Abstract
To better understand the functions and fidelity of DNA polymerase ε (Pol ε), we report here on the fidelity of yeast Pol ε mutants with leucine, tryptophan or phenylalanine replacing Met644. The Met644 side chain interacts with an invariant tyrosine that contacts the sugar of the incoming dNTP. M644W and M644L Pol ε synthesize DNA with high fidelity, but M644F Pol ε has reduced fidelity resulting from strongly increased misinsertion rates. When Msh6-dependent repair of replication errors is defective, the mutation rate of a pol2-M644F strain is 16-fold higher than that of a strain with wild-type Pol ε. In conjunction with earlier studies of low-fidelity mutants with replacements for the homologous amino acid in yeast Pol α (L868M/F) and Pol δ (L612M), these data indicate that the active site location occupied by Met644 in Pol ε is a key determinant of replication fidelity by all three B family replicative polymerases. Interestingly, error specificity of M644F Pol ε is distinct from that of L868M/F Pol α or L612M Pol δ, implying that each polymerase has different active site geometry, and suggesting that these polymerase alleles may generate distinctive mutational signatures for probing functions in vivo.
INTRODUCTION
DNA polymerase ε (Pol ε) is a central player in many DNA transactions that determine the stability of the nuclear genome. Current evidence indicates that Pol ε participates in replicating and recombining the nuclear genome, in modulating cellular responses to DNA damage, and in excision repair of damaged and mismatched bases [reviewed in (1–5)]. Despite these extensive responsibilities, Pol ε's precise roles in these processes in vivo remain somewhat uncertain. This uncertainty partly derives from the fact that Pol ε is but one of many eukaryotic polymerases whose functions may overlap. For example, Pol ε is one of four eukaryotic members of the B family, sharing homology and certain biochemical properties with DNA polymerases α, δ and ζ. The latter polymerases, like Pol ε, are also suggested to participate to varying degrees in replication, recombination, excision repair and/or DNA damage responses. Pol ε is also a large and complex DNA polymerase, comprising four subunits (6,7). At 256 kDa, the Pol ε catalytic subunit encoded by the POL2 gene is the largest of eight DNA polymerases in Saccharomyces cerevisiae (5). The non-catalytic C-terminal region of this large subunit contains residues that modulate cell cycle checkpoint responses to DNA damage, and deletion of this C-terminal region is lethal (8). It is the N-terminal region of the large Pol ε subunit that contains the DNA polymerase activity, as well as a 3′ exonuclease activity that can proofread DNA synthesis errors (9). Deletion of the N-terminal region of the large subunit of the S. cerevisiae protein causes a very severe growth defect (10,11), while mutation of the catalytic aspartic acids to alanines causes lethality, implying that Pol ε catalytic activity is critical for efficient nuclear DNA replication (12).
Several ideas have been put forth regarding Pol ε's biosynthetic role in replication, including replication of the leading strand (3), replication of the lagging strand (4) and a particularly important role in replication of heterochromatic DNA late in S phase (13).
To probe these ideas, we have been searching for polymerases that retain normal polymerization activity yet have reduced DNA synthesis fidelity, such that they result in a mutator phenotype in yeast cells. Our attention is focused on polymerases with replacements for amino acids in highly conserved sequence motif A, which along with motifs B and C form the active site of multiple polymerases. This includes B family enzymes like Pol ε, Pol α, Pol δ, Pol ζ, and their viral homologs, bacteriophage T4 and RB69 Pols. In the crystal structure of RB69 Pol (14), an invariant tyrosine in motif A interacts with the sugar of the incoming dNTP in the polymerase active site. Immediately adjacent to this tyrosine is a hydrophobic amino acid, usually a leucine. In T4 DNA polymerase, substituting Leu412 with methionine yielded bacteriophage that replicated efficiently but had an elevated mutation rate (15). Subsequent studies indicated that this mutator effect results from inefficient proofreading due to defective movement of mismatches generated by the polymerase into the exonuclease active site (16,17). In yeast Pol α, studies of L868F and L868M mutants (18,19) reveal enzymes with normal polymerase-specific activity, enhanced mismatch extension efficiency (L868M) and reduced DNA synthesis fidelity in vitro. Yeast strains harboring the L868F and L868M Pol α mutant alleles have elevated spontaneous mutation rates that are strongly enhanced when mismatch repair is inactivated, indicating that the mutator effect is due to reduced replication fidelity in vivo. The mutator effect of the Pol α L868M mutant allele was also elevated by inactivating the 3′ exonuclease activity of Pol δ (18), suggesting that the 3′ exonuclease activity of Pol δ may excise errors made by Pol α, a type of extrinsic proofreading (20). Other studies (21–23) have shown that replacing Leu612 in yeast Pol δ with other amino acids also yields strains with mutator phenotypes that are increased when mismatch repair is inactivated. This suggests reduced replication fidelity in vivo, which is supported by the observation that purified yeast L612M Pol δ has reduced DNA synthesis fidelity, despite retaining normal 3′ exonuclease activity (22). The properties of L612M Pol δ led us to suggest that, like L412M T4 Pol (17), L612M Pol δ sometimes fails to partition mismatched termini to the 3′ exonuclease active site. Analogous to the promiscuous mismatch extension ability of L868M Pol α (18), we suggested that the partitioning defect of L612M Pol δ could be due to promiscuous mismatch extension.
Sequence alignments reveal that a methionine (Met644 in yeast) is present in Pol ε at the position corresponding to the leucine in T4 Pol (Leu412), yeast Pol α (Leu868) and yeast Pol δ (Leu612). Based on those earlier studies, we purified Pol ε mutants containing M644L, M644W and M644F changes and present here an analysis of their fidelity, with three goals in mind. First, we wished to determine if Met644 in Pol ε, like its homologs in Pol α and Pol δ, has an important role in determining the fidelity of DNA synthesis. This is of interest because these three polymerases are not equally accurate. For example, Pol α is naturally exonuclease deficient and is therefore less accurate than proofreading-proficient Pol δ and Pol ε [see (24) and references therein]; Pol δ generates single-base deletions at higher rates than does Pol ε (24,25); and others have reported that Pol δ generates base substitutions at a 20-fold higher rate than does Pol ε (26). Anticipating that a change at Met644 would reduce fidelity, a second motive was to determine if reduced fidelity reflected lower selectivity, inefficient proofreading or both. In our earlier study of L612M Pol δ, we could not assess these parameters quantitatively because we were unable to obtain the vector needed to express and purify exonuclease-deficient L612M Pol δ, perhaps due to error catastrophe in yeast cells (22). In the present study, we have been able to express and purify the yeast M644F Pol ε mutant in both exonuclease-proficient and exonuclease-deficient forms. This permits a comparison of the fidelity of these two mutant enzymes in a forward mutation assay to ascertain the extent by which Met644 modulates single-base deletion and insertion fidelity, the extent of reduction in nucleotide selectivity for each of the 12 possible base–base mismatches, and the contribution of proofreading to these error rates. A third goal is to identify a Pol ε mutant that retains robust polymerization activity yet is sufficiently inaccurate to generate a mutator phenotype that can be used to probe Pol ε functions in yeast. Each of these goals addresses the mechanisms for accurate functions of Pol ε, perhaps the most complex, yet least studied of the three major replicative polymerases in eukaryotes.
MATERIALS AND METHODS
Materials
Materials for the fidelity assay were from previously described sources (27).
Construction of vectors to express 152-kDa Pol ε
A plasmid for expressing the polymerase domain of Pol2 was created by digestion of the pJL1 plasmid with the restriction endonuclease NdeI. Next, a linker containing a His-tag followed by a stop codon was ligated to the linearized fragment creating plasmid pJL152. To obtain an exonuclease-deficient variant of the proteolytic fragment, pJL1-4 (24,28) and pJL152 was digested with BglII. A 2.9-kb DNA fragment containing the mutated exonuclease motif replaced the corresponding DNA fragment in the pJL152 plasmid, resulting in the plasmid pJL152-4. The M644W, M644L and M644F active site mutations were introduced by site-directed mutagenesis of the two plasmids pJL152 and pJL152-4. The entire coding regions of all constructs were verified to be correct by sequence analysis.
Expression and purification of 152-kDa Pol ε
All purification steps were carried out at 4°C. The following buffers were used. Buffer A: 150 mM Tris-acetate pH 7.8, 50 mM sodium acetate, 2 mM EDTA, 1 mM EGTA, 10 mM NaHSO3, 1 mM dithiothreitol, 5 µM pepstatin A, 5 µM leupeptin, 0.3 mM p-phenylmethylsulfonyl fluoride and 5 mM benzamidine. Buffer B: 25 mM HEPES–NaOH pH 7.6, 10% glycerol, 1 mM EDTA, 0.5 mM EGTA, 0.01% Nonidet P-40, 3 mM dithiothreitol, 2 µM pepstatin A, 2 µM leupeptin and 5 mM NaHSO3. The concentration of sodium acetate is indicated as suffix, for example buffer B100 = buffer B with 100 mM sodium acetate. All variants of the 152-kDa fragment of Pol2 were over-expressed and purified by conventional chromatography essentially as previously described (28). The following modifications were made during the purification process. The cells were lysed using a freezer mill 6850 (Spex Certiprep) running the following program: pre-cooling time: 5 min, 7 cycles (grinding time: 2 min, re-cooling time: 2 min), impact frequency rate 12. The cell lysate was thawed on ice in Buffer A, followed by an ammonium sulfate cut, dialysis and loading of the proteins on a phosphocellulose column (P11) as previously described (28,29). After washing the column with B300, the bound proteins were eluted with B700. The eluted proteins were diluted with B0 to a conductivity corresponding to B300, followed by centrifugation using a Beckman JA25.5 rotor at 17 000 r.p.m. for 60 min. The cleared protein solution was loaded on a 1 ml MonoQ column equilibrated in B300, washed with B300 and the bound protein was eluted with a 20 ml linear gradient from B300 to B700. Next the peak fractions were pooled, diluted with B0 to a conductivity corresponding to B250, followed by centrifugation using a Beckman JA25.5 rotor at 17 000 r.p.m. for 60 min. The cleared protein solution was loaded on a 1 ml MonoS column equilibrated in B250, washed with B250 and the bound protein was eluted with a 20 ml linear gradient from B250 to B1000. Finally, the peak fractions were concentrated in a spin column, followed by gel filtration over a Superose12 column equilibrated in B400. Note that the four-subunit Pol ε elutes from the MonoQ column at B1000, whereas the 152-kDa fragment elutes from the column at B550, such that this column separates any endogenous wild-type four-subunit DNA Pol ε from the purified 152-kDa fragment.
Measuring polymerase specific activity
Polymerase-specific activity was measured using two different DNA templates, either a oligo(dT) primed poly(dA) template or activated salmon sperm DNA (29). A typical reaction contained 20 mM Tris-HCl pH 7.8, 4% glycerol, 0.1 mg/ml bovine serum albumin, 5 mM dithiothreitol, 8 mM MgAc2, 80 μM dATP, dCTP and dGTP, 25 μM [3H]dTTP (371 c.p.m./pmol), 1 mM spermidine, 0.01 unit poly(dA)·oligo(dT) or 50 µg of activated salmon sperm DNA, and DNA polymerase, either 0.27 nmol or 1 nmol in a reaction volume of 50 µl. Reactions were assembled on ice and incubated for 1–16 min at 30°C. The reactions were stopped by the addition of 150 µl of stop solution (50 mM sodium pyrophosphate, 25 mM EDTA and 50 μg/ml salmon sperm DNA). Insoluble material was precipitated by adding 1.5 ml of 10% trichloroacetic acid and incubating on ice for 30 min. The insoluble material was filtered on GF/C filters, washed three times with 2 ml 1 M HCl in 0.05 M sodium pyrophosphate, rinsed with ethanol, dried and counted by liquid scintillation. One unit of the enzyme activity incorporates 1 nmol of total nucleotide/hr (30).
Measuring exonuclease specific activity
Single-stranded exonuclease activity was measured in a reaction mixture (20 µl) containing 50 mM Tris (pH 7.4), 100 μg/ml BSA, 2 mM DTT, 8 mM MgCl2, 10% glycerol and 50 nM 32P-labeled single-stranded oligonucleotide (5′-CATCACAGTGAGTAC-3′). Reactions were initiated by adding 2.5 nM wild-type or M644F Pol ε. Reaction mixtures were incubated at 30°C and time points were taken at 1, 2, 5 and 10 min, with reactions stopped by adding an equal volume of formamide loading dye and were analyzed by electrophoresis on a denaturing 16% polyacrylamide gel. Products were detected and quantified using a PhosphorImager and ImageQuaNT software (Molecular Dynamics).
Gap-filling DNA synthesis reactions and product analysis for determining fidelity
DNA polymerase fidelity was measured using the lacZ forward mutation assay as described (27). Briefly, Pol ε was used to copy a single-stranded region of the M13 lacZα-complementation gene. Gap-filling reaction mixtures (25 µl) contained 50 mM Tris (pH 7.4), 2 mM DTT, 8 mM MgCl2, 100 µg/ml BSA, 10% glycerol, 250 μM dNTPs, 1.5 nM of gapped M13mp2 DNA and 15 nM Pol ε. Gap-filling reactions were incubated at 30°C for 30 min and complete gap filling was monitored by agarose gel electrophoresis. Products containing completely filled gaps were introduced by electroporation into the host strain, and replication errors were scored by plating as described (27). M13 DNA from independent mutant M13 plaques was isolated and sequenced to determine the types of polymerization errors. Error rates were calculated as described (27). Briefly, the error rate = (number of mutations of a particular type ÷ total number of mutations) × (total mutant frequency ÷ probability of expressing the (−) strand ÷ number of sites where the particular mutation is detectable). The statistical significance of differences was calculated using Fisher's Exact Test (31).
Yeast strains
Strains used were isogenic derivatives of yeast strain Δ|(-2)|-7B-YUNI300 (MATa his7-2 leu2Δ::kanMX ura3Δ lys2-ΔGG2899-2900 trp1-289 ade2-1 CAN1) described in (32). DNA polymerase ε mutations were introduced via integration-excision method using a plasmid, p173, encoding a portion of POL2 (33) with a targeted change. Mutations in POL2 were introduced via site-directed mutagenesis using the QuickChange Mutagenesis kit from Stratagene (La Jolla, CA). Primers used were 5′-GAT GTC GCC TCT TTT TAC CCA AAC ATC-3′ and 3′-GAT GTT TGG GTA AAA AGA GGC GAC ATC-5′ for pol2-M644F. MSH6 was disrupted by PCR-based targeted gene disruption. pRS304 was used as a template to generate PCR fragments containing the TRP1 gene flanked by sequence homologous to MSH6 (32). After transformation, disruption of MSH6 was verified by growth in the absence of tryptophan and by PCR across the disrupted region of genomic DNA.
Mutation rate determination
Spontaneous forward mutation rates were determined as described (34). Briefly, each rate was determined using at least 12 independent cultures. Single colonies were grown on YPDA for 2 days at 30°C and used to inoculate 10 ml YPDA. Cultures were grown to stationary phase at 30°C and then appropriate dilutions were plated out onto complete medium or complete medium lacking arginine and supplemented with 60 μg/ml canavinine. Mutation rates were calculated as described (35).
RESULTS
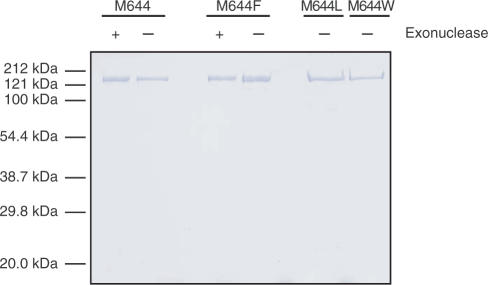
The large size of the catalytic subunit and the multi-subunit nature of Pol ε make expression and purification of the four-subunit holoenzyme labor intensive. To reduce the effort needed to characterize the biochemical properties of multiple Pol ε derivatives, we began this study by constructing a yeast expression vector for a catalytically active 152-kDa fragment of POL2 corresponding to amino acids 1–1351. This fragment approximately corresponds to the 140 kDa Pol ε fragment initially purified by Hamatake and co-workers (30), and it contains all the domains needed for DNA polymerase and 3′ to 5′ exonuclease activity. The 152-kDa fragment expressed well (greater than 100-fold over-expression) in yeast and was obtained in highly purified form (Figure 1). The wild-type 152-kDa protein is highly active on synthetic and natural DNA templates, with a specific polymerization activity using a poly (dA)·oligo (dT) substrate of 18 000 U/mg, a value only slightly lower than that of Pol ε holoenzyme (7,28) and similar to that of wild-type 140-kDa Pol ε purified from yeast (30). The mutant Pol ε variants were created as described (see Materials and methods section) and then expressed and purified (Figure 1) in the same fashion as the wild-type enzyme.
Figure 1.
Purification of Pol ε-N152 Met644 derivatives. Here, ∼1 μg of the indicated purified proteins was loaded onto a 10 % SDS-polyacrylamide gel, which was subsequently stained with Coomassie. The exonuclease proficiency of each polymerase is indicated by + or −. M644 denotes the wild-type, Met644, polymerase. Molecular weight standards are indicated.
Fidelity of Pol ε 152-kDa fragment
Before characterizing the fidelity of Pol ε mutants, we determined how accurately the 152-kDa fragment of wild-type (i.e. Met644) Pol ε copies the lacZα-complementation sequence in a gapped circular M13mp2 DNA substrate. Both the exonuclease-proficient and exonuclease-deficient forms of Pol ε 152-kDa fragment filled the 407-nt gap used for the fidelity assay to apparent completion (data not shown, but see Materials and methods section). The DNA products of these reactions were introduced into a lacZα-complementation E. coli strain and the cells were plated to score DNA synthesis errors as light blue and colorless M13 plaques among greater numbers of dark blue plaques resulting from correct synthesis. The lacZ mutant frequencies generated by exonuclease-deficient Pol ε 152-kDa fragment [simply designated M (for Met644) in Table 1, Experiment 1] and exonuclease-proficient Pol ε 152-kDa fragment (Table 1, Experiment 2) were similar to those previously reported (24) for the corresponding holoenzyme forms of Pol ε that contain an intact 256 kDa catalytic subunit plus three accessory subunits (Table 1). Sequencing of the lacZα-complementation gene in DNA isolated from independent lacZ mutant plaques allowed calculation (see Materials and methods section) of the average rates at which the 152-kDa fragment enzymes generated single-base substitution, deletion and addition errors (black bars in Figure 2). These rates were not significantly different from those previously observed for the holoenzyme (24). While we cannot exclude possible differences of a few-fold in error rates for specific mismatches in specific sequence contexts, the data in Table 1 and Figure 2 validate using the Pol ε 152-kDa fragment as a surrogate for the holoenzyme in fidelity studies in vitro.
Table 1.
lacZ mutant frequency for products of gap filling by derivatives of DNA polymerase ε.
| Pol ε derivative | Plaques | Mutant frequency (×10−4) | |
|---|---|---|---|
| Total | Mutant | ||
| Experiment 1: Exonuclease-deficient Pol ε | |||
| Holoenzyme | – | – | 260 |
| M | 5160 | 84 | 160 |
| W | 2553 | 22 | 98 |
| L | 4466 | 64 | 140 |
| F | 2079 | 232 | 1100 |
| Experiment 2: Exonuclease-proficient Pol ε | |||
| Holoenzyme | – | – | 19 |
| M | 4528 | 7 | 16 |
| F | 16110 | 69 | 43 |
lacZ mutant frequencies were calculated as described in the Materials and methods section. Holoenzyme mutant frequencies were taken from (24). For each Pol ε derivative whose error specificity was determined, the following number of mutants were sequenced: Experiment 1: M—84; F—77; Experiment 2: M—7; F—69. The single letter refers to the amino acid at position 644 in each derivative tested. Mutant frequency measurements were obtained with a second, independent preparation of M13 gapped DNA substrate for those derivatives for which lacZ mutants were sequenced and found to be reproducible.
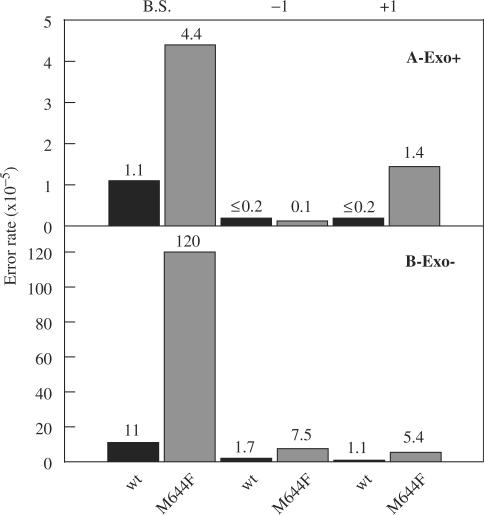
Figure 2.
Base substitution, −1, and +1 frameshift error rates for wild-type and M644F Pol ε. Error rates for base substitution (B.S.), single-base deletions (−1) and single-base additions (+1) for exonuclease-proficient (left) and exonuclease-deficient Pol ε were calculated as described (27). Error rates are shown for both wild-type Pol ε (black bars) and M644F Pol ε (gray bars).
Fidelity of M644W, M644L and M644F Pol ε mutants
Similar fidelity analyses were then performed with exonuclease-deficient and exonuclease-proficient forms of Pol ε 152-kDa fragment containing Trp, Leu and Phe replacements for Met644. All enzymes filled the gap in the lacZ substrate (data not shown). The DNA products of reactions generated by exonuclease-deficient M644W and M644L mutants yielded lacZ mutant frequencies similar to the frequency for synthesis by the exonuclease-deficient ‘wild-type’ (i.e. M) Pol ε fragment (Table 1, Experiment 1). This suggests that Pol ε fidelity is largely insensitive to the presence of methionine, tryptophan or leucine at this location in the polymerase active site. (Again, we cannot exclude possible differences of a few-fold in error rates for specific mismatches in specific sequence contexts. Further, we cannot exclude the possibility that, while the overall mutant frequencies are similar to the wild-type enzyme, the error specificities of M644W and M644L may differ from wild type.) The observation that all three derivatives have similar fidelity differs from observations with yeast Pol α (18–20) and Pol δ (21–23) derivatives (see Discussion). Since the M644W and M644L polymerases were not obviously less accurate than wild-type Pol ε, these mutant enzymes were not investigated further.
The exonuclease-deficient M644F derivative was 6.9-fold less accurate than wild type (Table 1, Experiment 1, compare 1100 × 10−4 to 160 × 10−4), clearly indicating that M644F Pol ε has reduced fidelity. Moreover, the lacZ mutant frequency for the products of gap filling by the exonuclease-proficient M644F derivative was 2.7-fold higher than for the exonuclease-proficient wild-type fragment (Table 1, Experiment 2, compare 43 × 10−4 to 16 × 10−4), suggesting that M644F Pol ε has reduced fidelity even when the protein retains an intact active site for 3′ exonuclease activity. Also of note, the mutant frequency of exonuclease-proficient M644F is 26-fold lower than that of exonuclease-deficient M644F (Table 1, compare 43 × 10−4 to 1100 × 10−4), indicating that exonuclease-proficient M644F Pol ε retains the ability to proofread most errors.
Specific polymerase and exonuclease activities of the M644F Pol ε mutants
Before performing more detailed error specificity analysis or measurements of mutation rates in vivo, we next compared the polymerase and 3′ exonuclease activities of the wild-type and M644F Pol ε fragments. The specific activities of the exonuclease-deficient and exonuclease-proficient M644F mutant enzymes were 98 and 27%, respectively, of the wild-type Pol ε fragment, using poly dA·oligo dT, a preferred substrate for DNA synthesis by Pol ε (30). Thus, replacing Met644 with phenylalanine does not reduce the activity of the exonuclease-deficient polymerase, and only reduces the apparent polymerase activity of the exonuclease-proficient protein by about 3-fold. Using a single-stranded 18-mer oligonucleotide as a substrate for the 3′ exonuclease of Pol ε, no digestion was seen with either exonuclease-deficient Pol ε fragments, while the exonuclease-proficient wild-type and M644F mutant both readily degraded the DNA. The specific exonuclease activity of the M644F mutant was similar (110%) to that of the wild-type Pol ε fragment. Thus, replacing Met644 in the polymerase active site with phenylalanine does not reduce 3′ exonuclease activity.
Error specificity of the M644F Pol ε mutants
To determine the types and locations of errors made by M644F mutant Pol ε, and to calculate error rates, we sequenced the lacZα-complementation gene in DNA isolated from independent mutant plaques (see legend of Table 1). While a few lacZ mutants contained errors involving more than a single base change, the number of such events was too small to be informative. Most errors involved deleting, adding or substituting a single base pair (Figure 3). From the proportion of each type of error and the mutant frequencies (Table 1), the rates at which exonuclease-deficient and exonuclease-proficient wild-type and M644F mutant Pol ε generated each type of error were calculated (Figure 2).
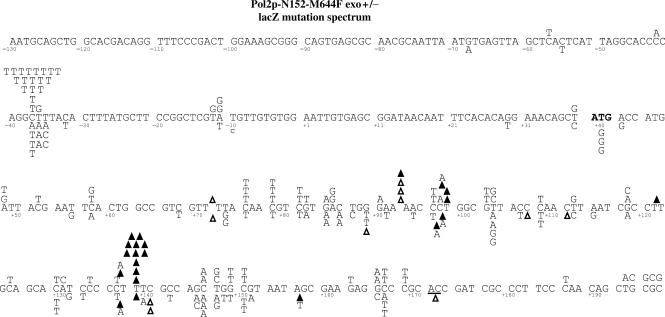
Figure 3.
Spectrum of single-base substitution and frameshift mutations made by M644F Pol ε in the lacZ gene. Errors made by the exonuclease-proficient M644F Pol ε are shown above the sequence, while errors made by the exonuclease-deficient M644F Pol ε are shown below the sequence. Deletions are denoted by an open triangle and insertions by a closed triangle. The lacZ numbering is in reference to the transcriptional start site (+1).
Single-base deletions
At 7.5 × 10−5, the average single-base deletion error rate of exonuclease-deficient M644F Pol ε (gray bars in Figure 2B) is 4.4-fold higher than that for exonuclease-deficient, wild-type Pol ε, (1.7 × 10−5, black bars in Figure 2B, P = 0.007). Thus, substituting phenylalanine for Met644 reduces discrimination against single-base deletion intermediates. Among seven deletions, three were within repetitive sequences and four were not (Figure 3, open triangles below each line of primary sequence). The overall average single-base deletion error rates of the exonuclease-proficient derivatives of M644F and wild-type Pol ε (Figure 2A) are both lower (0.1 × 10−5 and ≤0.2 × 10−5, and P < 0.001 and P = 0.002, respectively), indicating that both forms of Pol ε proofread the intermediates responsible for single-base deletions.
Single-base additions
The average single-base addition error rate of exonuclease-deficient M644F Pol ε is 5.4 × 10−5 (Figure 2B), which is 4.9-fold higher than that for exonuclease-deficient, wild-type Pol ε, (1.1 × 10−5, P = 0.01). Thus, substituting phenylalanine for Met644 also reduces discrimination against single-base addition intermediates. Four of the five additions (closed triangles in Figure 3) were of bases that were not identical to either the 3′ or 5′ neighbor, suggesting a mechanism other than classical strand slippage in repetitive sequences (36). Just as seen for deletions, the average single-base addition error rates of the exonuclease-proficient derivatives of M644F and wild-type Pol ε are both lower (1.4 × 10−5 and ≤0.2 × 10−5, and P = 0.02 and 0.07, respectively, Figure 2A), indicating that both forms of Pol ε proofread the intermediates responsible for the single-base additions.
Single-base substitutions
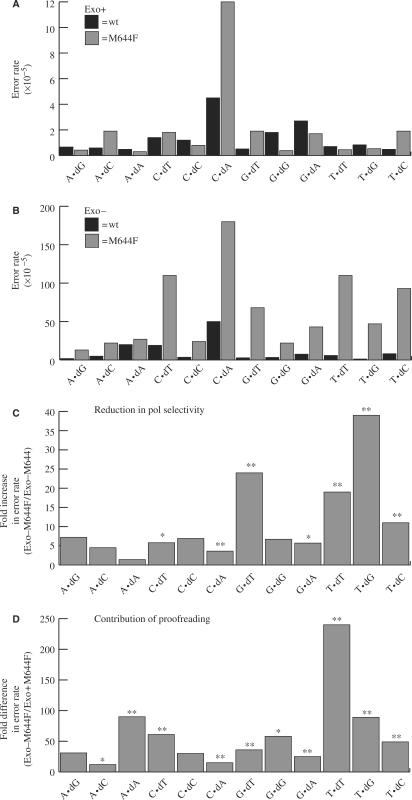
For the exonuclease-deficient enzymes, the majority of errors made by wild-type and M644F Pol ε were single-base substitutions (Figure 3). At 120 × 10−5, the average base substitution error rate of M644F Pol ε is 11-fold higher than that of wild-type Pol ε (Figure 2B, P < 0.001). Thus, substituting phenylalanine for Met644 strongly reduces the ability to prevent stable misincorporation of dNTPs. Error rates for most of the 12 mismatches are elevated (Figure 4B, C), i.e. replacing Met644 with phenylalanine reduces discrimination for errors involving each of the four incoming dNTPs (e.g. T-dGMP, G-dAMP, T-dCMP and G-dTMP), and it reduces discrimination against errors involving all four possible template bases. Nonetheless, some specificity is evident because discrimination is reduced more for some mismatches than for others (Figure 4C). For example, M644F Pol ε is less accurate than wild-type Pol ε by 24-fold for the G·dTMP mismatch and by 19-fold for the T·dTMP mismatch, while the two enzymes differ in error rate for several other mismatches (e.g. A·dAMP, A·dGMP) by 2-fold or less. The determinants of this specificity depend on the particular polymerase active site context in which the phenylalanine is located, because the error specificity of exonuclease-deficient M644F Pol ε is clearly different from that of Pol α containing the same amino acid at the homologous position, i.e. L868F Pol α (see Discussion).
Figure 4.
Effects on Pol ε fidelity of individual base substitutions due to M644F mutation. (A) Error rates for each of the 12 possible mispairs for the exonuclease-proficient holoenzyme (black bars; values taken from (24)) and M644F N152 Pol ε (gray bars, this study). (B) Error rates for each of the 12 possible mispairs for the exonuclease-deficient N152 Pol ε (black bars) and M644F Pol ε (gray bars). (C) Reduction in selectivity for each individual mispair due to M644F Pol ε mutation. Values were obtained by dividing the error rate of the exo-deficient M644F Pol ε by that of the exo-deficient wild type enzyme. (D) Contribution of proofreading to Pol ε M644F mutant fidelity. Values were obtained by dividing the error rate of the exo-deficient M644F Pol ε by that of the exo-proficient M644F enzyme. All error rates are × 10−5. The (≤) symbol indicates the error rate (or ratio) is less than or equal to the calculated value. The (≥) symbol indicates the error rate (or ratio) is greater than or equal to the calculated value. P values are shown above each comparison that was found to be statistically significant. The (* and **) symbols indicate P values where P ≤ 0.05 and 0.001, respectively.
Exonuclease-proficient wild-type and M644F Pol ε (Figures 2A and 4A) are both more accurate than their exonuclease-deficient counterparts (Figures 2B and 4B). As previously seen with wild-type Pol ε (24), the majority of base substitutions (43/75) seen with exonuclease-proficient M644F Pol ε are C to T transitions (Figures 4A and 3). Many of these events may result from ‘correct’ incorporation of dAMP opposite uracil resulting from rare cystosine deamination, i.e. they may not be true misincorporation events. For the 11 other substitutions, M644F Pol ε retains relatively high fidelity when its 3′ exonuclease remains functional. Assuming that differences in error rates for the exonuclease-proficient and exonuclease-deficient polymerases are due to the status of the 3′ exonuclease activity, then the observed 12- to ≥240-fold differences in error rates for various mismatches (Figure 4D) indicate that proofreading excises the majority of single-base mismatches generated by M644F Pol ε. Nonetheless, proofreading does not correct all these errors, as indicated by a 4-fold higher overall average error rate (Figure 2, 4.4 × 10−5 for M644F Pol ε as compared to 1.1 × 10−5 for wild-type Pol ε, P < 0.001), and by slightly higher error rates for certain mismatches, e.g. A-dCMP, G-dTMP, T-dCMP (Figure 4A, P = 0.09, 0.09 and 0.05, respectively). These results on the proofreading capacity of M644F Pol ε are interesting in light of earlier studies demonstrating that L412M T4 Pol (17) and yeast L612M Pol δ (22) have strongly reduced proofreading capacity (see Discussion).
Spontaneous mutator phenotype of pol2-M644F yeast strains
A haploid yeast strain harboring the exonuclease-proficient M644F Pol ε allele has a spontaneous mutation rate at the CAN1 locus that is not significantly different from that of a wild-type yeast strain (Table 2). When repair of single-base mismatches is inactivated in an msh6 background, the pol2-M644F strain has a spontaneous mutation rate at the CAN1 locus of 380 × 10−7, a 16-fold higher rate than in a msh6 strain with a wild-type POL2 allele (24 × 10−7, Table 2). This mutator effect is consistent with replication infidelity catalyzed in vivo by M644F Pol ε. A similar interpretation has been put forth for mutator effects (summarized in Table 2) of L868F and L868M Pol α (18,19), and L612M Pol δ (22,23,37).
Table 2.
Mutation rates at CAN1 in strains with mutations in Pols ε, α and δ
| Yeast strain | Mutation rate(×10−7) | 95% Confidence intervals | Relative rate | Reference |
|---|---|---|---|---|
| Wild type | 2.3 | 1.6–2.8 | 1.0 | This study |
| pol2-M644F | 2.9 | 2.6–3.5 | 1.3 | This study |
| Δmsh6 | 24 | 13–36 | 1.0 | This study |
| Δmsh6 pol2-M644F | 380 | 270–530 | 16 | This study |
| Wild type | 4.8 | 1.0 | (19) | |
| pol1-L868M | 14 | 2.9 | (19) | |
| 2.6 | (18) | |||
| pol1-L868F | 39 | 8.1 | (19) | |
| Wild type | 3.1 | 1.0 | (21) | |
| pol3-L612M | 11 | 3.5 | (21) | |
| 7.5 | (23) | |||
| 9.0 | (22) | |||
| Δmsh6 | 18 | 1.0 | (21) | |
| Δmsh6 pol3-L612M | 420 | 23 | (21) | |
| Wild type | 2.0 | 1.0 | (23) | |
| pol3-L612F | 7.0 | 3.5 | (23) |
Strains were constructed and mutation rates were measured as described (see Materials and methods section).
DISCUSSION
When compared to earlier studies of other B family polymerases with replacements for a homologous leucine, the properties described here for yeast Pol ε derivatives with replacements for Met644 permit several interesting conclusions to be drawn. First, the collective studies to date reveal an important general similarity among Pol ε and the two other yeast B family major replicative polymerases, Pol α (18,19) and Pol δ (22). In each case, replacing the native amino acid at the Met644 position, or the corresponding position in Pol α or Pol δ, with other amino acids, reduces the fidelity of DNA synthesis. This commonality strongly supports the hypothesis that a key determinant of the nucleotide selectivity of polymerases is correct base-pairing geometry. Based on the crystal structure of the B family homolog RB69 Pol (14), Met644 in Pol ε, Leu868 in Pol α and Leu612 in Pol δ are inferred to be immediately adjacent to an invariant tyrosine in motif A that interacts with the sugar of the incoming dNTP in the polymerase active site, and therefore forms part of the binding pocket for the nascent base pair. Earlier studies [reviewed in (38)] have shown that replacements for residues that form this pocket result in reduced fidelity. This includes one study showing that replacing the orthologous motif A residue in the large fragment of E. coli DNA polymerase I reduces DNA synthesis fidelity in vitro (39), and another showing that replacing the invariant tyrosine (Y869) in yeast Pol α reduces replication fidelity in vivo (40). On this basis, we speculate that replacing Met644 in Pol ε with phenyalanine may indirectly alter the geometry of the binding pocket. The consequence is reduced selectivity against dNTP misinsertion, as revealed in Figure 4C, as well as more efficient extension of aberrant primer termini, as suggested by the inability to proofread some errors (Figures 2A and 4A) and by increased rates for single-base deletion and addition errors (Figure 2), which require extension of misaligned template-primers.
Second, the data reveal a number of interesting differences in DNA synthesis fidelity in vitro when comparing the mutant derivatives of Pol α, Pol δ and Pol ε. Consider the consequences of changing conserved Leu868 in naturally exonuclease-deficient Pol α to those for changing Met644 in exonuclease-deficient Pol ε. In both enzymes, replacement with phenylalanine reduces fidelity [Pol α (18,19); this study for Pol ε], but the error specificity of these phenyalanine derivatives is different. L868F Pol α generates single-base deletions at much higher error rates than does M644F Pol ε, and L868F Pol α also has higher error rates for some single-base mismatches than does M644F Pol ε (Figure 5B). In contrast, M644F Pol ε is less accurate than L868F Pol α for T-dTMP mismatches (Figure 5B). Also note that substituting tryptophan for L868 in Pol α reduced fidelity (19) whereas substituting tryptophan for Met644 in Pol ε does not reduce fidelity (Table 1). Moreover, a L612M substitution in Pol δ reduced fidelity (22), whereas the reverse M644L substitution in Pol ε has little effect on overall average fidelity (Table 1). Collectively, these facts demonstrate that the fidelity of polymerization by B family polymerases depends on at least three interrelated variables: (i) the identity of the side chain at the Met644 or homologous position, (ii) the interactions that side chain has within the surrounding active site context, whether Pol α, Pol δ or Pol ε and (iii) the type of replication error that is considered. These variables in turn suggest that, despite the idea that all polymerases use a common catalytic mechanism, the geometry of the nascent base pair binding pocket of Pol α, Pol δ and Pol ε are different with respect to determinants of specificity.
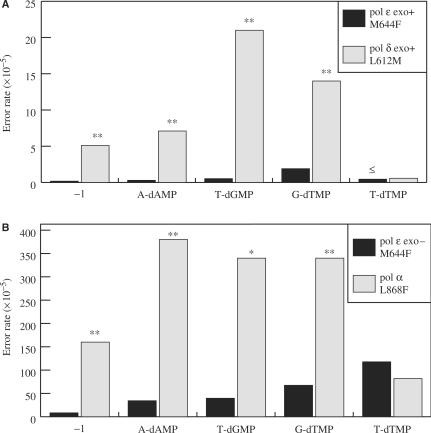
Figure 5.
Comparison of changes to a conserved motif A residue and the contributions to fidelity of B family replicative DNA polymerases. (A) Error rates are shown for exonuclease-proficient M644F Pol ε (black bars; this study) and L612M Pol δ [gray bars; (22)]. Values are for T·dTMP, A·dAMP, T·dGMP and G·dTMP base–base mismatches, as well as single-base deletions (−1). (B) Error rates are shown for exonuclease-deficient M644F Pol ε [black bars; this study) and L868F Pol α (gray bars; (19)]. Values are for T·dTMP, A·dAMP, T·dGMP and G·dTMP base–base mismatches, as well as single-base deletions (−1). The (≤) symbol indicates the error rate (or ratio) is less than or equal to the calculated value. P values are shown above each comparison that was found to be statistically significant. The (* and **) symbols indicate P values where P ≤ 0.01 and 0.001, respectively.
Third, comparisons between exonuclease-proficient Pol ε and Pol δ mutants are informative regarding proofreading potential (Figure 5A). Changing Leu612 to Met in Pol δ strongly elevated error rates, to levels observed with exonuclease-deficient Pol δ (22). In contrast, changing Met644 to Phe in Pol ε elevated error rates to a much lesser extent (Figures 2A and 4A and B), primarily due to relatively efficient proofreading of most errors (Figure 4D). The overall consequence is that exonuclease-proficient M644F Pol ε is considerably more accurate than exonuclease-proficient L612M Pol δ for a variety of errors (Figure 5A). This includes single-base deletions errors, with exonuclease-proficient M644F Pol ε being at least 28-fold more accurate than exonuclease-proficient L612M Pol δ. This strongly reinforces previous studies of wild-type enzymes showing that Pol ε proofreads frameshift intermediates much more efficiently than does Pol δ (25).
Finally, comparisons can now be made regarding the consequences of active site amino acid replacements in all three major replicative polymerases on spontaneous mutation rates in yeast (Table 2). The results (Table 2 and references therein) indicate that common mutant polymerase alleles, e.g. pol2-M644F, pol1-L868F, pol3-L612F, all result in elevated spontaneous mutation rates that depend on the status of post-replication repair by the Msh6-dependent mismatch repair system. When correlated with the fact that the corresponding purified polymerases (L612M for Pol δ) all synthesize DNA with reduced fidelity in vitro, the results clearly indicate that the spontaneous mutator phenotypes in vivo result from DNA replication errors by the mutant polymerases. On this basis, and as mentioned in the Introduction, we are currently using these mutator polymerase alleles in attempts to probe the roles of Pol α, Pol δ and Pol ε in DNA replication in vivo. We hope to use the different error specificities of the mutant enzymes [e.g. Figures 4 and 5 and (22)] as signatures to assign participation in specific biosynthetic reactions. For example, differences in the rates at which these mutator polymerases generate specific base substitutions (e.g. Figure 5, T-dTMP versus A-dAMP) may be helpful to investigate their roles in leading and lagging strand replication, and differences in error rates for frameshifts (Figure 5) may provide insights into the identity of the polymerases responsible for replication errors that lead to the microsatellite instability in mismatch repair defective tumors, or the instability associated with degenerative diseases characterized by repeat expansions.
ACKNOWLEDGEMENTS
We thank Dinh Nguyen for DNA substrate preparation and sequence analysis of lacZ mutants, and Stephanie Nick McElhinny and Katarzyna Bebenek for critically reading the manuscript. This work was funded in part by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences to T.A.K. and partly by the Swedish Research Council, The Swedish Cancer Society, and the Fund for Basic Science-Oriented Biotechnology and the Medical Faculty at Umeå University to E.J. Funding to pay the Open Access publication charges for this article was provided by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences to T.A.K.
REFERENCES
- 1.Hubscher U., Maga G., Spadari S. Eukaryotic DNA polymerases. Annu. Rev. Biochem. 2002;71:133–163. doi: 10.1146/annurev.biochem.71.090501.150041. [DOI] [PubMed] [Google Scholar]
- 2.Bebenek K., Kunkel T.A. Functions of DNA polymerases. Adv. Protein Chem. 2004;69:137–165. doi: 10.1016/S0065-3233(04)69005-X. [DOI] [PubMed] [Google Scholar]
- 3.Garg P., Burgers P.M. DNA polymerases that propagate the eukaryotic DNA replication fork. Crit. Rev. Biochem. Mol. Biol. 2005;40:115–128. doi: 10.1080/10409230590935433. [DOI] [PubMed] [Google Scholar]
- 4.Johnson A., O’Donnell M. Cellular DNA replicases: components and dynamics at the replication fork. Annu. Rev. Biochem. 2005;74:283–315. doi: 10.1146/annurev.biochem.73.011303.073859. [DOI] [PubMed] [Google Scholar]
- 5.Pavlov Y.I., Shcherbakova P.V., Rogozin I.B. Roles of DNA polymerases in replication, repair, and recombination in eukaryotes. Int. Rev. Cytol. 2006;255:41–132. doi: 10.1016/S0074-7696(06)55002-8. [DOI] [PubMed] [Google Scholar]
- 6.Morrison A., Araki H., Clark A.B., Hamatake R.K., Sugino A. A third essential DNA polymerase in S. cerevisiae. Cell. 1990;62:1143–1151. doi: 10.1016/0092-8674(90)90391-q. [DOI] [PubMed] [Google Scholar]
- 7.Asturias F.J., Cheung I.K., Sabouri N., Chilkova O., Wepplo D., Johansson E. Structure of Saccharomyces cerevisiae DNA polymerase epsilon by cryo-electron microscopy. Nat. Struct. Mol. Biol. 2006;13:35–43. doi: 10.1038/nsmb1040. [DOI] [PubMed] [Google Scholar]
- 8.Navas T.A., Zhou Z., Elledge S.J. DNA polymerase epsilon links the DNA replication machinery to the S phase checkpoint. Cell. 1995;80:29–39. doi: 10.1016/0092-8674(95)90448-4. [DOI] [PubMed] [Google Scholar]
- 9.Morrison A., Bell J.B., Kunkel T.A., Sugino A. Eukaryotic DNA polymerase amino acid sequence required for 3' – 5' exonuclease activity. Proc. Natl Acad. Sci. USA. 1991;88:9473–9477. doi: 10.1073/pnas.88.21.9473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kesti T., Flick K., Keranen S., Syvaoja J.E., Wittenberg C. DNA polymerase epsilon catalytic domains are dispensable for DNA replication, DNA repair, and cell viability. Mol. Cell. 1999;3:679–685. doi: 10.1016/s1097-2765(00)80361-5. [DOI] [PubMed] [Google Scholar]
- 11.Ohya T., Kawasaki Y., Hiraga S., Kanbara S., Nakajo K., Nakashima N., Suzuki A., Sugino A. The DNA polymerase domain of pol(epsilon) is required for rapid, efficient, and highly accurate chromosomal DNA replication, telomere length maintenance, and normal cell senescence in Saccharomyces cerevisiae. J. Biol. Chem. 2002;277:28099–28108. doi: 10.1074/jbc.M111573200. [DOI] [PubMed] [Google Scholar]
- 12.Dua R., Levy D.L., Campbell J.L. Analysis of the essential functions of the C-terminal protein/protein interaction domain of Saccharomyces cerevisiae pol epsilon and its unexpected ability to support growth in the absence of the DNA polymerase domain. J. Biol. Chem. 1999;274:22283–22288. doi: 10.1074/jbc.274.32.22283. [DOI] [PubMed] [Google Scholar]
- 13.Fuss J., Linn S. Human DNA polymerase epsilon colocalizes with proliferating cell nuclear antigen and DNA replication late, but not early, in S phase. J. Biol. Chem. 2002;277:8658–8666. doi: 10.1074/jbc.M110615200. [DOI] [PubMed] [Google Scholar]
- 14.Franklin M.C., Wang J., Steitz T.A. Structure of the replicating complex of a pol alpha family DNA polymerase. Cell. 2001;105:657–667. doi: 10.1016/s0092-8674(01)00367-1. [DOI] [PubMed] [Google Scholar]
- 15.Reha-Krantz L.J., Nonay R.L. Motif A of bacteriophage T4 DNA polymerase: role in primer extension and DNA replication fidelity. Isolation of new antimutator and mutator DNA polymerases. J. Biol. Chem. 1994;269:5635–5643. [PubMed] [Google Scholar]
- 16.Beechem J.M., Otto M.R., Bloom L.B., Eritja R., Reha-Krantz L.J., Goodman M.F. Exonuclease-polymerase active site partitioning of primer-template DNA strands and equilibrium Mg2+ binding properties of bacteriophage T4 DNA polymerase. Biochemistry. 1998;37:10144–10155. doi: 10.1021/bi980074b. [DOI] [PubMed] [Google Scholar]
- 17.Fidalgo da Silva E., Mandal S.S., Reha-Krantz L.J. Using 2-aminopurine fluorescence to measure incorporation of incorrect nucleotides by wild type and mutant bacteriophage T4 DNA polymerases. J. Biol. Chem. 2002;277:40640–40649. doi: 10.1074/jbc.M203315200. [DOI] [PubMed] [Google Scholar]
- 18.Pavlov Y.I., Frahm C., Nick McElhinny S.A., Niimi A., Suzuki M., Kunkel T.A. Evidence that errors made by DNA polymerase alpha are corrected by DNA polymerase delta. Curr. Biol. 2006;16:202–207. doi: 10.1016/j.cub.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 19.Niimi A., Limsirichaikul S., Yoshida S., Iwai S., Masutani C., Hanaoka F., Kool E.T., Nishiyama Y., Suzuki M. Palm mutants in DNA polymerases alpha and eta alter DNA replication fidelity and translesion activity. Mol. Cell. Biol. 2004;24:2734–2746. doi: 10.1128/MCB.24.7.2734-2746.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nick McElhinny S.A., Pavlov Y.I., Kunkel T.A. Evidence for extrinsic exonucleolytic proofreading. Cell Cycle. 2006;5:958–962. doi: 10.4161/cc.5.9.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li L., Murphy K.M., Kanevets U., Reha-Krantz L.J. Sensitivity to phosphonoacetic acid: a new phenotype to probe DNA polymerase delta in Saccharomyces cerevisiae. Genetics. 2005;170:569–580. doi: 10.1534/genetics.104.040295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nick McElhinny S.A., Stith C.M., Burgers P.M., Kunkel T.A. Inefficient proofreading and biased error rates during inaccurate DNA synthesis by a mutant derivative of Saccharomyces cerevisiae DNA polymerase delta. J. Biol. Chem. 2006 doi: 10.1074/jbc.M609591200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Venkatesan R.N., Hsu J.J., Lawrence N.A., Preston B.D., Loeb L.A. Mutator phenotypes caused by substitution at a conserved motif A residue in eukaryotic DNA polymerase delta. J. Biol. Chem. 2006;281:4486–4494. doi: 10.1074/jbc.M510245200. [DOI] [PubMed] [Google Scholar]
- 24.Shcherbakova P.V., Pavlov Y.I., Chilkova O., Rogozin I.B., Johansson E., Kunkel T.A. Unique error signature of the four-subunit yeast DNA polymerase epsilon. J. Biol. Chem. 2003;278:43770–43780. doi: 10.1074/jbc.M306893200. [DOI] [PubMed] [Google Scholar]
- 25.Fortune J.M., Pavlov Y.I., Welch C.M., Johansson E., Burgers P.M., Kunkel T.A. Saccharomyces cerevisiae DNA polymerase delta: high fidelity for base substitutions but lower fidelity for single- and multi-base deletions. J. Biol. Chem. 2005;280:29980–29987. doi: 10.1074/jbc.M505236200. [DOI] [PubMed] [Google Scholar]
- 26.Shimizu K., Hashimoto K., Kirchner J.M., Nakai W., Nishikawa H., Resnick M.A., Sugino A. Fidelity of DNA polymerase epsilon holoenzyme from budding yeast Saccharomyces cerevisiae. J. Biol. Chem. 2002;277:37422–37429. doi: 10.1074/jbc.M204476200. [DOI] [PubMed] [Google Scholar]
- 27.Bebenek K., Kunkel T.A. Analyzing fidelity of DNA polymerases. Methods Enzymol. 1995;262:217–232. doi: 10.1016/0076-6879(95)62020-6. [DOI] [PubMed] [Google Scholar]
- 28.Chilkova O., Jonsson B.H., Johansson E. The quaternary structure of DNA polymerase epsilon from Saccharomyces cerevisiae. J. Biol. Chem. 2003;278:14082–14086. doi: 10.1074/jbc.M211818200. [DOI] [PubMed] [Google Scholar]
- 29.Burgers P.M. DNA polymerase from Saccharomyces cerevisiae. Methods Enzymol. 1995;262:49–62. doi: 10.1016/0076-6879(95)62008-7. [DOI] [PubMed] [Google Scholar]
- 30.Hamatake R.K., Hasegawa H., Clark A.B., Bebenek K., Kunkel T.A., Sugino A. Purification and characterization of DNA polymerase II from the yeast Saccharomyces cerevisiae. Identification of the catalytic core and a possible holoenzyme form of the enzyme. J. Biol. Chem. 1990;265:4072–4083. [PubMed] [Google Scholar]
- 31.Fleiss J.L. Statistical Methods for Rates and Proportions. New York: John Wiley and Sons; 1973. [Google Scholar]
- 32.Pavlov Y.I., Mian I.M., Kunkel T.A. Evidence for preferential mismatch repair of lagging strand DNA replication errors in yeast. Curr. Biol. 2003;13:744–748. doi: 10.1016/s0960-9822(03)00284-7. [DOI] [PubMed] [Google Scholar]
- 33.Kirchner J.M., Tran H., Resnick M.A. A DNA polymerase epsilon mutant that specifically causes +1 frameshift mutations within homonucleotide runs in yeast. Genetics. 2000;155:1623–1632. doi: 10.1093/genetics/155.4.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shcherbakova P.V., Kunkel T.A. Mutator phenotypes conferred by MLH1 overexpression and by heterozygosity for mlh1 mutations. Mol. Cell. Biol. 1999;19:3177–3183. doi: 10.1128/mcb.19.4.3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Drake J.W. A constant rate of spontaneous mutation in DNA-based microbes. Proc. Natl Acad. Sci. USA. 1991;88:7160–7164. doi: 10.1073/pnas.88.16.7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garcia-Diaz M., Kunkel T.A. Mechanism of a genetic glissando: structural biology of indel mutations. Trends Biochem. Sci. 2006;31:206–214. doi: 10.1016/j.tibs.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 37.Hadjimarcou M.I., Kokoska R.J., Petes T.D., Reha-Krantz L.J. Identification of a mutant DNA polymerase delta in Saccharomyces cerevisiae with an antimutator phenotype for frameshift mutations. Genetics. 2001;158:177–186. doi: 10.1093/genetics/158.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kunkel T.A., Bebenek K. DNA replication fidelity. Annu. Rev. Biochem. 2000;69:497–529. doi: 10.1146/annurev.biochem.69.1.497. [DOI] [PubMed] [Google Scholar]
- 39.Minnick D.T., Liu L., Grindley N.D., Kunkel T.A., Joyce C.M. Discrimination against purine-pyrimidine mispairs in the polymerase active site of DNA polymerase I: a structural explanation. Proc. Natl Acad. Sci. USA. 2002;99:1194–1199. doi: 10.1073/pnas.032457899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pavlov Y.I., Shcherbakova P.V., Kunkel T.A. In vivo consequences of putative active site mutations in yeast DNA polymerases alpha, epsilon, delta, and zeta. Genetics. 2001;159:47–64. doi: 10.1093/genetics/159.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]