Abstract
The CAF1 protein is a component of the CCR4–NOT deadenylase complex. While yeast CAF1 displays deadenylase activity, this activity is not required for its deadenylation function in vivo, and CCR4 is the primary deadenylase in the complex. In order to identify CAF1-specific functional regions required for deadenylation in vivo, we targeted for mutagenesis six regions of CAF1 that are specifically conserved among CAF1 orthologs. Defects in residues 213–215, found to be a site required for binding CCR4, reduced the rate of deadenylation to a lesser extent and resulted in in vivo phenotypes that were less severe than did defects in other regions of CAF1 that displayed greater contact to CCR4. These results imply that CAF1, while affecting deadenylation through its contact to CCR4, has functions in deadenylation separate from its contact to CCR4. Synthetic lethalities of caf1Δ, but not that of ccr4Δ, with defects in DHH1 or PAB1, both of which are involved in translation, further supports a role of CAF1 separate from that of CCR4. Importantly, other mutations in PAB1 that reduced translation, while not affecting deadenylation by themselves or when combined with ccr4Δ, severely blocked deadenylation when coupled with a caf1 deletion. These results indicate that both CAF1 and factors involved in translation are required for deadenylation.
INTRODUCTION
In eukaryotes, the major pathway of mRNA degradation is initiated by poly(A) tail shortening (deadenylation) followed by removal of the 5′ cap structure (decapping) and 5′–3′ exonucleolytic cleavage of the mRNA body by the XRN1 exonuclease (1–3). Deadenylation, as compared to decapping and 5′–3′ nuclease digestion, has been shown to be most important in the control of mRNA degradation rates (4). The CCR4-NOT complex in responsible for the majority of the poly(A) shortening process in yeast (5–7), and its 1.0 MDa core complex is comprised of 10 components: CCR4, CAF1 (also known as POP2), five NOT proteins (NOT1–NOT5), CAF40, CAF130 and BTT1 (8–11). Of these proteins, CCR4 and CAF1 appear to be the most important players in the mRNA deadenylation process (5,6).
The CCR4 protein as a member of ExoIII family of nucleases/phosphatases (12,13) is the catalytic subunit responsible for degradation of the mRNA poly(A) tail, and point mutations in the predicted active site of CCR4 cause deadenylation defects like that of a ccr4 deletion (5,7). Similarly, a ccr4 mutant in Drosophila also displayed an in vivo deadenylation defect, as did reduction in CAF1 levels (14).
The role of the CAF1 protein, however, is less clear. Its principal role has been considered to be that of linking CCR4 to the remainder of the CCR4-NOT complex (8,9,15,16). Deletion of the CAF1 gene showed a dramatic deadenylation defect in vivo, albeit to a lesser extent than that of ccr4Δ (6), and this has been interpreted to mean that CCR4 must be part of the CCR4-NOT complex to function well in vivo. Although CAF1 is classified as a member of the DEDDh family of nucleases (17,18) and the polypeptide isolated from Escherichia coli can function as a 3′–5′ exonuclease with some preference for poly(A) sequences (19,20), inactivation of predicted key catalytic active sites of CAF1 did not affect in vivo deadenylation function (16). Moreover, over-expression of CCR4 could partially complement the deadenylation defect of a caf1 deletion, but over-expression of CAF1 did not rescue phenotypes associated with that of a ccr4 deletion (6,7). These results indicate that the deadenylase activity of CAF1 is not required for its in vivo deadenylation function. Moreover, CAF1 has been shown recently to interact with PUF5 and to be required for PUF5-induced deadenylation of mRNA (21), supporting other roles of CAF1 in mRNA deadenylation.
Since the predicted catalytic sites of CAF1 are not critical to its in vivo function, understanding the functional roles of CAF1 on mRNA turnover will require the identification of other important regions of the CAF1 protein. To identify in the CAF1 protein functional regions, which are critical for mRNA degradation, we conducted a deletion analysis of yeast CAF1 by targeting regions absolutely conserved among CAF1 orthologs. This mutation analysis identified four functional regions of CAF1, including those required for binding CCR4 and for deadenylation. Domains of CAF1 that are important to deadenylation in vivo included but are not limited to those that are required for CCR4 binding. Relatedly, defects in CAF1, but not that in CCR4, were lethal with defects in DHH1 (33) and PAB1, factors involved in translation, and over-expression of translation-associated proteins could suppress caf1Δ pab1 lethality. Moreover, we showed that a PAB1 translation defect severely reduced deadenylation when coupled with a caf1 deletion. CAF1, in addition to binding CCR4, may act on the deadenylation process in conjunction with factors affecting translation.
MATERIALS AND METHODS
Yeast strains and growth conditions
The background yeast strains used in this study are listed in Table 1. Yeast strains were grown in YEP medium (2% yeast extract, 1% Bacto peptone) or minimal medium supplemented with nutrients required for auxotrophic deficiencies and with 4% glucose or 2% galactose/2% raffinose unless otherwise indicated. YD plates consisted of YEP medium supplemented with 2% glucose and 2% agar and caffeine plates are YD plates with 5, 8 or 15 mM caffeine as indicated.
Table 1.
List of yeast strains
| Strain | Genotype |
|---|---|
| EGY188 | MATa ura3 his3 trp1 LexA–LEU2 |
| EGY188-c1 | Isogenic to EGY188 except caf1 :: URA3 |
| EGY188-c1-1 | Isogenic to EGY188 except caf1 :: ura3 |
| EGY188-1a-1-c1 | Isogenic to EGY188 except ccr4 :: ura3 caf1 :: LEU2 |
| EGY191 | MATα ura3 his3 trp1 LexA–LEU2 |
| EGY191-c1 | Isogenic to EGY191 except caf1 :: URA3 |
| KY803-c1 | MATa leu2-PET56 trp1-Δ1 ura3-52 gal2 gcn4-Δ1 caf1 :: LEU2 |
| AS319/YC504 | MATα ura3 leu2 his3 trp1 pab1 :: HIS3 [PAB1-TRP1] |
| AS319-1a-uN/YC504 | Isogenic to AS319 except ccr4 :: ura3 :: NEO |
| AS319-c1-lN/YC504 | Isogenic to AS319 except caf1 :: leu2 :: NEO |
| AS319-cl-lN/YC505/YC360 | Isogenic to AS319-cl-lN except [PAB1-ΔRRM1-TRP1] [PAB1-URA3] |
| AS319-cl-lN/YC506/YC360 | Isogenic to AS319-cl-lN except [PAB1-ΔRRM2-TRP1] [PAB1-URA3] |
| AS319-d1-uL/YC504 | Isogenic to AS319 except dhh1 :: ura3 :: LEU2 |
| 1743-2/YC504 | MATα ura3 leu2 his3 trp1 pab1 :: HIS3 [PAB1-TRP1] prt1-63 |
| 1881/YC504 | Isogenic to AS319 except cdc33-1 |
| A1385-uT-c1/TB3 | MATa ura3 leu2 his3 trp1 dhh1 :: ura3 :: TRP1 caf1 :: LEU2 [GAL1-CAF1-URA3] |
Library screening for high-copy suppressors and analysis of synthetic lethalities
Strain ASY319-c1-lN/YC360/YC506 was transformed with a YEp13 (2 μ LEU2) high-copy yeast genomic library and plated on minimal medium lacking leucine and tryptophan. Here, ∼15 000 Leu+ transformants were plated onto plates containing 5-fluoroorotic acid (FOA). The plasmids from yeast capable of growth on FOA plates were rescued, transformed into E. coli, rescued and retransformed into the original strain to confirm the high-copy suppression. Yeast genomic sequences within the plasmids were identified by DNA sequence analysis. In cases where more than two yeast genes were located on the plasmid insert, such as for DHH1 and CAF1, the MPT1 and MPT0 plasmids carrying only DHH1 and CAF1, respectively, were used to confirm that these genes were responsible for the suppression of the lethality (29). caf1Δ lethality with PAB1-ΔRRM2 or with PAB1-ΔRRM1 was determined by transforming strain AS319-c1-lN/YC360 with YC504, YC505 and YC506, respectively. Following selection of these YC504/YC505/YC506 transformants, their ability to survive in the presence of the caf1Δ allele was determined by the transformants’ abilities to lose the YC360 (URA3) plasmid and grow on FOA-containing plates. Testing for the lethality of deletions in other genes and PAB1-ΔRRM2 was done by a similar methodology.
Site-directed deletion mutagenesis
The MPT0 plasmid harboring the yeast CAF1 open reading frame was used as the template for the polymerase chain reaction (PCR). All PCR reactions were performed with Vent polymerase (New England Biolabs). In order to generate 173DVW175, 213FRS215, 255WQF257, 303SGL305, 329LMN331 and 340DFE342 deletions, we designed the following six pairs of oligonucleotide primers: caf1-1-f (5′-TATCTTTTCGTTCGCAAGTCCAACCTTTACAGTGAATTC-3′) and caf1-1-r (5′-GTAAAGGTTGGACTTGCGAACGAAAAGATAATTTGGGG-3′), caf1-2-f (5′-AGGCCGATCGGCACTAAGGTCGATTACCACTATCAGACA-3′) and caf1-2-r (5′-GTGGTAATCGACCTTAGTGCCGATCGGCCTAGCCAAAGT-3′), caf1-3-f (5′-AACGGTCCCTCAACGAATTTTGAATTTGACCCAAAGAAG-3′) and caf1-3-r (5′-GTCAAATTCAAAATTCGTTGAGGGACCGTTGTCAGGCTT-3′), caf1-4-f (5′-CAGCTTCTAATGGACATGATGGATGATTCTGTTACTTGG-3′) and caf1-4-r (5′-AGAATCATCCATCATGTCCATTAGAAGCTGCGAAAATTC-3′), caf1-5-f (5′-TTCCTGATCAACATTGACTCCATGCCCAACAACAAGGAG-3′) and caf1-5-r (5′-GTTGGGCATGGAGTCAATGTTGATCAGGAAACCTAGATC-3′), and caf1-6-f (5′-CCCAACAACAAGGAGTGGTGGGTCCATCAATACATGCCC-3′) and caf1-6-r (5′-TTGATGGACCCACCACTCCTTGTTGTTGGGCATGGAGTC-3′). These primers were used for the first PCR reaction in combination with primers containing BamHI and HindIII restriction sites Caf1-Bm-f (5′-AAAGGATCCATGCAATCTATGAATGTACAA) and caf1/2291-Hind (5′-TACATATAAAGCTTAAATGATCATTGGTCCC-3′). The first PCR products were used for amplifying final full-length CAF1 mutant alleles using Caf1-Bm-f and caf1/2291-Hind primers. The final PCR products were purified, digested with BamHI and HindIII, and inserted into BamHI- and HindIII-digested pET-23a(+) to generate plasmids pTB8a (CAF1), pTB8-1 (caf1-1), pTB8-2 (caf1-2), pTB8-3 (caf1-3), pTB8-4 (caf1-4), pTB8-5 (caf1-5) and pTB8-6 (caf1-6), respectively. All the sequences of the mutagenized CAF1 alleles were verified by sequencing. For further analysis, CAF1 open reading frames were subcloned into pLexA202-2 (22), pJG4-5 (22) and pJCN112 (7) to make N-terminal LexA, HA and N-terminal Flag with C-terminal 6His epitope tagged CAF1 proteins, respectively. pLexA202-2 fusions were constructed to allow constitutive high expression of CAF1 from an ADH1 promoter, and the other two fusions were to allow galactose-inducible expression of CAF1 from a GAL1 promoter. LexA-CAF1 fusions were also used in certain cases because they were carried on a HIS3-containing plasmid that allowed it to be co-transformed with URA3-based plasmids, such as those carrying Flag-CAF1, Flag-CCR4 and Flag-PAB1.
Flag pull-down analysis
The yeast cultures containing Flag-CAF1 fusions were grown to late log phase in 50 ml selective medium supplemented with 4% glucose, shifted to 100 ml of the same medium with 2% galactose/raffinose and grown for 16 h. The cells were washed and lysed by the glass bead method in extraction buffer (50 mM Tris–Cl [pH7.9], 150 mM NaCl, 0.1 mM MgCl2, 0.1% NP40, 20% glycerol) plus a protease inhibitor cocktail. After clarification of the crude cell lysates by centrifugation at 15 000 × g at 4°C for 15 min twice, the supernatants were incubated with 400 μl of anti-Flag M2 affinity agarose (Sigma) at 4°C overnight. The bound resins were washed with 10 ml wash buffer (extraction buffer with 5% glycerol) three times, transferred to microfuge tubes and washed with 1 ml wash buffer five times. The resultant precipitates were run subsequently on an 8% SDS-polyacrylamide gel, electro-transferred to polyvinylidene fluoride (PVDF) membrane (Immobilon™-P, Millipore) and analyzed by western blot using SuperSignal® West Pico Luminol/Enhancer Solution (Pierce).
Protein extraction, in vitro deadenylation assays and in vivo translation assays
Flag-CCR4 protein purification was performed as described above for purification of Flag-CAF1 with some modifications (23). The strain EGY188-c1-1a-1 harboring single copy of CCR4 (pYC343) and the LexA-CAF1 fusion variants were grown in 1 l of selective medium with 4% glucose to mid-log phase (OD600 of 0.7). Following lysis and incubation of supernatants with the anti-Flag M2 antibody-agarose (Sigma), the Flag agarose beads were washed with 10 ml of washing buffer three times, and the Flag fusion proteins were eluted twice with extraction buffer containing 200 μg/ml Flag peptide (Sigma). In vitro deadenylation assays were conducted as described (16,23). The rates of in vivo protein synthesis were determined by quantitating the amount of [35S]-methionine incorporation into protein as described (24).
RNA preparation and analyses
RNA samples were prepared using the hot acidic phenol method described in Ref. (25). Briefly, yeast cells were resuspended in TES (10 mM Tris–Cl [pH 7.5], 10 mM EDTA, 0.5% SDS) and the acidic phenol and incubated at 65°C for 45 min with frequent vortexing. Following centrifugation, the supernatants were re-extracted with acidic phenol and chloroform. RNA present in the supernatants was precipitated with 95% ethanol and dissolved in water containing DEPC.
To perform the transcriptional pulse-chase experiments, strain EGY188-cl carrying the individual LexA-caf1 variants was grown in 5 ml of synthetic medium supplemented with 2% raffinose, transferred and regrown in 100 ml fresh medium until mid-log phase (OD600 of 0.7). Cells were then harvested, washed once with fresh medium and transferred to 15 ml of fresh medium containing 2% raffinose and grown for 15 min. GAL1 transcription was activated by adding galactose at a 2% final concentration for 15 min and shut-off by adding glucose to a concentration of 4%. Since yeast cells harboring the LexA–caf1-2 protein resulted in poor GAL1 transcription, the GAL1 promoter was activated for 30 min prior to transcription shut-off.
The deadenylation rates and end points for GAL1 mRNA were determined following RNase H treatment of purified RNAs as described previously (26). Here, ∼4 μg of an oligonucleotide probe (5′-GCCATTTGGGCCCCCTGG-3) complementary to a segment 133 bp upstream of the GAL1 translational stop codon was hybridized with 12 μg of total RNA prior to RNase H digestion. The resultant GAL1 3′ polyadenylated species were separated on a denaturing polyacrylamide gel (6%/7.5 M urea) and detected by northern analysis using a probe complementary to the 3′ end of the GAL1 mRNA (5′-GCCCAATGCTGGTTTAGAGACGATGATAGCATTTTCTAGCTCAGCATCAGTGATCTTAGGG-3). The rate of deadenylation was determined for the shortest poly(A) tail as previously described (6,16). Calculating the rate of deadenylation using the average length of poly(A) gave similar results.
For steady-state MFA2pG mRNA analyses, strains were grown to exponential phase (OD600 of 0.4) in selective medium supplemented with 2% raffinose. Cells were then transferred to fresh medium for an hour, and the GAL1 promoter was induced for 3 h following the addition of galactose.
RESULTS
Mutagenesis of CAF1 in the regions that are highly conserved among CAF1 orthologs
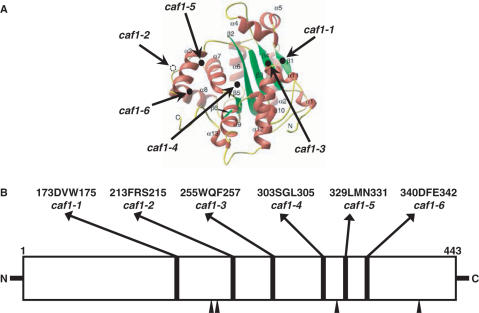
In order to identify CAF1-specific functional regions, we targeted for mutagenesis the regions of CAF1 that are common only to CAF1 orthologs and are not present in any other DEDDh nuclease family members (17,18). Six regions were identified as absolutely conserved among CAF1 orthologs (Figure 1B). Three amino acid deletions were made in each case to remove the region of homology, resulting in caf1 alleles 1 through 6. After these deletions were constructed and their phenotypes obtained, the X-ray crystallographic analysis of the C-terminal RNase D domain of CAF1 protein was published (19). The relative locations of each of the constructed caf1 alleles are shown in Figure 1A: caf1-1 (173DVW175) and caf1-3 (255WQF257) alleles are positioned in β-sheets β1 and β4 that hydrogen bond to each other, caf1-2 (213FRS215) is located between β2 and α3 in a crystallographically undefined loop region, and caf1-4 (303SGL305), caf1-5 (329LMN331) and caf1-6 (340DFE342), are located in a turn at the end of α6, just at the beginning of a turn after α7, and on α8, respectively.
Figure 1.
Mutagenesis of CAF1. (A) Location of CAF1 mutations are illustrated on the 3D structure of the nuclease domain of the CAF1 protein (19). (B) The six absolutely conserved regions targeted for mutagenesis are composed of three amino acids each as indicated above the schematic representation of CAF1 and are located in regions separate from the putative four key catalytic amino acids conserved among DEDDh ribonucleases (black arrows). The names of the mutant alleles are also shown beneath the deleted amino acids.
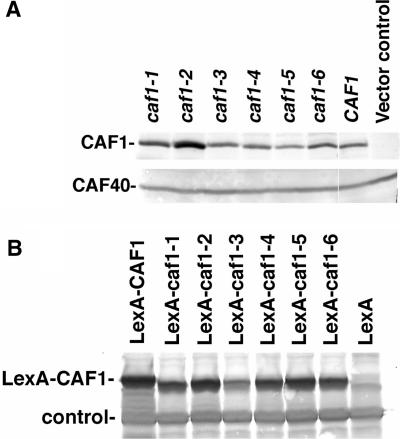
Each of the caf1 mutant alleles was subsequently cloned into three types of plasmid vectors, pLexA(202), pJG4-5 (HA-tagged) and pJCN112 (Flag-tagged), for further analysis (see Materials and Methods section). Each of the mutant proteins, regardless of the epitope attached to CAF1, was expressed to comparable levels as that found for the wild-type CAF1 protein, indicating that these small deletions did not affect the protein expression or stability of CAF1 (Figures 2A,B and 4A).
Figure 2.
Western blot analysis of CAF1 mutant proteins. (A) The HA-CAF1 proteins were expressed in the EGY191-c1 strain, protein extracts were made and the CAF1 proteins were detected by western analysis. The apparent increased abundance of caf1-2 protein in the gel that is presented is artifactual and no significant differences in the abundance of these proteins were observed in numerous other experiments. Vector control refers to pJG4-5 without the CAF1 protein. Equivalent amounts of protein were loaded in each lane as determined by Bradford assay, and western analysis using CAF40 (lower panel), CCR4 and NOT3 antibodies confirmed equivalent levels of CAF40, CCR4 and NOT3 proteins in each of the lanes. (B) The LexA-CAF1 proteins were expressed in EGY188-c1 and the CAF1 proteins were detected by western analysis using LexA antibody. The cause for the multiple bands present for CAF1 is not known, but they are not related to the phosphorylation of CAF1 that has previously been observed (27). The control band refers to a non-specific protein that interacts with the LexA antibody and indicates that nearly equivalent amounts of protein were loaded into each lane.
Figure 4.
caf1-2 is deficient in contacting CCR4 in vivo. (A) N-terminal Flag tagged CAF1 alleles were transformed into the caf1 deletion strain, KY803-c1. Crude extracts (5 mg) were immunoprecipitated with anti-FLAG M2 antibody and proteins were visualized by western blot analysis with antibodies directed against CCR4, NOT1 and CAF1 as indicated. (B) Plasmids expressing LexA-CAF1 were transformed into strain EGY188-1a-1-c1 containing FLAG-CCR4 expressed from its own promoter on a centromere-containing plasmid. Flag-CCR4 was purified as described for Flag-CAF1, and LexA antibody was used to identify CAF1 as indicated.
Phenotypic analysis of caf1 alleles
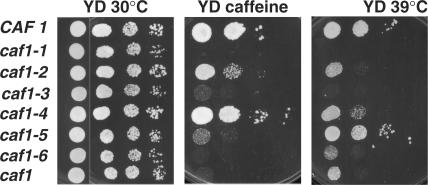
One of the most prominent phenotypes of a strain carrying a caf1 deletion is its caffeine sensitivity, although other phenotypes such as temperature sensitivity and slow growth have been observed (15,28,29). To examine the phenotypes of CAF1 mutant alleles, we tested the growth phenotypes of the caf1 alleles by using the KY803-c1 (caf1Δ) strain carrying a plasmid expressing an N-terminal Flag fusion with each of the mutant CAF1 proteins (Figure 3; summarized in Table 2). Of the six CAF1 mutants, caf1-1, caf1-3 and caf1-6 alleles resulted in an inability to grow on YD plates containing 5 mM caffeine. Interestingly, strains carrying the caf1-1 and caf1-3 alleles were observed to grow slower on rich medium lacking caffeine than a strain carrying the caf1 null allele (data not shown). The caf1-2 and caf1-5 alleles caused weak sensitivity to caffeine, and for the caf1-4 allele, we did not observe any difference in growth on caffeine plates as compared to the wild-type strain. The same results were obtained with LexA-CAF1 fusions (data not shown).
Figure 3.
Growth of strains carrying caf1 alleles. Yeast strains (KY803-c1 with Flag-CAF1 variants as indicated or with pJCN112, no Flag-CAF1, caf1) were grown on YD plates at 30°C, on YD plates with 5 mM caffeine, or on YD plates at 39°C. In each panel, moving from left to right 10-fold less cells were plated in each column. Initial concentrations of cells were ∼1 × 107 cells/ml for the left row and 4 μl of sample was placed in each spot.
Table 2.
Growth phenotypes of yeast cells carrying caf1 mutant alleles
| CAF1 allele | 5 mM caffeine | 39°C |
|---|---|---|
| Wild-type | + | + |
| caf1-1 | − | − |
| caf1-2 | w | w/− |
| caf1-3 | − | − |
| caf1-4 | + | w |
| caf1-5 | w/− | + |
| caf1-6 | − | w/− |
| caf1Δ | − | w/− |
| caf1-10 | w | w/− |
| caf1-11 | − | − |
KY803-c1 strains carrying FLAG-CAF1 proteins or pJCN112 vector alone (caf1Δ) were tested. Growth was detected on YEP plates containing 2% glucose (YD) and supplemented with 5 mM caffeine. Temperature sensitive growth was monitored on YD plates at 39°C. +, good growth; w, weak growth; −, no growth
In terms of temperature-sensitive growth, strains carrying the wild-type and caf1-5 alleles were able to grow at 39°C (Figure 3; Table 2), whereas those containing the caf1-1 and -3 alleles failed to grow at 39°C. The strain with caf1-4 grew weakly at 39°C, and strains carrying caf1-2 and -6 grew very weakly. In order to verify that the deletion of three consecutive amino acid residues did not dramatically affect the structure of the protein and result in the caffeine- and temperature-sensitive phenotypes that were observed, we constructed two additional caf1 alleles in which three alanine residues were substituted for the three residues deleted in caf1-2 and caf1-3. These two alleles were chosen in that results shown below indicated that the caf1-1 and caf1-6 alleles appeared phenotypically similar to caf1-3, and caf1-2 was unique for being most defective in CCR4 binding. Strains carrying caf1-10 (213AAA215) and caf1-11 (255AAA257) displayed the exact same phenotypes as did strains with their respective deletion alleles, caf1-2 and caf1-3 (Table 2). For the remainder of our analysis the original deletion mutations were used. It should also be noted that these CAF1 deletion derivatives, as has been previously observed for caf1 deletions (8–10), did not affect the relative protein amounts and, therefore, expression in the cell of other CCR4-NOT components, such as CCR4, NOT1, NOT3 or CAF40 (data not shown).
The caf1-2 protein is defective in binding with CCR4
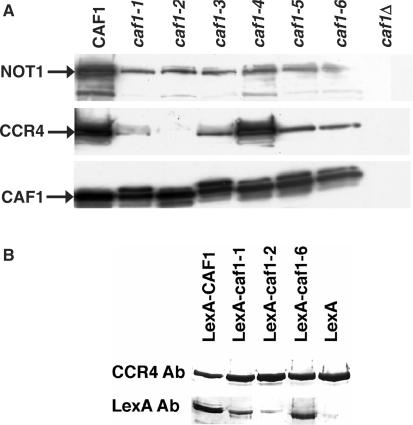
The relative locations of the proteins in the CCR4-NOT core complex have been determined (9–11) in which the CAF1 protein is critical for linking CCR4 to NOT1 and the remainder of the CCR4-NOT complex. We, therefore, assessed the ability of each of the CAF1 mutant proteins to bind CCR4 and NOT1, using Flag-tagged CAF1 protein variants expressed in a caf1 deletion background. Flag-CAF1 was used for this analysis as the Flag immunoprecipitations resulted in substantially cleaner backgrounds than immunoprecipitations with either HA- or LexA-CAF1 variants. After Flag immunoprecipitations were conducted, as shown in Figure 4A, we found that the caf1-4 protein was able to bind CCR4 as well as wild-type CAF1. The caf1-2 protein, in contrast, was totally defective in binding to CCR4 (Figure 4A). The slight band observed in the caf1-2 lane with CCR4 antibody does not migrate where CCR4 is located and is a result of a non-specific protein interaction. The caf1-1, -3, -5 and -6 proteins also displayed reduced binding to CCR4, but they still displayed significantly more association with CCR4 than did caf1-2. While all of the mutant proteins were still capable of binding NOT1, they did display reduced binding to NOT1 as compared to wild-type CAF1.
In order to verify these results, we also conducted the immunoprecipitation in the reverse direction for several of the key CAF1 variants. Using a Flag-tagged CCR4 and LexA-fused versions of the CAF1 variants, we isolated Flag-CCR4 and determined the abundance of LexA-CAF1 that was co-immunoprecipitated. As shown in Figure 4B, LexA-caf1-2 was most severely deficient in binding CCR4. In contrast, LexA-caf1-1 and LexA-caf1-6 proteins bound significantly better to CCR4 than did LexA-caf1-2. However, as demonstrated in Figure 4A, both the LexA-caf1-1 and LexA-caf1-6 proteins were still defective in binding CCR4 as compared to wild-type CAF1 (Figure 4B).
These above results suggest several conclusions. First, the immunoprecipitation data indicate that the differences in the growth and caffeine phenotypes described above do not result solely from differences in CAF1 binding NOT1 or CCR4. The severe phenotypes displayed with the caf1-1, -3 and -6 alleles were not due solely to loss of CCR4 binding, as each of these corresponding proteins could bind CCR4 as well as caf1-5, whose allele did not display as severe growth defects. The caf1-2 protein was most defective for binding CCR4, and yet the strain carrying caf1-2 grew better than did those carrying caf1-1, -3 or -6, indicating that loss of CCR4 binding alone was not the cause for the caf1 deletion phenotypes. Therefore, while CAF1 binding to CCR4 contributes to part of the defects observed with caf1Δ, as exemplified by caf1-2, other CAF1 interactions must also be important to its function. Third, the ratio of NOT1-CCR4 in the caf1-1, -3, -5 and -6 immunoprecipitations was similar to that found in the wild-type strain, arguing that some of the CCR4-NOT complex was intact in the cell. The expression and abundance of the individual CCR4-NOT proteins in the cell were not affected by the caf1 variants as determined by western analysis (data not shown), suggesting that the each of the caf1 variants except for caf1-4 was causing some destabilization of the CCR4-NOT complex. Yet, as the strain carrying caf1-5 was similar to wild type in terms of its growth at high temperature, the reduced abundance of the CCR4-NOT complex per se is not the sole cause for all of the severe growth defects observed with caf1-1, -3 and -6. These observations also suggest that the severe growth defects of caf1-1, -3 and -6 relative to that of caf1-2 and caf1-5 are not specifically due to the caf1-1, -3 and -6 proteins being completely misfolded and hence comparable to the caf1Δ allele, as some of these caf1-1, -3 and -6 proteins are still capable of binding CCR4 and NOT1.
We also assayed the purified CCR4-NOT complex from each of the mutated strains for in vitro CCR4 deadenylation activity (23). CAF1 is not required for CCR4 in vitro deadenylation function (7,30), and, as expected, the six caf1 alleles did not significantly alter CCR4-NOT in vitro deadenylation activity following purification of Flag-CCR4 (Supplementary Figure 1). We were not able to assay, however, for CAF1 deadenylase activity in vitro since we have not been able to demonstrate enzyme activity for yeast CAF1 (6,16).
Identification of separable functional regions required for mRNA deadenylation
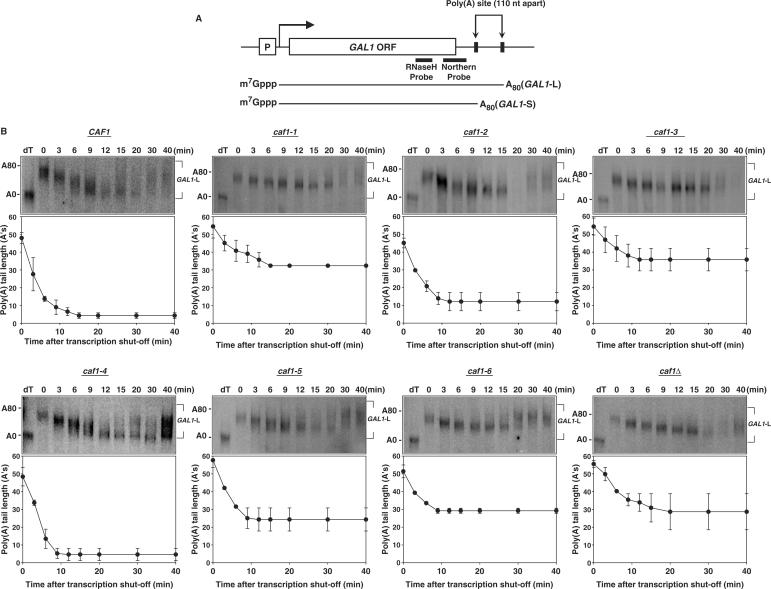
In order to determine if the caf1 mutant alleles affected mRNA deadenylation in vivo, transcriptional pulse-chase experiments were performed on the GAL1 mRNA. In this experiment, transcription of GAL1 mRNA was induced by adding galactose and then repressed by adding glucose to the growing culture. Following a 15 min induction, GAL1 mRNA synthesis was shut-off at time zero and aliquots were prepared for northern analysis at each time point. As shown in Figure 5A, GAL1 mRNA was shortened with a 3′ oligonucleotide probe and RNase H prior to separation on a denaturing polyacrylamide gel for size resolution of the poly(A) tail. Due to two polyadenylation sites in the GAL1 gene, GAL1 mRNAs consists of two forms: GAL1-L and GAL1-S in which the long GAL1-L has an additional 110 nt in its 3′-UTR segment as compared to GAL1-S (Figure 5A) (31,32). However, as GAL1-S mRNA is less abundant and therefore more difficult to detect and quantitate than that of GAL1-L mRNA, we present only the data for GAL1-L. Also, for this experiment LexA fusions to the CAF1 variants were used to transform a caf1 deletion yeast strain so that CAF1 would be constitutively expressed from its ADH1 promoter.
Figure 5.
Mutagenesis of CAF1 identifies separable functional regions involved in deadenylation. (A) Diagram for the GAL1 gene and its two mRNAs. (B) Transcriptional pulse-chase analyses on GAL1-L mRNA were conducted with EGY188-c1 harboring each LexA–caf1 fusion or the LexA vector as the negative control. Following a 15–30-min induction of the GAL1 gene by addition of galactose, transcription was shut-off at time zero by adding glucose. Northern blot analyses were conducted with RNA samples prepared from yeast cells harvested at the time points indicated. dT refers to the RNA sample probed with oligo (dT) followed by RNase H digestion to remove the poly(A) tail and represents 0 adenosines (A's). The 80 A's marker size was determined from the known GAL1-L and GAL1-S deadenylated lengths and from comparing GAL1-L poly(A) tail lengths against those of MFA2pG in other experiments (16). It should also be noted that at later time points the GAL1 mRNA synthesis is incompletely turned off by glucose and that at the 30 min time point it is often observed that newly synthesized mRNA with long poly(A) tails become present. This new mRNA does not significantly obscure the data for the original pulse of mRNA synthesis, and their poly(A) tail lengths were not used in the determinations of the poly(A) tail length of the original pulse of mRNA synthesis.
The deadenylation rate analysis of GAL1-L mRNA identified several functional regions in CAF1 that are important for normal deadenylation in vivo. Of the six caf1 alleles, caf1-1, caf1-3 and caf1-6 alleles displayed severe deadenylation rate defects similar to that displayed with a caf1 null allele. For the wild-type strain (Figure 5B), GAL1-L poly(A) tails were shortened to 8–12 nt within 9 min. In contrast, in the caf1-1, -3 and -6 allele background, GAL1-L was only deadenylated to ∼30–35 A's within the first 9 min (Figure 5B). Similar defects in GAL1-S mRNA deadenylation were also observed for caf1-1, -3 and -6 as compared to wild type (data not shown). The average rate of deadenylation for GAL1-L in wild type was 4.9 A's/min in which the rate of deadenylation was based on the lower end of the poly(A) distribution (26) (Table 3). In the caf1-1, -3 and -6 mutated strains, the range of the deadenylation rates for GAL1-L mRNAs were from 2.0 to 2.9 nt/min (Table 3). For caf1-2 and caf1-5, in contrast, the deadenylation rates for GAL1-L were in the 3.4–4.4 A's/min range, indicating that they were defective in deadenylation but not to the extent observed for the caf1-1, -3 and -6 alleles (Table 3; Figure 5B). caf1-4 displayed a faster deadenylation rate than wild type. In addition, it should be observed that the caf1-1, -3 and -6 proteins failed to allow complete deadenylation to 8–10 A's even at the 40-min time point. caf1-2 allowed deadenylation to proceed much further to nearly wild-type lengths, whereas caf1-5 displayed a more severe deadenylation endpoint defect with GAL1-L. Given that the phenotypic analysis of caf1-2 and caf1-5 showed that their temperature and caffeine sensitivities were halfway between wild type and that of the caf1 deletion, these in vivo deadenylation data agree well with their in vivo phenotypes.
Table 3.
Deadenylation rates of GAL1 and endpoints of MFA2pG mRNA
| CAF1 allele | Deadenylation rate (A's/min ± SEM) | Deadenylation endpoint range [oligo(A) shortest to longest ± SEM for longest lengths] |
|---|---|---|
| GAL1-L | MFA2pG | |
| Wild type | 4.9 ± 0.5 | 0–11 ± 1.9 |
| caf1-1 | 2.0 ± 0.2 | 5.3–29 ± 3.4 |
| caf1-2 | 3.4 ± 0.7 | 1.9–15 ± 2.1 |
| caf1-3 | 2.1 ± 0.5 | 5.2–29 ± 3.4 |
| caf1-4 | 6.1 ± 1.6 | 0–11 ± 2.1 |
| caf1-5 | 4.4 ± 0.6 | 0–12 ± 4.3 |
| caf1-6 | 2.9 ± 0.7 | 2.2–24 ± 2.0 |
| caf1Δ | 2.4 ± 0.5 | 3.5–26 ± 2.7 |
caf1 deletion strains (EGY188-c1) harboring LexA versions of wild-type, caf1 mutants and vector (LexA202-2) were used for transcriptional pulse chase analysis. Rates of deadenylation were measured by the change in the length of the shortest poly(A) tail as a function of time following transcriptional shut-off (Figure 4B). Similar results were obtained using the mean poly(A) tail length. EGY188-c1-1 strains harboring LexA-caf1 variants and pRP485 (GAL1-MFA2pG) were used for analyzing steady-state MFA2pG mRNA. The range in oligo(A) lengths for the pG fragments from northern analysis was taken from deadenylation endpoint measurements. Deadenylation rate values represent the average of three separate experiments and the standard error of the mean (SEM), and the deadenylation endpoints are the average of two separate experiments and the SEM for the longest length poly(A) segments.
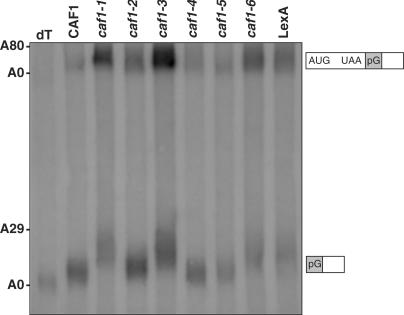
To more precisely analyze deadenylation endpoints, the steady-state RNA levels of MFA2pG were studied (6). In this case, the pG mRNA fragments that results from deadenylation, decapping and 5′ nuclease digestion of MFA2pG allows a better estimate of the deadenylation endpoint than does pulse-chase analysis. Previously, a caf1 deletion has been shown to result in median endpoints of ∼16–20 A's as compared to 8–12 A's for wild-type CAF1 (6). We found that caf1-1, -3 and -6 alleles resulted in a longer poly(A) tail length for the pG fragment that results after deadenylation, decapping and 5′–3′ nuclease action on MFA2pG than for wild-type CAF1: 24–29 A's for the longest A lengths as compared to 11 A's for wild-type (Figure 6, lanes 2, 4, 7 compared to lane 1; summarized in Table 3). In the caf1-2 and caf1-5 alleles, the deadenylation endpoints of the pG fragment were not significantly affected as compared to wild-type (Figure 6, lanes 3, 6; Table 3). It should also be noted that the rate of MFA2pG deadenylation appeared slower in the caf1-1, -3 and -6 backgrounds as compared to caf1-2, -5, and CAF1. In the top part of Figure 6, it can be observed that full-length MFA2pG mRNA poly(A) tails are on average significantly longer for caf1-1, -3, and -6 than that of caf1-2, -5 and CAF1, indicative of slowed deadenylation. Moreover, the ratio of full-length MFA2pG RNA to that of the pG fragment was significantly greater for caf1-1, -3 and -6 as compared to caf1-2, -5, and CAF1, indicating that the rate of decapping and degradation of the MFA2pG mRNA was being slowed, most likely due to a slowed rate of deadenylation (6). These overall results indicate that the severe growth phenotypes of caf1-1, -3 and -6 probably result from the inability of the CCR4-NOT complex to properly deadenylate in vivo and that CAF1 must also interfere with the deadenylation process separately from its contacts to CCR4 and NOT1.
Figure 6.
caf1-1, -3 and -6 alleles are defective in the deadenylation endpoint of the MFA2pG mRNA. Northern analysis of steady-state MFA2pG mRNA was conducted in a caf1Δ background (EGY188-c1-1) in which LexA-CAF1 fusion proteins were expressed as indicated. dT refers to removal of oligo(A) by incubation with oligo(dT) followed by RNase H digestion. The approximate lengths of the poly(A) tail and migration positions of the MFA2pG and pG fragments are indicated to the left and right, respectively. LexA refers to the LexA vector alone expressed in strain EY188-c1-1.
Analysis of the role of CAF1 in deadenylation separate from contacting CCR4
The above results support the model that CAF1 has an effect on the mRNA degradation process in addition to that derived from its contact to CCR4. In this regard, the caf1 deletion has previously been shown to be lethal with a deletion in the decapping/translational regulator DHH1 whereas the ccr4 deletion is not lethal with that of dhh1Δ (33). In order to determine which CAF1 allele can confer lethality with dhh1Δ, we constructed a strain carrying both a dhh1Δ and a caf1Δ that was covered by a plasmid expressing a GAL1-CAF1 gene. This strain was unable to grow on glucose growth conditions when GAL1-CAF1 expression was shut-off (Supplementary Figure 2). Following transformation of LexA-CAF1 plasmids encoding each of the above mutagenized CAF1 alleles, the caf1-1, -3 and -6 alleles did not allow growth on glucose growth conditions whereas caf1-2 and -5 alleles allowed growth on glucose (Supplementary Figure 2). These data suggest that caf1-1, -3 and -6 alleles are also defective in a non-deadenylation function.
caf1Δ dhh1Δ lethality could result from their combined effects on deadenylation that is separate from CCR4 effects on deadenylation. However, this model is unlikely in that lethality does not occur under circumstances when deadenylation in the cell is completely blocked (6). An alternative model would be that as DHH1 is known to be involved in both decapping (34) and translation (35), CAF1 could be involved separately from CCR4 in either or both of these functions leading to a caf1Δ dhh1Δ lethality. This model would further suggest that CAF1 effects on deadenylation that are separate from binding CCR4 would result from its involvement in this other function. That is, CAF1 interaction with the decapping/translation processes may affect deadenylation. However, CAF1 is not probably involved in decapping separately from CCR4. Studies that measured the relative decapping rate using the MFA2pG RNA have suggested that the slowed deadenylation observed with a caf1 deletion does not result in a more severe decapping defect than does ccr4Δ (5,6; data not shown).
In an analysis of the genetic interactions of poly(A)-binding protein (PAB1) with CCR4 and CAF1, we observed that deleting the RRM2 or RRM1 domain of PAB1 was lethal with a caf1 deletion but not with ccr4Δ. The RRM2 domain of PAB1 is important both for contacting the poly(A) tail and the translation initiation factor eIF4G, and both RRM1 and RRM2 are required for poly(A) stimulated in vitro translation (36–38). This observation suggests that CAF1 has functions separate from CCR4 that involve some role of PAB1. We subsequently analyzed whether deletion of other components of the CCR4-NOT complex or those of the PAN2/3 deadenylase were lethal with these PAB1 variants. We found that the not4Δ, not5Δ and caf40Δ, as well as pan2Δ and pan3Δ, were not lethal with these PAB1 mutants (data not shown), implying that it was a specific property of CAF1 that resulted in the lethality. This observed caf1Δ pab1-ΔRRM2 lethality suggests that CAF1 may functionally interact with factors affecting translation, which in turn may affect deadenylation. The RRM1 and RRM2 deletions of PAB1 also reduced the in vivo rate of protein synthesis by 28 and 15%, respectively, whereas deletion of RRM3, RRM4 or of the C-terminal region of PAB1 had <6% effects on the rate of protein translation (data not shown). Deletions of RRM3, RRM4 or the C-terminus of PAB1 were correspondingly not lethal with caf1Δ (data not shown). We also found that the caf1-1, -3 and -6 alleles but not those of caf1-2 or caf1-5 were lethal with PAB1-ΔRRM2 (data not shown). These genetic interactions support the possibility that CAF1 is functionally involved with factors that affect the translation process.
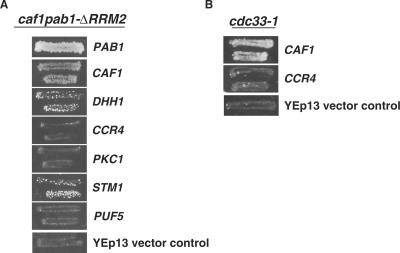
To clarify the genetic interaction between caf1Δ and PAB1-ΔRRM2, we conducted a genetic screen to identify genes whose over-expression could rescue this synthetic lethality. A high-copy YEp13-LEU2 yeast genomic library was transformed into a strain carrying the caf1Δ PAB1-ΔRRM2 allele and the YC360 plasmid [PAB1-URA3]. Eleven plasmids that suppressed the caf1Δ PAB1-ΔRRM2 lethality were identified, as evidenced by their ability to support loss of the YC360 plasmid following growth on 5′-FOA plates. Five of these plasmids contained CAF1 and one plasmid carried PAB1. All of these plasmids suppressed the lethality strongly (Figure 7). Of the remaining five plasmids, two displayed moderate suppression, and of these one was found to carry the DHH1 gene and the other the STM1 gene. The remaining three weakly suppressing plasmids will be reported on separately.
Figure 7.
High-copy suppressors of caf1Δ PAB1-ΔRRM2 lethality. (A) Over-expression of DHH1 and STM1 suppress the synthetic lethality of the caf1Δ PAB1-ΔRRM2 double mutant. Strain ASY319-c1-lN/YC360 (PAB1-URA3)/YC506 (PAB1-ΔRRM2-TRP1) was transformed with YEp13 plasmids carrying MPT0 (CAF1), MPT1 (DHH1), MPT2 (CCR4), MPT3 (PKC1), MPT4 (STM1), MPT5 (PUF5) (29) and PAB1, respectively. Transformants were selected on minimal medium lacking leucine and tryptophan and replica plated to medium containing FOA to lose the YC360 plasmid. For the DHH1 and MPT4 complementation of caf1Δ PAB1-ΔRRM2 lethality, the papillation of colonies that is observed are not revertants as they represent cells that begin growth at the same time that cells begin to grow in the control PAB1 or CAF1 situations. Similar papillations are typically observed for growth on 5′-FOA plates during the selection for loss of YC360 plasmids in many other experiments. (B) Over-expression of CAF1 can suppress the caffeine sensitivity associated with the cdc33-1 allele. Strain 1881/YC360 was transformed with MPT0, MPT2 or YEp13. The YD plates contained 8 mM caffeine.
Because previous analysis has identified several multi-copy suppressors of a caf1 deletion, including that of DHH1, CCR4, PKC1, STM1 and PUF5 (29), we tested whether over-expression of each of these different genes in a strain carrying caf1Δ PAB1-ΔRRM2 and PAB1-URA3 could suppress the caf1Δ PAB1-ΔRRM2 lethality and allow loss of PAB1-URA3 and subsequent growth on 5′-FOA media. As shown in Figure 7A, we failed to observe any high-copy suppression of caf1Δ PAB1-ΔRRM2 by CCR4, PKC1 or PUF5. Since over-expression of CCR4 can complement caf1Δ deadenylation defects (6), this last result confirms that caf1Δ pab1-ΔRRM2 lethality is not due solely to a deadenylation defect. Moreover, it indicates that the ability of over-expressed DHH1 and STM1 to complement the caf1Δ PAB1-ΔRRM2 lethality does not result from simply the ability to complement caf1Δ defects. These findings provide additional genetic evidence supporting the functional participation of CAF1 in a process that is separate from the role involving CCR4.
One additional model to explain the role of DHH1 in suppressing the caf1Δ PAB1-ΔRRM2 lethality would be that over-expression of DHH1 could compensate for the deadenylation defect caused by caf1Δ. In analyzing the steady-state MFA2pG levels, over-expression of DHH1, however, had no effect on the deadenylation defects observed with the caf1-1, -6 and caf1Δ alleles (data not shown). In each of these strains, the MFA2pG mRNA did not deadenylate completely and had longer poly(A) tail lengths than the wild-type regardless of whether DHH1 was over-expressed or not. We additionally tested whether DHH1 association with the CAF1 variants was altered, as it has been shown that NOT1 and CAF1 can associate with DHH1 (33,34). DHH1 interaction with the CCR4-NOT complex for each of the CAF1 variants, however, was unaffected (Supplementary Figure 3), in agreement with previous data that showed that a caf1 deletion had little effect on DHH1 association with NOT1 (33).
As DHH1 is involved in translation processes (34,35) and STM1 can interact with the ribosome and eIF4E and is important for protein synthesis (39–43), we determined whether over-expression of CAF1 or CCR4 could in turn complement a defect in translation factor eIF4E (cdc33-1 allele). As shown in Figure 7B, over-expression of CAF1 but not that of CCR4 could complement the caffeine sensitivity displayed in a strain carrying the cdc33-1 allele. As cdc33-1 is specifically defective in binding the 5′ mRNA cap (44), these results provide further support for a functional interaction of CAF1 with translational factors distinct from the role of CCR4 in deadenylation. However, as over-expression of CAF1 was found neither to rescue the cdc33-1 defect in in vivo protein translation nor did it affect in vivo protein translation in a wild-type background (data not shown), over-expression of CAF1 does not probably restore eIF4E function completely.
Because of the genetic interaction between CAF1 and that of cdc33-1, we also assessed whether a caf1Δ could affect translation directly. Two aspects were analyzed. First, we conducted polysome profiles of mRNA isolated from wild-type and caf1Δ strains. No differences between the polysome profiles were observed (data not shown). Second, we assayed if a caf1Δ affected the rate of in vivo protein translation by quantitating the rate of incorporation of [35S]-methionine into newly synthesized protein. A caf1Δ was found to reduce the rate of in vivo translation to 35% of wild type. In contrast, a ccr4Δ reduced in vivo translation only to 55% of wild type. SEMs for these series of experiments were <5% of the values. Similarly, in a cdc33-1 background, a caf1Δ deletion reduced the rate of in vivo protein synthesis from 35% (cdc33-1) to that of 13% (cdc33-1 caf1Δ); the rate of protein synthesis was reduced to 24% by combining cdc33-1 with ccr4Δ. These results indicate that a caf1Δ was displaying an additional effect on protein translation than observed through its possible effects on CCR4.
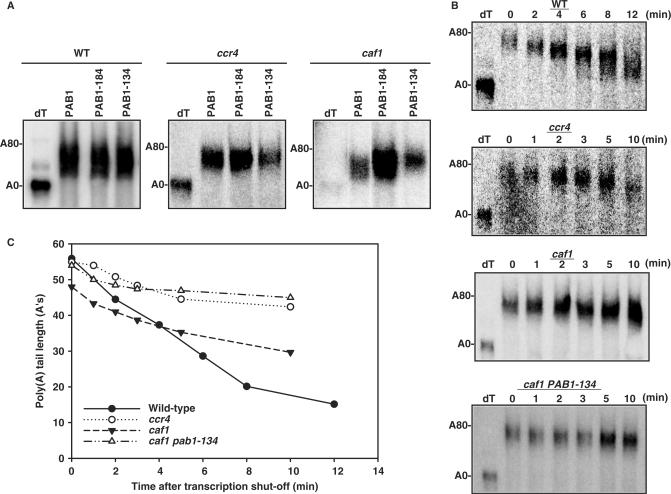
If CAF1 were to functionally interact with PAB1 and/or the translation process, it might be expected that defects in PAB1 could, in turn, when combined with caf1Δ, specifically reduce deadenylation. We subsequently analyzed whether other less severe defects in the RRM2 domain of PAB1 in conjunction with a caf1 deletion affected the deadenylation process. Two PAB1 variants are known to affect different PAB1 translational functions (38). PAB1-184 and PAB1-134 carry three amino acids substitutions each: PAB1-184 affects eIF4G binding in vitro and PAB1-134 has an undefined in vitro translation defect involving 5′-cap-dependent translation (38). We first analyzed the effect of these variants on the steady-state distribution of poly(A) tail lengths for GAL1-L mRNA in conjunction with a caf1 deletion. caf1Δ PAB1-134 resulted in significantly longer steady-state GAL1-L poly(A) tail lengths as compared to caf1Δ alone, indicative of a slowed deadenylation rate (Figure 8A, right panel; data not shown), whereas PAB1-184 had no effect on poly(A) tail lengths in conjunction with caf1Δ. This result suggests that both CAF1 and the PAB1-134 mutated residues are required for complete deadenylation. In contrast, PAB1-134 in combination with ccr4Δ did not affect the steady-state distribution of GAL1-L poly(A) tail length as compared to ccr4Δ alone (Figure 8A; middle panel) nor did it affect the GAL1-L poly(A) tail lengths with wild-type PAB1 (Figure 8A, left panel). These latter results demonstrate that PAB1-134 is not affecting PAN2/3 deadenylase activity, which is the only deadenylase activity present in a strain lacking CCR4 (6).
Figure 8.
PAB1-134 blocks deadenylation in a caf1Δ background. (A) Steady-state mRNA analysis of GAL1 was conducted as described in the Materials and Methods section. wt- AS319; ccr4Δ- AS319-1a-uN; caf1Δ-AS319-c1-lN. (B) Pulse-chase analysis of GAL1-L mRNA deadenylation was conducted as described in Figure 4. PAB1-134 refers to AS319/YC545 and PAB1-184 refers to AS319/YC538. YC545 expresses the PAB1 protein with residues 134HPD137 replaced with DKS and YC538 has residues 184DAL187 of PAB1 replaced with EKM (38). PAB1-134 and PAB1-184 were expressed in yeast to levels comparable to that of wild-type PAB1 (Supplementary Figure 4). (C) The graphical representation of the deadenylation rate analyses from (B).
Analysis of the actual deadenylation rate for GAL1-L showed that a caf1Δ PAB1-134 background caused a severe slowing of the rate of deadenylation (0.7 A's/min compared to 4.0 A's/min for wild-type PAB1 and 2.2 A's/min for caf1Δ) (Figure 8B). This slowing of the rate of deadenylation by caf1Δ PAB1-134 was as severe as that observed in a ccr4 deletion (Figure 8C; 1.1 A's/min). These observations suggest that defects in translation caused by PAB1-134 can cause corresponding defects in deadenylation and that proper translation in conjunction with CAF1 is required for complete deadenylation by CCR4 (see Discussion). Because the PAB1-134 protein had only been assessed for effects on in vitro translation (38), it remains possible that PAB1-134 does not affect in vivo translation. We consequently determined if PAB1-134 was defective for in vivo translation by quantitating the rate of incorporation of [35S]-methionine into newly synthesized protein. PAB1-134 did not affect the rate of in vivo translation as compared to PAB1 wild type (data not shown). However, when we combined PAB1-134 with a defect in eIF3 (prt1-63 allele) that has previously been shown to reduce the rate of in vivo translation (24), we found that PAB1-134 prt1-63 reduced the rate of in vivo translation significantly compared to prt1-63 alone: prt1-63 PAB1-134 (31 ± 5.0%), prt1-63 (45 ± 3.0%), and wild type (100%). In contrast, PAB1-184 did not affect in vivo translation alone or in conjunction with prt1-63 (48 ± 7.5%). These results indicate that the PAB1-134 protein does display defects for in vivo translation.
DISCUSSION
It has been shown that the CAF1 C-terminal domain purified from E. coli exhibits an exonuclease activity with a poly(A) preference (19,23). Our previous mutation studies of CAF1 established, however, that its RNase function is not required for the in vivo deadenylation process (16). This discrepancy prompted us to identify the functional domains that are critical for CAF1 to perform its role in mRNA turnover. By targeting only those sequences absolutely conserved amongst all CAF1 orthologs but not present in other members of the DEDDh family of nucleases, we sought to identify CAF1-specific functions. This methodology led us to define several separable functional regions in the CAF1 protein.
Immunoprecipitation analyses with CAF1 mutants used in this study showed that three amino acids (213FRS215) deleted in caf1-2 are most required for the CAF1 interaction with CCR4. As mammalian CAF1 can also bind yeast CCR4, it would be expected that the CAF1 residues involved in this contact have been retained evolutionarily. Residues 213–215 are found in a loop region whose structure was not fixed enough to be determined in the X-ray analysis of CAF1 (Figure 1A) (19). Other residues of CAF1 might also be involved in contacting CCR4 as the caf1-1, -3, -5 and -6 proteins also displayed weakened binding to CCR4. The caf1-2 protein displayed a weak caffeine-sensitive phenotype and intermediate effects on deadenylation in vivo as compared to a caf1 deletion or to mutations in other regions of the CAF1 protein that were capable of binding CCR4 to a much greater extent. It appears, therefore, that CAF1 binding to CCR4 is not the only function of CAF1 for deadenylation. This conclusion extends the results from our previous analysis wherein mutations in CCR4 that significantly reduced, albeit did not eliminate, CCR4 binding to CAF1 appeared phenotypically wild type (30).
In contrast to caf1-2, the most important regions for CAF1 function in vivo were identified by the caf1-1, -3 and -6 alleles. These three caf1 mutant alleles displayed severe defects in poly(A) shortening like that of a caf1 deletion and were as caffeine sensitive as the caf1 deletion. Three observations suggest that caf1-1 and caf1-3 alleles are equivalent physically and functionally. First, the X-ray crystal structure showed that caf1-1 and caf1-3 alleles are located in the same region where two parallel β strands (β1 and β4) are aligned side by side (Figure 1A) (19). The exact locations of caf1-1 and caf1-3 in these β strands indicate that each triplet of residues do not actually H-bond with each other, but lie contiguous along the β sheet structure (19). Each defect would be expected to partially disrupt this region of the β sheet structure. Second, yeast cells carrying either of the two alleles grew more slowly than did a caf1 deletion. Third, both alleles showed similar GAL1 deadenylation rates and defects in MFA2pG endpoints. These observations indicate that the caf1-1 and caf1-3 alleles define a novel functional region of yeast CAF1 required for its cellular activity. The caf1-6 allele, deleting residues 340DFE342, defines a third functional region of CAF1. Phenotypes associated with caf1-6 are nearly identical to those observed for caf1-1 and -3, although the location of the caf1-6 mutation is on the opposite site of the CAF1 protein as that of caf1-1 and -3. It is unlikely, however, that the putative contacts made by the caf1-6 region (α8) are to the same factors as presumed to contact the β1 and β4 sheets. At least, both regions and their putative contacts are central to CAF1 in vivo function.
In regards to the location of the six mutations in the structure of CAF1, the following observations can be made. First, none of the mutations are either in the presumed catalytic area (α4 and α5 and the loop between the β2 and β3 strands) or in the negatively charged cavity of CAF1 that coincides with the active site found in the structurally and evolutionarily related ε subunit of DNA polymerase III (polIII) or DNA polymerase I (polI) (19). Second, the β1 strand of CAF1 (the site of the caf1-1 alteration) is not found in polIII, although it is present in polI. The sequence features of β4 that were mutated in caf1-3 are alternating non-polar/polar/non-polar residues, a pattern retained by both polIII and polI, albeit with distinctly different residues for the polar residues and different sizes for the non-polar amino acids. Third, the whole protein region corresponding to α8, the turn region after α7, and the loop between β2 and α3, the sites of the caf1-6, -5 and -2 mutations, respectively, is not present in either polIII or polI. These three sites are all close to one another and are located on the exterior of the protein. The evolutionary conservation of this region across the CAF1 protein family argues for unique contacts made by it, including but not limited to binding CCR4.
In that we also observed that none of our CAF1 alleles severely affected in vitro CCR4 deadenylation activity regardless of whether the alleles blocked CCR4 binding to CAF1 (caf1-2) or not, we conclude that CAF1 does not directly regulate CCR4 activity in vitro. This is in agreement with our previous observations (7,30). Also, as mentioned above each of the CAF1 alleles interacted equivalently well with NOT1, further suggesting that the identified roles for CAF1 in vivo are not solely defined through this interaction either. Yet, because binding to NOT1 was reduced by each of the defective CAF1 proteins, it remains possible that disruptions in the CAF1-NOT1 interaction are responsible for at least part of the reductions in deadenylation that were observed. Moreover, in that DHH1 is known to contact the CCR4-NOT complex through the N-terminal part of NOT1 (33), the genetic interactions observed between DHH1 and CAF1 could be mediated in part by NOT1.
Because the caf1-1, -3 and -6 alleles resulted in more significant defects in deadenylation than did the caf1-2 allele, CAF1 appears to function to control the deadenylation process through non-CCR4 contacts in addition to its contact to CCR4. As mentioned above, disrupting the NOT1 contact does not seem to be solely responsible for this phenotype and we showed that all of the CAF1 variants interacted equivalently with DHH1. The genetic analyses demonstrating caf1Δ but not ccr4Δ lethality with the decapping/translation regulator dhh1Δ and with PAB1-ΔRRM2/ΔRRM1 is consistent with CAF1 exerting effects separate from CCR4. However, neither of these lethalities is probably due solely to deadenylation defects that are more severe than those observed in a ccr4Δ background in that the total lack of deadenylation in yeast is not lethal (6; data not shown). As it has been observed that a caf1 deletion does not result in more severe decapping defects than does ccr4Δ (5,6; data not shown), it is also unlikely that CAF1 is exerting these other effects through decapping (6). Rather, it appears that CAF1 in conjunction with DHH1 and with PAB1 is required for some other essential process.
In terms of the role of CAF1 in deadenylation, CAF1 appears to play at least two roles. First, through its contact to CCR4 it aids CCR4 association to the CCR4-NOT complex that is important for CCR4 deadenylation. Second, it may affect some aspect of translation that in turn interferes with deadenylation and/or act on deadenylation in conjunction with factors such as PAB1 and DHH1 that play roles in translation. Several observations support this inference. Deleting those domains of PAB1 that are known to affect in vivo and in vitro translation were lethal with deleting CAF1. In addition to the observed lethalities described above, we found that over-expression of translational regulator DHH1 and ribosome-associated protein STM1 can specifically suppress the caf1Δ PAB1-ΔRRM2 lethality. Other suppressors of caf1Δ not as directly involved in translation, such as CCR4, PKC1 and PUF5, did not display this phenotype. We also showed that over-expression of CAF1 but not that of CCR4 could suppress phenotypes associated with a defect in translation factor eIF4E. In addition, a caf1Δ slowed in vivo protein synthesis, although how it does this is not clear. Taken together, these data point to a role for CAF1 in functionally interacting with translationally important proteins.
A possible role for translation or factors involved in translation in being required for deadenylation was supported by the observation that the PAB1-134 variant slowed the rate of deadenylation when combined with a caf1 deletion. PAB1-134 did not affect deadenylation by itself and had no effect on deadenylaton when combined with a ccr4 deletion. The PAB1-134 protein is known to be defective for in vitro translation (38), and we showed that it reduced in vivo translation when combined with defects in translation initiation factors. Previous studies have indicated that defects in translation initiation factors accelerated the CCR4 deadenylation process (24). In these cases, interference with translation initiation would appear to predispose the mRNP for deadenylation and should not necessarily inhibit it. In contrast, the PAB1-134 caf1Δ effect on deadenylation implies that both PAB1 and CAF1 are required for deadenylation. As PAB1 has been suggested to both inhibit and be required for deadenylation (5,45,46), additional studies are necessary to clarify how other factors of the mRNP, such as PAB1, actually affect CCR4 deadenylation. Their analysis may shed light on the means by which CAF1 affects the mRNA degradation process.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
ACKNOWLEDGEMENTS
We would like to thank R. Parker, D. Mangus and D. Belostotsky for plasmids used in this study. This research was supported by NIH grants GM41215 and GM78078, and by Hatch award H291. This is Scientific Contribution Number 2277 from the New Hampshire Agricultural Experiment Station. Funding to pay the Open Access publication charges for this article was provided by the New Hampshire Agricultural Experiment Station.
Conflict of interest statement. None declared.
REFERENCES
- 1.Parker R., Song H. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- 2.Beelman C.A., Parker R. Degradation of mRNA in eukaryotes. Cell. 1995;81:179–183. doi: 10.1016/0092-8674(95)90326-7. [DOI] [PubMed] [Google Scholar]
- 3.Wilusz C.J., Wormington M., Peltz S.W. The cap-to-tail guide to mRNA turnover. Nat. Rev. Mol. Cell Biol. 2001;2:237–246. doi: 10.1038/35067025. [DOI] [PubMed] [Google Scholar]
- 4.Cao D., Parker R. Computational modeling of eukaryotic mRNA turnover. RNA. 2001;7:1192–1212. doi: 10.1017/s1355838201010330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tucker M., Staples R.R., Valencia-Sanchez M.A., Muhlrad D., Parker R. Ccr4p is the catalytic sub-unit of a Ccr4/Pop2p/Notp mRNA deadenylase complex in Saccharomyces cerevisiae. EMBO J. 2002;21:1427–1436. doi: 10.1093/emboj/21.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tucker M., Valencia-Sanchez M.A., Staples R.R., Chen J., Denis C.L., Parker R. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell. 2001;104:377–406. doi: 10.1016/s0092-8674(01)00225-2. [DOI] [PubMed] [Google Scholar]
- 7.Chen J., Chiang Y.-C., Denis C.L. CCR4, a 3′-5′ poly(A) RNA and ssDNA exonuclease, is the catalytic component of the cytoplasmic deadenylase. EMBO J. 2002;21:1414–1426. doi: 10.1093/emboj/21.6.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu H.-Y., Badarinarayana V., Audino D.C., Rappsilber J., Mann M., Denis C.L. The NOT proteins are part of the CCR4 transcriptional complex and affect gene expression both positively and negatively. EMBO J. 1998;17:1096–1106. doi: 10.1093/emboj/17.4.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bai Y., Salvadore C., Chiang Y.-C., Collart M., Liu H.-Y., Denis C.L. The CCR4 and CAF1 proteins of the CCR4-NOT complex are physically and functionally separate from NOT2, NOT4, and NOT5. Mol. Cell. Biol. 1999;19:6642–6651. doi: 10.1128/mcb.19.10.6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen J., Rappsilber J., Chiang Y.-C., Russell P., Mann M., Denis C.L. Purification and characterization of the 1.0 MDa CCR4-NOT complex identifies two novel components of the complex. J. Mol. Biol. 2001;314:683–694. doi: 10.1006/jmbi.2001.5162. [DOI] [PubMed] [Google Scholar]
- 11.Denis C.L., Chen J. The CCR4-NOT complex plays diverse roles in mRNA metabolism. Nucl. Acids Res. Mol. Biol. 2003;73:221–250. doi: 10.1016/s0079-6603(03)01007-9. [DOI] [PubMed] [Google Scholar]
- 12.Dlakic M. Functionally unrelated signaling proteins contain a fold similar to Mg2+-dependent endonucleases. Trends Biochem. Sci. 2000;25:272–273. doi: 10.1016/s0968-0004(00)01582-6. [DOI] [PubMed] [Google Scholar]
- 13.Dupressoir A., Barbot W., Loireau M.P., Hedmann T. Characterization of a mammalian gene related to the yeast CCR4 general transcription factor and revealed by transposon insertion. J. Biol. Chem. 1999;274:31068–31075. doi: 10.1074/jbc.274.43.31068. [DOI] [PubMed] [Google Scholar]
- 14.Temme C., Zaessinger S., Meyer S., Simonelig M., Wahle E. A complex containing the CCR4 and CAF1 proteins is involved in mRNA deadenylation in Drosophila. EMBO J. 2004;23:2862–2871. doi: 10.1038/sj.emboj.7600273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Draper M.P., Salvadore C., Denis C.L. Identification of a mouse protein whose homolog in Saccharomyces cerevisiae is a component of the CCR4 transcriptional regulatory complex. Mol. Cell. Biol. 1995;15:487–495. doi: 10.1128/mcb.15.7.3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Viswanathan P., Ohn T., Chiang Y.-C., Chen J., Denis C.L. Mouse CAF1 can function as a processive deadenylase/3′-5′-exonuclease in vitro but in yeast the deadenylase function of CAF1 is not required for mRNA poly(A) removal. J. Biol. Chem. 2004;279:23988–23995. doi: 10.1074/jbc.M402803200. [DOI] [PubMed] [Google Scholar]
- 17.Moser M.J., Holley W.R., Chatterjee A., Mian I.S. The proofreading domain of Escherichia coli DNA polymerse I and other DNA and/or RNA exonuclease domain. Nucleic Acids Res. 1997;25:5110–5118. doi: 10.1093/nar/25.24.5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zuo Y., Deutscher M.P. Exoribonuclease superfamilies: structural analysis and phylogenetic distribution. Nucleic Acids Res. 2001;29:1017–1026. doi: 10.1093/nar/29.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thore S., Mauxion F., Séraphin B., Suck D. X-ray structure and activity of the yeast Pop2 protein: a nuclease subunit of the mRNA deadenylase complex. EMBO Rep. 2003;4:1150–1155. doi: 10.1038/sj.embor.7400020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Daugeron M.-C., Mauxion F., Séraphin B. The yeast POP2 gene encodes a nuclease involved in mRNA deadenylation. Nucleic Acids Res. 2001;29:2448–2455. doi: 10.1093/nar/29.12.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldstrohm A.C., Hook B.A., Seay D.J., Wickens M. PUF proteins bind Pop2p to regulate messenger RNAs. Nat. Struct. Mol. Biol. 2006;13:533–539. doi: 10.1038/nsmb1100. [DOI] [PubMed] [Google Scholar]
- 22.Cook W.J., Chase D., Audino D.C., Denis C.L. Dissection of the ADR1 protein reveals multiple, functionally redundant activation domains interspersed with inhibitory regions: evidence for a repressor binding to the ADR1c region. Mol. Cell. Biol. 1994;14:629–640. doi: 10.1128/mcb.14.1.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Viswanathan P., Chen J., Chiang Y.-C., Denis C.L. Identification of multiple RNA features that influence CCR4 deadenylation activity. J. Biol. Chem. 2003;278:14949–14955. doi: 10.1074/jbc.M211794200. [DOI] [PubMed] [Google Scholar]
- 24.Schwartz D.C., Parker R. Mutations in translation initiation factors lead to increased rates of deadenylation and decapping of mRNAs in Saccharomyces cereviseae. Mol. Cell. Biol. 1999;19:5247–5256. doi: 10.1128/mcb.19.8.5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cook W.J., Denis C.L. Identification of three genes required for the glucose-dependent transcription of the yeast transcriptional activator ADR1. Curr. Genet. 1993;23:192–200. doi: 10.1007/BF00351495. [DOI] [PubMed] [Google Scholar]
- 26.Muhlrad D., Parker R. Mutations affecting stability and deadenylation of the yeast MFA2 transcript. Genes Dev. 1992;6:2100–2111. doi: 10.1101/gad.6.11.2100. [DOI] [PubMed] [Google Scholar]
- 27.Moriya H., Shimizu-Yoshida Y., Omori A., Iwashita S., Katoh M., Sakai A. Yak1p, a DYRK family kinase, translocates to the nucleus and phosphorylates yeast Pop2p in response to a glucose signal. Genes Dev. 2001;15:1217–1228. doi: 10.1101/gad.884001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sakai A., Chibazakura T., Shimizu Y., Hishinuma F. Molecular analysis of POP2 gene, a gene required for glucose-derepression of gene expression in Saccharomyces cerevisiae. Nucleic Acids Res. 1992;20:6227–6233. doi: 10.1093/nar/20.23.6227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hata H., Mitsui H., Liu H.-Y., Bai Y., Denis C.L., Shimizu Y., Sakai A. Dhh1p, a putative RNA helicase, associates with the general transcription factors Pop2p and Ccr4p from Saccharomyces cerevisiae. Genetics. 1998;148:571–579. doi: 10.1093/genetics/148.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clark L.C., Viswanathan P., Quigley G., Chiang Y.-C., McMahon J.S., Yao G., Chen J., Nelsbach A., Denis C.L. Systematic mutagenesis of the leucin-rich repeat (LRR) domain of CCR4 reveals specific sites for binding to CAF1 and a separate critical role for the LRR in CCR4 deadenylase activity. J. Biol. Chem. 2004;279:13616–13623. doi: 10.1074/jbc.M313202200. [DOI] [PubMed] [Google Scholar]
- 31.Cui Y., Denis C.L. In vivo evidence that defects in the transcriptional elongation factors RPB2, TFIIS, and SPT5 enhance upstream poly(A) site utilization. Mol. Cell. Biol. 2003;23:7887–7910. doi: 10.1128/MCB.23.21.7887-7901.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miyajima A., Nakayama N., Miyajima I., Arai N., Okayama H., Arai K. Analysis of full-length cDNA clones carrying GAL1 of Saccharomyces cerevisiae: a model system for cDNA expression. Nucleic Acids Res. 1984;12:6397–6414. doi: 10.1093/nar/12.16.6397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maillet L., Collart M.A. Interaction between Not1p, a component of the Ccr4 complex, a global regulator of transcription, and Dhh1p, a putative RNA helicase. J. Biol. Chem. 2002;277:2835–2842. doi: 10.1074/jbc.M107979200. [DOI] [PubMed] [Google Scholar]
- 34.Coller J.M., Tucker M., Sheth U., Valencia-Sanchez M.A., Parker R. The DEAD box helicase, Dhh1p, functions in mRNA decapping and interacts with both the decapping and deadenylase complexes. RNA. 2001;7:1717–1727. doi: 10.1017/s135583820101994x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Coller J.M., Parker R. General translational repression by activators of mRNA decapping. Cell. 2005;122:875–886. doi: 10.1016/j.cell.2005.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deardorff J.A., Sachs A.B. Differential effects of aromatic and charged residue substitutions in the RNA binding domains of the yeast poly(A)-binding protein. J. Mol. Biol. 1997;269:67–81. doi: 10.1006/jmbi.1997.1013. [DOI] [PubMed] [Google Scholar]
- 37.Kessler S.H., Sachs A.B. RNA recognition motif 2 of yeast Pab1p is required for its functional interaction with eukaryotic translation initiation factor 4G. Mol. Cell. Biol. 1998;18:51–57. doi: 10.1128/mcb.18.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Otero L.J., Ashe M.P., Sachs A.B. The yeast poly(A)-binding protein Pab1p stimulates in vitro poly(A)-dependent and cap-dependent translation by distinct mechanisms. EMBO J. 1999;18:3153–3163. doi: 10.1093/emboj/18.11.3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Dyke N., Baby J., Van Dyke M.W. Stm1p, a ribosome-associated protein, is important for protein synthesis in Saccharomyces cerevisiae under nutritional stress conditions. J. Mol. Biol. 2006;358:1023–1031. doi: 10.1016/j.jmb.2006.03.018. [DOI] [PubMed] [Google Scholar]
- 40.Van Dyke M.W., Nelson L.D., Weilbaecher R.G., Mehta D.V. Stm1p, a G4 quadruplex and purine motif triplex nucleic acid-binding protein, interacts with ribosomes and subtelomeric Y’ DNA in Saccharomyces cerevisiae. J. Biol. Chem. 2004;279:24323–24333. doi: 10.1074/jbc.M401981200. [DOI] [PubMed] [Google Scholar]
- 41.Gavin A.C., Bosche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J.M., Michon A.-M., et al. Functional organization of the yeast proteome by systemic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 42.Inada T., Winstall E., Tarun S.Z., Jr, Yates J.R., III, Schieltz D., Sachs A.B. One-step affinity purification of the yeast ribosome and its associated proteins and mRNAs. RNA. 2002;8:948–956. doi: 10.1017/s1355838202026018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takaku H., Mutoh E., Horiuchi H., Ohta A., Takagi M. Ray38p, a homolog of a purine motif triple-helical DNA-binding protein, Stm1p, is a ribosome-associated protein and dissociated from ribosomes prior to the induction of cycloheximide resistance in Candida maltosa. Biochem. Biophys. Res. Commun. 2001;284:194–202. doi: 10.1006/bbrc.2001.4951. [DOI] [PubMed] [Google Scholar]
- 44.Altmann M., Trachsel H. Altered mRNA cap recognition activity of initiation factor 4E in the yeast cell cycle division mutant cdc33. Nucleic Acids Res. 1989;17:5923–5931. doi: 10.1093/nar/17.15.5923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Caponigro G., Parker R. Multiple functions for the poly(A)-binding protein in mRNA decapping and deadenylation in yeast. Genes Dev. 1995;9:2421–2432. doi: 10.1101/gad.9.19.2421. [DOI] [PubMed] [Google Scholar]
- 46.Morrissey J.P., Deardorff J.A., Hebron C., Sachs A.B. Decapping of stabilized, polyadenylated mRNA in yeast pab1 mutants. Yeast. 1999;15:687–702. doi: 10.1002/(SICI)1097-0061(19990615)15:8<687::AID-YEA412>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.