very 3′ end, recognition of the 2′ and 3′ OH groups could be mediated by the backbone atoms of F53 and G55 as well as the side chain of the active-site residue E52, which can provide H-bonds to both OH groups (Figure 3C). These amino acids are conserved in Pop2p in most other species and in both PARN and Rrp6p have been shown to recognize the 2′ and 3′ OH groups of the terminal nucleotide (17,31). The phosphate group between the 3′ nucleotide (N) and the penultimate nucleotide (N–1) is clearly kept in place by the two metal ions in the pre-hydrolysis state as described above (Figure 3A), while the penultimate phosphate, linking N–1 and N–2, may make contacts to the backbone amino group of L219 and the side chain of K206 (Figure 3C). These residues are likely to be involved in keeping the phosphate in place like the homologous residues (L343 and K326) in PARN, and consistently, the two amino acids are conserved in Pop2p proteins from most species. In conclusion, it appears that Pop2p is able to engage in a range of specific interactions with the RNA backbone allowing the protein to place the substrate optimally for cleavage to take place.
On the other hand, the PARN-RNA structure did not provide an explanation of the basis of sequence specificity, as no discriminative base atoms are contacted by the protein. However, as PARN apparently functions as a homodimer, the situation is complicated by the fact that the specificity could be contributed by residues on the neighbouring molecule that could potentially reach the nucleotide bases (17). In contrast to PARN, there is no evidence that Pop2p should function in an oligomeric state and the basis of sequence-specific RNA recognition may therefore differ significantly between the two proteins. However, based on the position of the bases in the PARN poly-A RNA, we can identify a few potential sites in Pop2p that could mediate direct protein–base contacts, such as the conserved E121, S122 and several residues in the Pop2-specific loop, 53–67 (see below), which may show a different conformation in vivo or when bound to substrate. Previous experiments with Pop2p from different organisms have shown varying results concerning the sequence specificity of the protein (6–9) and our in vitro experiments show that the enzyme can degrade non-poly-A substrates under some conditions. This means that a strict sequence discrimination may not be an intrinsic property of the enzyme.
Structural alignment of S. pombe Pop2p suggests a common ancestral fold
As indicated by the modelling of RNA from the PARN-RNA structure into the active site groove, it appears that Pop2p is structurally very similar to PARN. High structural similarity between proteins is often indicative of a functional relationship, whereas structural differences may imply functional divergence. Structural comparison can therefore indicate which parts of the proteins are important for shared function, but also point out those that have diverged to serve unique roles. To compare S. pombe Pop2p to structurally similar proteins, a DALI search (27) was carried out and the resulting structural alignment used to identify regions of similarity and divergence. The results of the analysis are shown colour coded in Figure 1A and B with blue representing regions found in most or all of the structures and red the structural features found solely in S. pombe Pop2p. Other colours represent regions of intermediate conservation as indicated. The highly conserved core domain consists of the mixed five-stranded β-sheet with alpha helices on both sides (blue) that contains the four active site residues of the DEDD nucleases. Pop2p has a one-strand extension to the β-sheet of the core motif (β1 in Figure 1A and B) and additional helices and loops surrounding the core nuclease region (green, yellow, orange and red). In general, the least structurally conserved region of Pop2p is situated on one side of the active site and is composed of helices α2 and α3 and the loop spanning amino acids 53–67. Whereas α3 is shared with a number of other DEDDh-type nucleases and thus may be a part of a general DEDDh exonuclease architecture, the N-terminal extension of α2 (orange) is currently only found in the structures of human PARN (17) and yeast Pop2p. The conformation observed for the loop 53–67 in the present Pop2p structure is unique, as the corresponding region is absent from the S. cerevisiae protein and located differently in PARN. This region, as well as part of α2 that also shows great variation between the homologous structures (Figure 1A and B) are therefore likely to be involved in Pop2p or deadenylase-specific functions. Consistently, mutation of residues in this loop results in failure of the enzyme to bind Ccr4p in S. cerevisiae (42) indicating its importance for protein–protein interactions inside the Ccr4-Not complex.
Eukaryotic DEDD nucleases differ markedly in their sequence organization
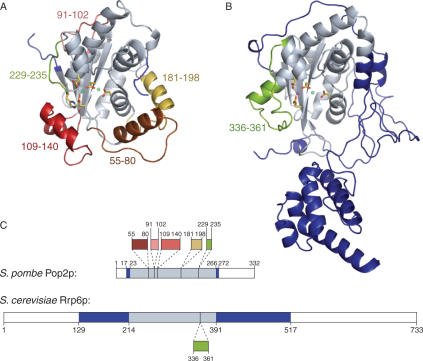
To reveal how the different DEDD-type nucleases have adapted structurally to their specific functions, we then compared the DEDDh-type Pop2p to a protein from the DEDDy group. With a DALI Z-score of 10.2, the recent structure of the nuclear exonuclease Rrp6p from S. cerevisiae is the closest structural neighbour to Pop2p in this group (31). Structural alignment shows that the two proteins share a common core nuclease region surrounded by several unique structural expansion elements presumably required for the specific function of each protein (Figure 4). However, sequence analysis reveals that these features arise in different ways. In Pop2p, the expansion elements are mostly found as insertions in the core exonuclease sequence (Figure 4A and C), whereas in Rrp6p, they mainly come from the extended N-terminal sequence (Figure 4B and C) (31). A single internal extension element is found in the exonuclease domain of Rrp6p (residues 336–361), but this element harbours the active site tyrosine (DEDDy) and may therefore be a part of a general subgroup architecture since it is also found in a number of other DEDDy-type proteins. It thus seems that Pop2p has added specialized functions to the common DEDD nuclease architecture by insertions into the primary exonuclease sequence, whereas Rrp6p has a rather undisturbed exonuclease motif and uses flanking sequences to add unique features to the core nuclease. Pop2p and Rrp6p therefore represent two different architectural approaches to adaptation of a common ancestral nuclease to changes in function.
Figure 4.
Sequence organization of DEDD nucleases. (A) Schizosaccharomyces pombe Pop2p colour coded according to structural similarity with S. cerevisiae Rrp6p (PDB entry 2HBL). Grey regions show parts of the structure homologous to regions in Rrp6p, whereas coloured regions illustrate structural elements not found in Rrp6p. The associated numbers correspond to the amino acid residues flanking the element. Active site ions and side chains of active site residues are shown as balls and sticks. (B) Saccharomyces cerevisiae Rrp6p (PDB entry 2HBL) colour coded according to structural similarity with S. pombe Pop2p following the same scheme as in (A). (C) Graphical representation of the S. pombe Pop2p and S. cerevisiae Rrp6p sequences using the same colour code as in (A) and (B). Structural elements unique to one of the proteins (coloured) found within the otherwise common core nuclease region (grey) are pulled out of the sequences with dashed lines illustrating the position of these in the core sequence.
DISCUSSION
Here, we have presented the 1.4-Å crystal structure of the S. pombe Pop2p deadenylase subunit that together with functional data clearly shows that the protein is a competent 3′–5′ DEDD-type riboexonuclease with a tunable active site specificity. Superposition of RNA and nucleotides into the active site shows that the mode of substrate binding is very likely to be conserved as well and that the reaction pathway most likely follows the same conserved hydrolytic mechanism. The protein contains insertions in the nuclease core domain that surround the conserved active site, which presumably provide deadenylase-specific structural features, however, these elements may not provide the enzyme with a strict poly-A specificity.
It is interesting to note that although RNA–backbone contacts seem to be somewhat conserved between Pop2p and PARN, base interactions most likely are not. If an inherent poly-A specificity were a key requirement for the function of these proteins, a stronger conservation would be anticipated. The lack of a strict poly-A specificity has been confirmed for different Pop2p enzymes by in vitro experiments showing that although the enzymes generally have a subtle poly-A preference, they are also capable of degrading non-poly-A substrates (6–9). Our in vitro results show that while the enzyme is able to degrade a non-poly-A substrate, the identity and relative amounts of divalent metal ions present can modulate this behaviour. Thus, Mn2+ alone causes the enzyme to be fast and non-specific, while Zn2+ in combination with Mn2+ or Mg2+ appears to be required for specificity. With the physiological mix of three ions, it is likely that the 100-fold higher Mg2+ concentration over Mn2+ (7.1 mM versus 75 μM) completely competes out Mn2+ from the active site and therefore that the observed poly-A specificity arises from the precise combination of Mg2+ and Zn2+ in the active site. However, future structural studies with defined ions and substrates will hopefully reveal the details of these subtle differences. One possibility is that the more promiscuous zinc ion gives more flexibility with respect to substrate binding than the strictly octahedral ions Mg2+ and Mn2+, and thereby allowing higher selectivity.
In any case, substrate selectivity is likely also defined by the cellular context of the protein. In this respect, we note that residues in the Pop2-specific loop (residues 53–67) appear to be involved in the interaction with Ccr4p. The proximity of this loop to the active site raises the possibility that Ccr4p may influence the entry of substrates into the active site of Pop2p and thereby affect the catalytic activity and possibly specificity of the enzyme. In any case, it is reasonable to believe that the control with deadenylation and thereby the overall rate of mRNA turnover requires more complexity than can be achieved with single nucleases specific for poly-A RNA, and that the process therefore is controlled at several levels. In the larger cellular context, RNA turnover has been shown to be regulated through localization of the mRNAs, for example via the so-called P-bodies that harbour a number of degradative enzymes involved in 5′–3′ decay (32). The Ccr4-Not complex is also found in the P-bodies, but not exclusively, as there appears to be a large cytoplasmic population as well, suggesting that localization to the P-bodies is not the main control point in RNA turnover (32,33).
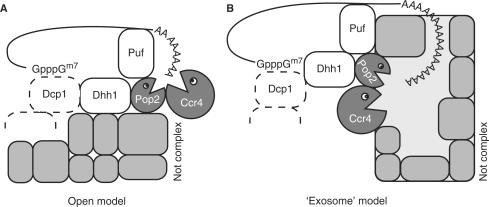
Several key questions about deadenylation still remain to be addressed: How is degradation regulated, what structural features of Pop2p and the Ccr4-Not complex provide the specificity for poly-A and why does the complex contain two nucleases? As for the role of Pop2p, there now appears two likely scenarios: One possibility is that the active site of Pop2p is suppressed or obscured inside the Ccr4-Not complex and that Ccr4p is the only active nuclease in vivo. That would reduce the role of Pop2p to an inactive, architectural protein as seen for some of the nuclease subunits in the exosome (34). Pop2p has been shown to constitute the physical link between Ccr4p and the rest of the Ccr4-Not complex (35,36), and its presence would then be required to maintain the overall structure of the complex and mediate interactions with external partners, such as the Puf proteins (37). These observations have resulted in a conceptual ‘open model’ for the structure of the Ccr4-Not complex (Figure 5A), in which the two nucleases are protruding from the core Not complex with Pop2p bridging between Ccr4p and Not proteins. This model could explain the reduced evolutionary pressure on the Pop2p to maintain its DEDD signature in S. cerevisiae.
Figure 5.
Models for organization of the Ccr4-Not complex. (A) Open model in which the nucleases Pop2p and Ccr4p are protruding from the rest of the Not complex. (B) The ‘exosome’ model with the active sites of Pop2p and Ccr4p contained within the complex.
But the structure of the S. pombe Pop2p and the functional data provided here have shown that this subunit of the Ccr4-Not complex at least in some organisms is a functional 3′–5′ exonuclease with poly-A specificity under the right conditions. Furthermore, in vitro degradation experiments show that even the aberrant S. cerevisiae Pop2p can act as an active nuclease (7,8). Therefore, it is more likely that both the Pop2p and Ccr4p nucleases are active at certain times or under certain conditions inside the cell, and that perhaps both are required to degrade all substrates. The situation is again reminiscent of the exosome, where several phosphorolytic and hydrolytic nucleases form a ring that enclose and regulate entry to some of the active sites at the centre. This has already turned out to be a general theme for degradative complexes in the cell, which in line with the proteasome protect the reactive and potentially harmful active sites within a larger structure. In the ‘exosome model’ for the Ccr4-Not complex, both active sites are regulated, not so much by RNA binding by the individual nucleases, but through controlled entry to the central cavity (Figure 5B). The targeting of mRNAs to the nuclease core is most likely controlled by external factors. An example of this is the Puf proteins, which bind to the 3′ untranslated region (UTR) of specific mRNAs and guide these to the Ccr4-Not complex by interacting with Pop2p (37).
We thus suggest that the Ccr4-Not complex may form a contained structure that assists in the substrate-screening process and protects the cell from unwanted RNA degradation. Overall, the emerging picture is that of a complex deadenylation machinery, which is activated by and interacts with many other cellular factors, such as RNA-binding proteins and helicases (37,38). These mechanisms may also partly ensure that deadenylation stops at the correct sites and does not proceed through non-poly-A sequence. The Ccr4-Not complex may thus form a central platform for directing deadenylase activity through the interaction with external factors, thereby making strict poly-A specificity of the nucleases superfluous.
COORDINATES
The structure of wt S. pombe Pop2p has been deposited along with structure factors in the RCSB Protein Data Bank (www.rcsb.org) with entry 2P51.
ACKNOWLEDGEMENTS
The authors would like to thank Prof. Clyde Denis, University of New Hampshire, Durham, NH, USA for critical reading of the manuscript and helpful suggestions, Dr J. Preben Morth for advise on heavy atom soaking experiments and help during data collection, and to John Woodward at The Wellcome Trust Sanger Institute, Cambridge, UK, for kindly providing the S. pombe contig clone. The work was supported by the Novo Nordisk Foundation (NNF) and a Human Frontier Science Programme Career Development Award (CDA) to D.E.B. A.T.J. was supported by a post-doctoral fellowship from the Lundbeck Foundation and K.R.A. by a Ph.D. programme grant from The Danish National Research Foundation (Danmarks Grundforskningsfond). Funding to pay the Open Access publication charge was provided by Novo Nordisk Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1.Parker R., Song H. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- 2.Cao D., Parker R. Computational modeling of eukaryotic mRNA turnover. RNA. 2001;7:1192–1212. doi: 10.1017/s1355838201010330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tucker M., Valencia-Sanchez M.A., Staples R.R., Chen J., Denis C.L., Parker R. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell. 2001;104:377–386. doi: 10.1016/s0092-8674(01)00225-2. [DOI] [PubMed] [Google Scholar]
- 4.Chen J., Chiang Y.C., Denis C.L. CCR4, a 3′-5′ poly(A) RNA and ssDNA exonuclease, is the catalytic component of the cytoplasmic deadenylase. EMBO J. 2002;21:1414–1426. doi: 10.1093/emboj/21.6.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tucker M., Staples R.R., Valencia-Sanchez M.A., Muhlrad D., Parker R. Ccr4p is the catalytic subunit of a Ccr4p/Pop2p/Notp mRNA deadenylase complex in Saccharomyces cerevisiae. EMBO J. 2002;21:1427–1436. doi: 10.1093/emboj/21.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bianchin C., Mauxion F., Sentis S., Seraphin B., Corbo L. Conservation of the deadenylase activity of proteins of the Caf1 family in human. RNA. 2005;11:487–494. doi: 10.1261/rna.7135305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daugeron M.C., Mauxion F., Seraphin B. The yeast POP2 gene encodes a nuclease involved in mRNA deadenylation. Nucleic Acids Res. 2001;29:2448–2455. doi: 10.1093/nar/29.12.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thore S., Mauxion F., Seraphin B., Suck D. X-ray structure and activity of the yeast Pop2 protein: a nuclease subunit of the mRNA deadenylase complex. EMBO Rep. 2003;4:1150–1155. doi: 10.1038/sj.embor.7400020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Viswanathan P., Ohn T., Chiang Y.C., Chen J., Denis C.L. Mouse CAF1 can function as a processive deadenylase/3′-5′-exonuclease in vitro but in yeast the deadenylase function of CAF1 is not required for mRNA poly(A) removal. J. Biol. Chem. 2004;279:23988–23995. doi: 10.1074/jbc.M402803200. [DOI] [PubMed] [Google Scholar]
- 10.Yamashita A., Chang T.C., Yamashita Y., Zhu W., Zhong Z., Chen C.Y., Shyu A.B. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat. Struct. Mol. Biol. 2005;12:1054–1063. doi: 10.1038/nsmb1016. [DOI] [PubMed] [Google Scholar]
- 11.Korner C.G., Wahle E. Poly(A) tail shortening by a mammalian poly(A)-specific 3′-exoribonuclease. J. Biol. Chem. 1997;272:10448–10456. doi: 10.1074/jbc.272.16.10448. [DOI] [PubMed] [Google Scholar]
- 12.Lai W.S., Kennington E.A., Blackshear P.J. Tristetraprolin and its family members can promote the cell-free deadenylation of AU-rich element-containing mRNAs by poly(A) ribonuclease. Mol. Cell. Biol. 2003;23:3798–3812. doi: 10.1128/MCB.23.11.3798-3812.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zuo Y., Deutscher M.P. Exoribonuclease superfamilies: structural analysis and phylogenetic distribution. Nucleic Acids Res. 2001;29:1017–1026. doi: 10.1093/nar/29.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dlakic M. Functionally unrelated signalling proteins contain a fold similar to Mg2+-dependent endonucleases. Trends Biochem Sci. 2000;25:272–273. doi: 10.1016/s0968-0004(00)01582-6. [DOI] [PubMed] [Google Scholar]
- 15.Beese L.S., Steitz T.A. Structural basis for the 3′-5′ exonuclease activity of Escherichia coli DNA polymerase I: a two metal ion mechanism. EMBO J. 1991;10:25–33. doi: 10.1002/j.1460-2075.1991.tb07917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steitz T.A., Steitz J.A. A general two-metal-ion mechanism for catalytic RNA. Proc. Natl Acad. Sci. USA. 1993;90:6498–6502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu M., Reuter M., Lilie H., Liu Y., Wahle E., Song H. Structural insight into poly(A) binding and catalytic mechanism of human PARN. EMBO J. 2005;24:4082–4093. doi: 10.1038/sj.emboj.7600869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hertz-Fowler C., Peacock C.S., Wood V., Aslett M., Kerhornou A., Mooney P., Tivey A., Berriman M., Hall N., et al. GeneDB: a resource for prokaryotic and eukaryotic organisms. Nucleic Acids Res. 2004;32:D339–D343. doi: 10.1093/nar/gkh007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tholey G., Ledig M., Mandel P., Sargentini L., Frivold A.H., Leroy M., Grippo A.A., Wedler F.C. Concentrations of physiologically important metal ions in glial cells cultured from chick cerebral cortex. Neurochem. Res. 1988;13:45–50. doi: 10.1007/BF00971853. [DOI] [PubMed] [Google Scholar]
- 20.Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 21.Collaborative Computational Project. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. number 4. [DOI] [PubMed] [Google Scholar]
- 22.delaFortelle E., Bricogne G. Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Method Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 23.Perrakis A., Morris R., Lamzin V.S. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 24.Jones T.A., Zou J.Y., Cowan S.W., Kjeldgaard Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47(Pt. 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 25.Brunger A.T., Adams P.D., Clore G.M., DeLano W.L., Gros P., Grosse-Kunstleve R.W., Jiang J.S., Kuszewski J., Nilges M., et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 26.Sheldrick G.M., Schneider T.R. SHELXL: High-resolution refinement. Macromol. Crystallogr., Pt. B. 1997;277:319–343. [PubMed] [Google Scholar]
- 27.Holm L., Sander C. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 28.Cheng Y., Patel D.J. Crystallographic structure of the nuclease domain of 3′hExo, a DEDDh family member, bound to rAMP. J. Mol. Biol. 2004;343:305–312. doi: 10.1016/j.jmb.2004.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hamdan S., Carr P.D., Brown S.E., Ollis D.L., Dixon N.E. Structural basis for proofreading during replication of the Escherichia coli chromosome. Structure. 2002;10:535–546. doi: 10.1016/s0969-2126(02)00738-4. [DOI] [PubMed] [Google Scholar]
- 30.Brautigam C.A., Steitz T.A. Structural principles for the inhibition of the 3′-5′ exonuclease activity of Escherichia coli DNA polymerase I by phosphorothioates. J. Mol. Biol. 1998;277:363–377. doi: 10.1006/jmbi.1997.1586. [DOI] [PubMed] [Google Scholar]
- 31.Midtgaard S.F., Assenholt J., Jonstrup A.T., Van L.B., Jensen T.H., Brodersen D.E. Structure of the nuclear exosome component Rrp6p reveals an interplay between the active site and the HRDC domain. Proc. Natl Acad. Sci. USA. 2006;103:11898–11903. doi: 10.1073/pnas.0604731103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sheth U., Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cougot N., Babajko S., Seraphin B. Cytoplasmic foci are sites of mRNA decay in human cells. J. Cell Biol. 2004;165:31–40. doi: 10.1083/jcb.200309008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lorentzen E., Walter P., Fribourg S., Evguenieva-Hackenberg E., Klug G., Conti E. The archaeal exosome core is a hexameric ring structure with three catalytic subunits. Nat. Struct. Mol. Biol. 2005;12:575–581. doi: 10.1038/nsmb952. [DOI] [PubMed] [Google Scholar]
- 35.Bai Y., Salvadore C., Chiang Y.C., Collart M.A., Liu H.Y., Denis C.L. The CCR4 and CAF1 proteins of the CCR4-NOT complex are physically and functionally separated from NOT2, NOT4, and NOT5. Mol. Cell Biol. 1999;19:6642–6651. doi: 10.1128/mcb.19.10.6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu H.Y., Badarinarayana V., Audino D.C., Rappsilber J., Mann M., Denis C.L. The NOT proteins are part of the CCR4 transcriptional complex and affect gene expression both positively and negatively. EMBO J. 1998;17: 1096–1106. doi: 10.1093/emboj/17.4.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goldstrohm A.C., Hook B.A., Seay D.J., Wickens M. PUF proteins bind Pop2p to regulate messenger RNAs. Nat. Struct. Mol. Biol. 2006;13:533–539. doi: 10.1038/nsmb1100. [DOI] [PubMed] [Google Scholar]
- 38.Hata H., Mitsui H., Liu H., Bai Y., Denis C.L., Shimizu Y., Sakai A. Dhh1p, a putative RNA helicase, associates with the general transcription factors Pop2p and Ccr4p from Saccharomyces cerevisiae. Genetics. 1998;148:571–579. doi: 10.1093/genetics/148.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brodersen DE. A Program For Presentation of Protein Sequence Alignments and Secondary Structure. SecSeq. 2001. http://www.bioxray.dk/~deb/secseq.
- 40.DeLano WL. The PyMOL Molecular Graphics System. 2002. http://www.pymol.org.
- 41.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ohn T, Chiang Y-C, Lee DJ, Yao G, Zhang Chongx, Denis CL. Nucleic Acids Res. doi: 10.1093/nar/gkm196. [DOI] [PMC free article] [PubMed] [Google Scholar]