Abstract
The actin-related proteins (Arps) comprise a conserved protein family. Arp4p is found in large multisubunits of the INO80 and SWR1 chromatin remodeling complexes and in the NuA4 histone acetyltransferase complex. Here we show that arp4 (arp4S23A/D159A) temperature-sensitive cells are defective in G2/M phase function. arp4 mutants are sensitive to the microtubule depolymerizing agent benomyl and arrest at G2/M phase at restrictive temperature. Arp4p is associated with centromeric and telomeric regions throughout cell cycle. Ino80p, Esa1p and Swr1p, components of the INO80, NuA4 and SWR1 complexes, respectively, also associate with centromeres. The association of many kinetochore components including Cse4p, a component of the centromere nucleosome, Mtw1p and Ctf3p is partially impaired in arp4 cells, suggesting that the G2/M arrest of arp4 mutant cells is due to a defect in formation of the chromosomal segregation apparatus.
INTRODUCTION
Actin-related proteins (Arps) constitute an evolutionarily conserved family of proteins that share significant primary sequence similarity with a larger family of conventional actins, all of which appear to have descended from a single ancestral molecule (1,2). Arps of the budding yeast Saccharomyces cerevisiae are classified as Arps 1–10, where Arp1p is the most similar, and Arp10p the least similar, to actin (1). While Arps 1–3 and Arp10p are located in the cytoplasm, the other six, Arps 4–9, are nuclear proteins (3–5).
Arp4p, which was the first reported example of a nuclear Arp, is an essential protein in budding yeast (6). The identification of actin and/or Arps in multicomponent enzymes specifically involved in chromatin metabolism suggested that these proteins perform important nuclear functions related to their participation in chromatin-remodeling processes. Phenotypic analysis of arp4 mutants has revealed defects consistent with a function in transcriptional regulation and chromatin structure (7,8), and the purified Arp4 protein has been shown to bind histones in vitro (7,9).
Nucleosome-based chromatin structures lie at the heart of DNA-dependent cellular activities, such as DNA repair and replication, gene expression and chromosome segregation, and they are controlled by various chromatin regulators. In general, regulators such as chromatin remodeling and chromatin modifying complexes induce conformational alterations. Arp4p associates with Arp5p and Arp8p in the INO80 complex, and with Arp6p in the SWR1 complex (10). Moreover, Arp4p and actin are also components of Esa1-histone acetyltransferase (HAT) complexes (11). The INO80 chromatin remodeling complex has been implicated in both transcription and DNA repair (12–15). The SWR1 complex has an ATP-driven histone exchange activity that replaces the histone H2A/H2B dimer with a variant dimer, H2A.Z (Htz1)/H2B, and the incorporation of H2A.Z is reduced in the absence of Swr1p. Consistent with a specialized role for the SWR1 complex in H2A.Z deposition, most of the genome-wide transcriptional defects seen in swr1 cells are also found in htz1 cells (16–19). The human Arp4p homolog BAF53 was originally identified as a BRG1-associated factor—BRG1 is a SWI/SNF family ATPase (20–22)—and it was later found to be a component of the Tip60 complex as well (23). Esa1p and Tip60p are the catalytic subunits of the nucleosome acetyltransferase of the NuA4–HAT complex, which acetylates histone H4 (and H2A to some extent) (7,11,24).
Among the nuclear ARPs so far identified, Arp4p is vital importance in understanding the cellular role of nuclear ARPs, because it is an essential ARP in S. cerevisiae (6). However, the mechanisms underlying its essential functions are not yet fully understood. In this study, we examined the roles of Arp4p, especially in G2/M phase with special reference to the assembly of kinetochores.
MATERIALS AND METHODS
Yeast strains
Yeast strains used in this study are listed in Supplementary Table S1. Null mutants, Myc-tagged, HA-tagged and FLAG-tagged alleles were made using standard PCR-based gene disruption and insertion methods, as previously described (25–27). Deletion mutants were obtained by amplification of the KANMX6, HPHMX4 and CgTRP1 constructs from pFA6aKANMX6, pAG32 and SHB1805, respectively, with gene-specific primers consisting of 40–45 nucleotides. The resulting PCR fragments were transformed into yeast cells and colonies were selected on YPAD plates containing G418 or hygromycin B or on SC-Trp plates. Gene disruption was confirmed by genomic PCR. The arp4S23A/D159A mutant was constructed as previously described (28). The sequences of the primers used to generate DNA constructs used for gene disruption, or for checking gene disruption, and details of the yeast strains used in this study will be provided upon request.
Synchronization of cells
Cells were grown to early log phase, and cultures were harvested and resuspended in fresh YPAD medium to a concentration of 107 cells/ml. Cells were treated with 100 ng/ml α-factor (Sigma, St. Louis, MO, USA) or with 15 μg/ml nocodazole (Sigma) for 4 h at 23°C. After confirmation of G1 or G2/M arrest by microscopic examination, the α-factor or nocodazole was washed away, and the cells were resuspended in fresh YPAD.
Flow cytometry and cytological methods
Cells were taken at the indicated time points, fixed with 70% ethanol overnight at 4°C and analyzed with a Becton-Dickinson FACScan flow cytometer system. For cytological analyses, cells were fixed for 15 min in 3.7% formaldehyde, and DNA was visualized by DAPI staining. Cell morphology was examined by light microscopy.
Cell fractionation
Whole-cell extracts and chromatin pellets were prepared as previously described (29), except that spheroplasts were lysed in buffer containing 0.25% Triton X-100 (EBX). The extracts were then centrifuged through a sucrose cushion, and the resulting chromatin pellet was examined by immunoblot analysis for the presence of Cdc45-3HA. Histone H3 was used as a loading control for protein levels in whole-cell extracts and pellet fractions.
ChIP chip analysis
Arp4p was tagged at the carboxy terminus with the Flag epitope, which was detected with the monoclonal anti FLAG M2 antibody (Sigma Aldrich). Immunoprecipitation and amplification of DNA were performed as described (30), and the amplified DNA was hybridized to a high-resolution tiling array (SC3456a520015F, Affymetrix) (31). All experiments were performed twice. Graphics were created with Genome Shovel software (http://www.sequence.info/3456chip/). The raw data of ChIP chip analysis obtained here were deposited at Gene Expression Omnibus database as an accession number, GSE7276.
Chromatin immunoprecipitation (ChIP)
Chromatin immunoprecipitation was carried out as previously described, with minor modifications (32). Briefly, cells were harvested and incubated in 1% formaldehyde for 15 min to cross-link proteins to DNA, and the reaction was quenched by incubating cells in 125 mM glycine for 5 min. Cells were lysed with glass beads, and extracts were sonicated to shear DNA to an average size of 0.5 kb. Extracts were then divided into two aliquots: input DNA, and IP DNA (1:20, respectively). Immunoprecipitation was carried out using a monoclonal anti-Myc antibody (9E10) (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) a monoclonal anti-FLAG antibody (M2) (Sigma) or a monoclonal anti-HA antibody (12CA5) (Roche, Nutley, NJ, USA), and immune complexes were captured using Dynabeads Protein G (Dynal Biotech, Oslo, Norway) for 4 h at 4°C. After a series of washes, proteins were released from the beads by incubation for 6 h at 65°C. The samples were treated with proteinase K, and DNA was purified for PCR analysis by phenol extraction, followed by ethanol precipitation. The primers used for the ChIP assay are listed in Supplementary Table S2. PCR products were resolved on 2.5% agarose gels in 1 × TAE buffer with 0.5 μg/ml ethidium bromide. Band intensities were quantified by using Scion Image software. Quantitative data were obtained by real-time PCR.
Micrococcal nuclease digestion assay
Spheroplasts were prepared as previously described (33,34). Briefly, yeast cells were synchronized in G2/M phase with nocodazole for 3 h at 23°C and cultured for 1 h at non-permissive temperature (37°C), and then 1 × 109 cells were harvested. Cells were washed in water and then in 1 M sorbitol, resuspended in 800 μl YLE buffer [1 mg/ml Zymolyase, 1 M sorbitol, 0.2% (v/v) 2-mercaptoethanol] and incubated for 1 h at 37°C. The resulting spheroplasts were collected by centrifugation, gently washed twice in 950 μl 1 M sorbitol and resuspended in 750 μl suspension buffer [1 M sorbitol, 50 mM NaCl, 10 mM Tris–HCl pH 7.5, 5 mM MgCl2, 0.04% (v/v) 2-mercaptoethanol, 0.5 mM spermidine, 0.075% Nonidet P40]. After transferring 200 μl of suspension to a tube, 8 μl of micrococcal nuclease (Sigma, 200 U/ml) (final conc. 2 U/ml) was added and the tube was incubated at 37°C. At the indicated time points, 208 μl aliquots were removed and transferred to tubes containing 20 μl stop solution (5% SDS, 250 mM EDTA). The DNA was then purified by phenol/chloroform extraction and ethanol precipitation, analyzed in a 1.5% agarose 0.5 × TBE gel and visualized with ethidium bromide.
RESULTS
The arp4 mutant arrests in G2/M phase at restricted temperature
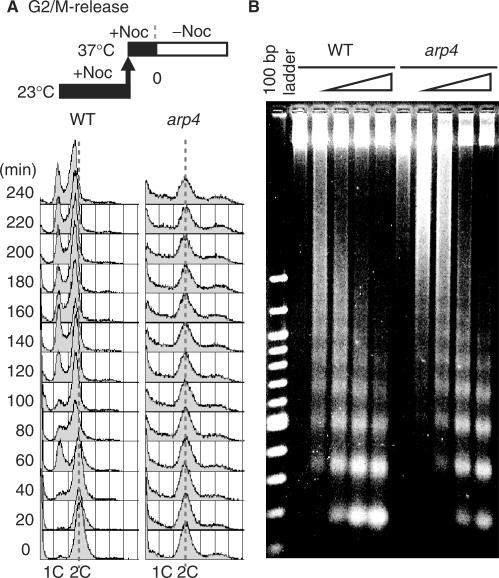
To investigate the roles of Arp4p in the cell cycle, we used a temperature sensitive arp4 mutant (arp4S23A/D159A) (Figure 1A), which bears mutations that affect residues in the ATP-binding pocket (28). The arp4 cells arrested in G2/M phase at non-permissive temperature (Figure 1B). Analyses of cell shape and spindle morphology revealed that the proportion of the population in telophase was decreased and the proportion in metaphase was increased when arp4 cells were cultured at restricted temperature, as compared with wild-type cells (Figure 1C), indicating that progression through metaphase is inhibited. Interestingly, the incidence of spindle abnormalities, including spindle elongation and spindle breakage associated with lagging chromosomes in mother cells, and of nuclear abnormalities, was increased in arp4 cells (Figure 1D and E, Supplementary Figure S1A). Arrest in G2/M phase is often due to activation of the DNA damage checkpoint or spindle assembly checkpoint (35,36). However, the G2/M arrest of arp4 cells was not released by deleting MAD2 or RAD9, which are involved in the spindle assembly checkpoint and the DNA damage checkpoint, respectively (Supplementary Figure S1B). Therefore, the ARP4 mutation may cause perturbations that activate both checkpoints. In this context, it is interesting that arp4 cells were hypersensitive to hydroxyurea (HU), an inhibitor of DNA synthesis, and to benomyl, a microtubule depolymerizing agent (Figure 1F and G). Although we observed a defect in the transition from G1 to S phase (Supplementary Figure S2) and a defect in recovery from HU arrest (Supplementary Figure S3), the most prominent phenotype of arp4 cells is the G2/M phase arrest. Thus, we focused on the molecular mechanism behind this phenomenon.
Figure 1.
arp4S23A/D159A mutants arrest in metaphase at restrictive temperature. (A) Temperature sensitivity of arp4 mutants. Wild-type (YHO800) and arp4S23A/D159A mutant (YHO820) cells were spotted on YPAD plates and incubated at 23 or 37°C for 3 days. (B) Cell cycle profiles of wild-type and arp4S23A/D159A cells at restrictive temperature. Logarithmically growing wild-type (YHO800) and arp4S23A/D159A (YHO820) cells were cultured in fresh prewarmed YPAD medium at 37°C for 12 h. Samples were analyzed by flow cytometry. (C) Large-budded wild-type (YHO800) and arp4S23A/D159A (YHO820) cells were classified with respect to spindle morphology (telophase, anaphase or metaphase). Logarithmically growing wild-type (YHO800) and arp4S23A/D159A (YHO820) cells were cultured in fresh prewarmed YPAD medium at 37°C for 12 h. Samples were fixed with formaldehyde, stained to visualize microtubules and scored (wild-type, n = 218; arp4S23A/D159A, n = 519). (D) The percentage of wild-type and arp4 large-budded cells described in Supplementary Figure S1A having an elongated spindle in the mother cell or having a broken spindle. Cells with an elongated microtubule spindle in the mother cell were visible as those having a single decondensed DNA mass and an intermediate spindle that extended beyond the center of either mother or bud. Cells with a broken spindle were visible as those having one or two DNA masses and a broken spindle. (E) Percentage of large-budded cells with lagging chromosomes, as indicated by the presence of two DNA masses. (F and G) Sensitivity of arp4 mutants to hydroxyurea (F) and benomyl (G). Wild-type (YHO800) and arp4S23A/D159A (YHO820) cells were spotted on YPAD plates containing 75 mM hydroxyurea (HU) and incubated at 30°C for 3 days or spotted on YPAD plates containing 15 μg/ml benomyl and incubated at 23°C for 3 days.
Arp4 is essential for recovery from G2/M arrest induced by nocodazole
Since arp4 cells were benomyl-sensitive, even at permissive temperature (Figure 1G), and arrested in G2/M phase with abnormal spindle morphology at restrictive temperature (Figure 1D, Supplementary Figure S1A), we next examined cell cycle progression of arp4 cells after release from a G2/M arrest. Cells were arrested in G2/M phase with nocodazole at 23°C, the temperature was up-shifted to 37°C to inactivate Arp4p, and the cell cycle was monitored by flow cytometry. As shown in Figure 2A, wild-type cells entered G1 phase 60 min after release from the nocodazole block, but arp4 cells remained in G2/M phase. Furthermore, the G2/M peak gradually decreased with a concomitant increase in the proportions of the population having a DNA content of less than 1C or greater than 2C during incubation up to 240 min, suggestive of chromosomal missegregation.
Figure 2.
The progression of mitotic phase in arp4S23A/D159A cells is impaired. (A) Wild-type (YHO800) and arp4S23A/D159A (YHO820) cells were grown in YPAD at 23°C for 3 h with 15 μg/ml nocodazole. The culture was shifted to 37°C and incubated in the presence of nocodazole for 1 h. Cells were released from the block by washing in prewarmed (37°C) YPAD and incubated in fresh prewarmed YPAD medium at 37°C. Samples were taken at the time points indicated and analyzed by flow cytometry. (B) Wild-type (YHO800) and arp4S23A/D159A (YHO820) cells were synchronized in G2/M phase with nocodazole for 3 h at 23°C and cultured for 1 h at non-permissive temperature (37°C). Equal quantities of yeast spheroplasts were digested with 2 U/μl micrococcal nuclease, which preferentially digests DNA in linker regions between nuclesomes, for 0, 2, 5, 15 and 30 min. The purified DNA was resolved on a 1.5% agarose 0.5 × TBE gel and visualized by ethidium bromide staining.
To investigate the status of chromatin in wild-type and arp4 cells arrested by nocodazole at restrictive temperature, chromatin was isolated, digested with micrococcal nuclease, and subjected to electrophoresis. The digestion pattern indicated that the chromatin in arp4 cells has a more highly condensed structure compared to that of wild-type cells (Figure 2B). Similar results were obtained with asynchronous cells cultured for 1 and 24 h at the restrictive temperature (Supplementary Figure S4).
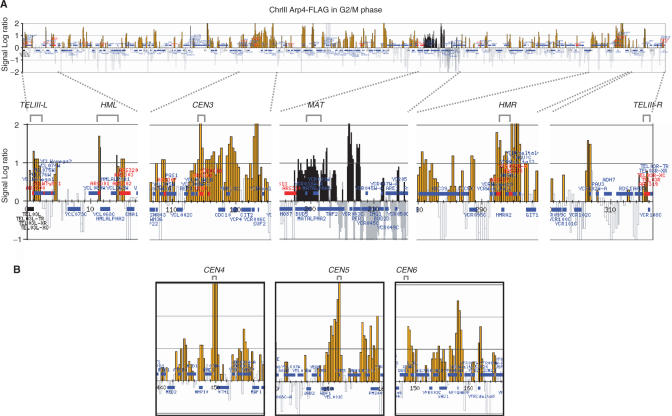
Genome-wide localization of Arp4p
Since the overall chromatin structure of arp4 cells appeared to be more highly condensed than that of wild-type cells (Figure 2B), we determined the localization of Arp4p protein on chromosomes III, IV and V, and on the right arm of chromosome VI by the ChIP chip method (30). Figure 3A shows that Arp4p was distributed broadly throughout chromosome III in G2/M phase-arrested cells. As expected from the involvement of Arp4p in transcription, many Arp4p peaks were detected in intergenic regions, which generally correspond to promoter regions. In addition, we detected binding of Arp4p to chromosome III centromeric sequences (CEN3), to the MAT, HML and HMR loci and to telomeric sequences (Figure 3A, lower panel). Localization of Arp4p in centromeric regions was confirmed for all other chromosomes tested (CEN4, CEN5 and CEN6) (Figure 3B). The distribution of Arp4p on chromosomes IV and V and on the right arm of chromosome VI is shown in a Supplementary Figure S5.
Figure 3.
Localization of Arp4p on Chromosome III in G2/M phase. Cells expressing Flag-tagged Arp4 (YHO311) were arrested in G2/M at 30°C. Samples were fixed and subjected to ChIP chip analysis with ChrIII-VI arrays. Blue horizontal lines indicate open reading frames, and orange peaks indicate significant chromosomal binding of Arp4p. CEN denotes the position of the centromere, and the red lines and numbers indicate the positions of autonomous replication origins (ARS ). The vertical lines indicate signal strength in one-log increments, and the horizontal scale bar indicates chromosomal coordinates in kb. (A) Arp4p binding across the entire chromosome III. (B) Arp4p binding to chromosomal regions around CEN4, CEN5 and CEN6.
Arp4p binds to specific regions throughout the cell cycle
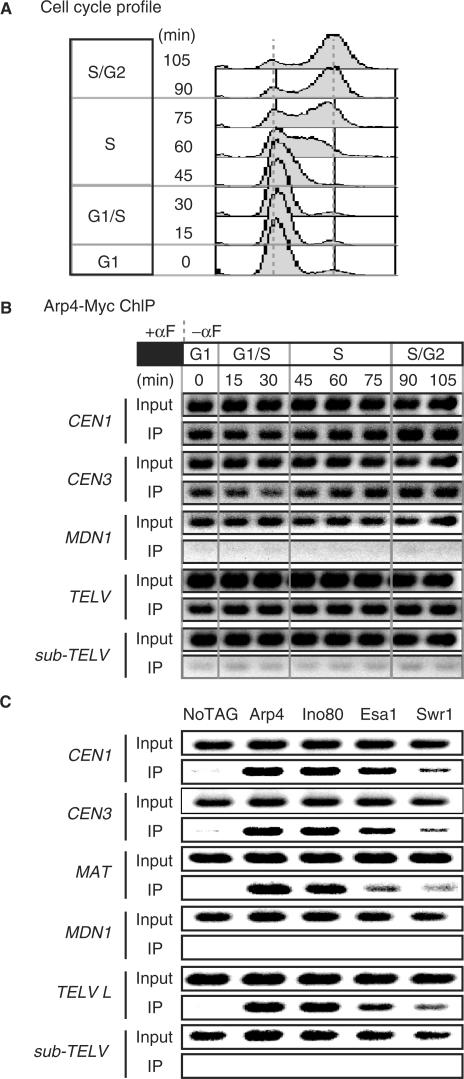
The arp4 cells showed a defect in entry into S phase after release from G1 arrest (Supplementary Figure S2), in progression through S phase after release from a HU block (Supplementary Figure S3) and in progression through G2/M phase (Figure 2). To determine whether the distribution of Arp4p on chromatin changes during the cell cycle, we analyzed the chromatin binding of Arp4p to specific regions in cells released from a G1 block (Figure 4A) by the ChIP method (Figure 4B). Arp4p associated with the centromeric regions CEN1 and CEN3 and with the telomeric region of chromosome V (TELV) throughout the cell cycle, but not with the large MDN1 ORF.
Figure 4.
Arp4p and Arp4 containing complexes associate with the centromere and telomere. Cells with 13Myc-tagged Arp4 (YHO312) were grown in YPAD at 20°C for 3 h with 100 ng/ml α-factor. Cells were released by washing in YPAD and incubated in fresh YPAD medium at 20°C. Samples were taken at the time points indicated and analyzed by flow cytometry (A) and ChIP (B). (A) Flow cytometry analysis of cell cycle profiles. (B) Input DNA and DNA coimmunoprecipitated with the anti-Myc antibody (IP) were amplified with primer sets corresponding to sequences around centromeres (CEN1 and CEN3), the inner region of a large ORF (MDN1), a telomere (TELV), and a sub-telomeric region (sub-TELV). To ensure the linearity of the PCR signal, appropriate dilutions of IP samples were used in PCR amplifications. ChIP PCR products were separated by agarose gel electrophoresis. Representative data are shown. (C) Arp4p, Ino80p, Esa1p and Swr1p interact with and localize to CEN1, CEN3, MAT and TELV, but not to MDN1 and a subtelomeric region. Flag-tagged Arp4 (YHO311), Ino80 (YHO313), Esa1 (YHO314), Swr1 (YHO315) or untagged (YK402) cells were arrested in G2/M by treatment with nocodazole at 30°C. Cells were fixed with 1% formaldehyde for 15 min and subjected to ChIP. Input DNA and DNA coimmunoprecipitated with the anti-FLAG antibody (IP) were amplified with primer sets corresponding to sequences around CEN1, CEN3, MAT, MDN1, TELV and a subtelomeric region. The templates used were total chromatin (Input) or immunoprecipitate (IP).
Arp4p is included in the NuA4 HAT histone H4 acetyltransferase complex, the INO80 chromatin remodeling complex, and the SWR1 H2A.Z deposition complex (7,12,13,17,18,24). However, it is not known whether these complexes colocalize on chromatin. Therefore, we examined whether these complexes associate with specific chromosomal regions in G2/M phase-arrested cells using Ino80p, Esa1p and Swr1p as representatives of the INO80, NuA4 HAT and SWR1 complexes, respectively (Figure 4C). Although the Swr1p signal was weaker than those of the other two proteins, these proteins associated with CEN1, CEN3, MAT and TEL-V but not with MDN1 or a sub TELV (9.7 kb). Thus, all of the INO80, NuA4 HAT and SWR1 complexes localize in the CEN1, CEN3, MAT and TEL-V regions.
Arp4p is required for the assembly of kinetochores
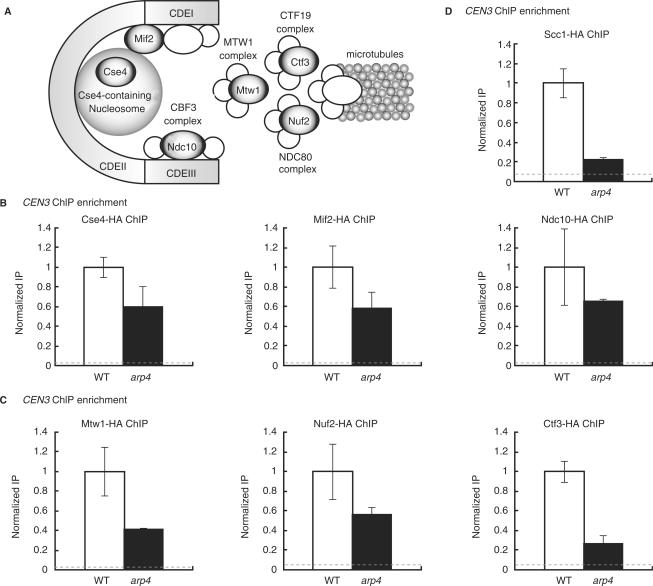
The centromere is the assembly site of the kinetochore complex, which consists of a large number of proteins and links chromosomes with spindle microtubles (MTs) (37,38). If kinetochores are not attached by spindle MTs, the spindle checkpoint is activated and cells arrest in metaphase (39). Since the benomyl sensitivity and defect in the recovery from nocodazole observed in arp4 cells are common features of kinetochore mutants, and since Arp4p associates with centromeric regions, it is likely that arp4 cells are defective in kinetochore function. The budding yeast kinetochore is the most fully characterized, and more than 65 components that constitutively localize to the kinetochore have been identified (37,40). Most yeast kinetochore proteins are found in several distinct complexes known as the CBF3, CTF19/COMA, MTW1, NDC80 and DAM1 complexes, which appear to assemble on a single centromeric nucleosome (Figure 5A) (41). Although the exact architecture of the kinetochore is not known, it is divided into inner, central and outer domains. The inner kinetochore contains the CBF3 complex (Ndc10p, Cep3p, Skp1p and Ctf13p), the DNA binding proteins Mif2p and Cbf1p, and the centromeric histone H3 variant Cse4p, which is associated with the nucleosome. The central kinetochore contains the MTW1 (Mtw1p, Dsn1p, Nnf1p and Nsl1p) and CTF19/COMA (Ctf19p, Mcm16p, Mcm19p, Mcm21p, Mcm22p, Ctf3p, Chl4p, Okp1p, Ame1p, Iml3p, Nkp1p and Nkp2p) complexes. The outer kinetochore includes the NDC80 (Ndc80p, Spc24p, Spc25p and Nuf2p) and DAM1 complexes (Dam1p, Ask1p, Duo1p, Dad1p, Dad2p, Dad3p, Dad4p, Spc19p and Spc34p); the DAM complex is considered to be the most outlying because microtubules and all other complexes are required for its localization in the kinetochore (42–44).
Figure 5.
The centromeric binding of kinetochore components is partially impaired in arp4S23A/D159A mutants. 3HA-tagged or untagged wild-type and arp4S23A/D159A cells were grown in YPAD at 23°C for 3 h with 15 μg/ml nocodazole to ensure that both populations had an equivalent cell cycle distribution since a higher proportion of arp4S23A/D159A cells are in G2/M phase. The culture was shifted to 37°C and incubated in the presence of nocodazole for 1 h. Cells were fixed with 1% formaldehyde for 15 min and subjected to ChIP. Input DNA and DNA coimmunoprecipitated with the anti-HA antibody (IP) were amplified with primer sets corresponding to sequences around centromeres (CEN3). Quantitative data were obtained by real-time PCR. To ensure the linearity of the PCR signal, appropriate dilutions of IP samples were used in PCR amplifications. In each case, CEN3 ChIP enrichment is expressed relative to that for a subtelomeric region of chromosome V (9716–9823). Results are expressed as the mean and SD of two independent ChIP experiments. Dashed lines indicate the background level of ChIP signal intensity in an untagged strain. (A) Schematic of kinetochore components. (B) The centromere-specific histone H3 variant Cse4p (wild-type cells: YHO805; arp4S23A/D159A cells: YHO825), a representative protein of the inner kinetochore Mif2p (wild-type cells: YHO806; arp4S23A/D159A cells: YHO826) and Ndc10p (wild-type cells: YHO807; arp4S23A/D159A cells: YHO827) were analyzed by ChIP at CEN3. (C) A representative protein of the outer kinetochore Mtw1p (wild-type cells: YHO808; arp4S23A/D159A cells: YHO828), Nuf2p (wild-type cells: YHO809; arp4S23A/D159A cells: YHO829) and Ctf3p (wild-type cells: YHO810; arp4S23A/D159A cells: YHO830) were analysed by ChIP at CEN3. (D) The cohesin component Scc1p (wild-type cells: YHO811; arp4S23A/D159A cells: YHO831) was analysed by ChIP at CEN3.
To address the role of Arp4p in kinetochore architecture, we examined the association of representative proteins in these complexes with CEN3 in G2/M phase-arrested wild-type and arp4 cells. Both cell types were synchronized in G2/M phase with nocodazole, and the culture temperature was up-shifted to 37°C to inactivate Arp4p. Under this condition, Cse4p, Mif2p and Ndc10p (members of the CBF3 complex) were enriched at CEN3 in both wild-type and arp4 cells. However, the level of all three proteins that were associated with CEN3 was slightly reduced in arp4 compared with wild-type cells (Figure 5B). We extended this analysis to the outer kinetochore components Mtw1p (MTW1 complex), Nuf2p (NDC80 complex) and Ctf3p (CTF19/COMA complex). A reduction of the CEN3 association was observed for all three of these proteins, most notably Ctf3, in arp4 mutants (Figure 5C). Thus, Arp4p would appear to be implicated in the G2/M phase transition through its participation in kinetochore assembly. Cohesin association with centromeres was found to be dependent on inner kinetochore proteins such as Mif2p (45). If the structure of the kinetochore is indeed perturbed in arp4 cells, the centromeric binding of cohesin might also be impaired in arp4 cells. Indeed, we observed a dramatic reduction in the binding of Scc1p, one of the components of cohesin, to CEN3 in arp4 cells (Figure 5D).
DISCUSSION
The roles of Arp4p in cell cycle
Arp4p is an essential component of chromatin modulating complexes, including NuA4 HAT, INO80 and SWR1 complexes (12,13,17,18,24). In this study, we found at least three defects in arp4 cells: entry into S phase, progression through S phase after release from a HU block, and progression through G2/M phase, suggesting the requirement of Arp4p throughout cell cycle. Since Arp4p is involved in global gene expression (8,22,28,46), all of these defects may be indirect consequences of transcriptional deregulation. However, genome-wide analyses of the distribution of Arp4p revealed that it associates with centromeres, and some defects in kinetochore architecture were observed in arp4 mutants, suggesting involvement of Arp4p in the assembly of kinetochores. We therefore focus on how the Arp4p defect leads to G2/M arrest at the molecular level.
The roles of Arp4p in kinetochore–spindle attachment
The arp4 mutant cells arrested in metaphase and could not recover from a nocodazole block at non-permissive temperature. Interestingly, a higher incidence of spindle abnormalities and of nuclear abnormalities was observed in arp4 cells. In addition, arp4 cells showed increased sensitivity to the microtubule depolymerizing agent benomyl, even at permissive temperature. Many kinetochore mutants also show metaphase arrest and benomyl sensitivity (47). Minoda et al. (48) observed that a fission yeast alp5-1134 mutant, which is defective in an ARP4 ortholog, is not sensitive to microtubule-depolymerizing agents unlike other mitotic mutants that are hypersensitive to these agents. It was hypothesized that Alp5p is involved in mitotic events in an indirect manner via the transcriptional regulation of genes involved in microtubule dynamics. In contrast, our results indicate that budding yeast Arp4p is directly involved in the assembly of kinetochores by regulating chromatin structure. Arp4p associates with all centromeres tested, including CEN1, CEN3, CEN4, CEN5 and CEN6. Moreover, a reduction in the centromere association of various kinetochore-associated proteins was observed in arp4 cells, and this was especially the case for Ctf3p, a component of a kinetochore outer complex. Since the presence of a Cse4p-containing nucleosome at the centromere is a prerequisite for the efficient loading of outer but not inner kinetochore proteins onto the centromere (49), it is likely that the reduced binding of Mtw1p, Muf2p and Ctf3p in arp4 cells is due to the inefficient binding of Cse4p. That is, the inefficient formation of Cse4p-containing nucleosomes is probably due to the inefficient binding of inner kinetochore protein complexes containing Mif2p and Ndc10p. Moreover, a dramatic reduction in the binding of Scc1p, one of the components of cohesin, to CEN3 was observed in arp4 cells. Cohesin reportedly accumulates around centromeres depending on the nature of the inner kinetochore proteins (45). Taken together, it is likely that proper regulation of chromatin structure by Arp4p-containing complexes is necessary for the efficient binding of inner kinetochore proteins to assemble kinetochores.
Interplay between INO80, NuA4 HAT and SWR1 complexes in chromosome segregation
If Arp4p is involved in the assembly of kinetochores, arp4 mutations should show a synthetic lethal phenotype in combination with mutations in genes encoding kinetochore proteins. Krogan et al. (50) showed that defects in various components of Arp4 containing complexes, such as NuA4 HAT and SWR1, lead to synthetic lethal phenotypes when combined with mutations in kinetochore components. In addition, increased chromosome missegregation is observed in NuA4 HAT and SWR1 related mutants. Thus, Arp4-containing NuA4 HAT and SWR1 complexes function at the centromere during chromosomal segregation (19,50,51). In the current study, we observed that the Ino80p, Esa1p and Swr1p components of INO80, NuA4 HAT and SWR1 complexes, respectively, localize to centromeres like Arp4p (Figure 5). Taken together, these results suggest that Arp4p functions in the assembly of kinetochores via these three Arp4p-containing complexes.
The next question concerns how the three Arp4-containing complexes interact in the assembly of kinetochores. Recently, interplay between NuA4 HAT complex and SWR1 complex was observed as a robust HAT activity of NuA4 HAT toward Htz1-K14 (52,53). However, there is no apparent abnormality in the association of kinetochore proteins with centromeres in htz1 null mutant cells or htz1–K14R mutant cells, which harbor an unacetylatable version of Htz1p (53). Hence, it is not likely that the NuA4 HAT and SWR1 complexes interact via the acetylation of Htz1p. Interestingly, the temperature sensitivity of the fission yeast alp5-1134 mutant is suppressed by mutation of class I type histone deacetylase (HDAC) (48). Similarly, disruption of the budding yeast class I type HDAC gene RPD3 also partially suppresses the benomyl sensitivity and temperature sensitivity of arp4 cells (Supplementary Figure S6). Thus, one function of Arp4p in the assembly of kinetochores seems to be exerted by the HAT activity of the NuA4 HAT complex.
Although the roles played by individual Arp4p containing complexes in kinetochore assembly remain to be clarified, our study opens an alternative way to understand the regulation of chromatin structure in the process of kinetochore assembly.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank U. Wintersberger for plasmids used in this study. We thank all members of the Enomoto lab for their support. This work was supported by Grants-in-Aid for Scientific Research on Priority Areas from The Ministry of Education, Science, Sports and Culture of Japan, and by Health Sciences Research Grants from the Ministry of Health and Welfare of Japan. Funding to pay the Open Access publication charges for this article was provided by Grants-in-Aid for Scientific Research on Priority Areas from The Ministry of Education, Science, Sports and Culture of Japan.
Conflict of interest statement. None declared.
REFERENCES
- 1.Poch O., Winsor B. Who's who among the Saccharomyces cerevisiae actin-related proteins? A classification and nomenclature proposal for a large family. Yeast. 1997;13:1053–1058. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1053::AID-YEA164>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 2.Schafer D.A., Schroer T.A. Actin-related proteins. Annu. Rev. Cell Dev. Biol. 1999;15:341–363. doi: 10.1146/annurev.cellbio.15.1.341. [DOI] [PubMed] [Google Scholar]
- 3.Weber V., Harata M., Hauser H., Wintersberger U. The actin-related protein Act3p of Saccharomyces cerevisiae is located in the nucleus. Mol. Biol. Cell. 1995;6:1263–1270. doi: 10.1091/mbc.6.10.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grava S., Dumoulin P., Madania A., Tarassov I., Winsor B. Functional analysis of six genes from chromosomes XIV and XV of Saccharomyces cerevisiae reveals YOR145c as an essential gene and YNL059c/ARP5 as a strain-dependent essential gene encoding nuclear proteins. Yeast. 2000;16:1025–1033. doi: 10.1002/1097-0061(200008)16:11<1025::AID-YEA602>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 5.Harata M., Oma Y., Tabuchi T., Zhang Y., Stillman D.J., Mizuno S. Multiple actin-related proteins of Saccharomyces cerevisiae are present in the nucleus. J. Biochem. (Tokyo) 2000;128:665–671. doi: 10.1093/oxfordjournals.jbchem.a022799. [DOI] [PubMed] [Google Scholar]
- 6.Harata M., Karwan A., Wintersberger U. An essential gene of Saccharomyces cerevisiae coding for an actin-related protein. Proc. Natl. Acad. Sci. USA. 1994;91:8258–8262. doi: 10.1073/pnas.91.17.8258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galarneau L., Nourani A., Boudreault A.A., Zhang Y., Heliot L., Allard S., Savard J., Lane W.S., Stillman D.J., Cote J. Multiple links between the NuA4 histone acetyltransferase complex and epigenetic control of transcription. Mol. Cell. 2000;5:927–937. doi: 10.1016/s1097-2765(00)80258-0. [DOI] [PubMed] [Google Scholar]
- 8.Jiang Y.W., Stillman D.J. Epigenetic effects on yeast transcription caused by mutations in an actin-related protein present in the nucleus. Genes Dev. 1996;10:604–619. doi: 10.1101/gad.10.5.604. [DOI] [PubMed] [Google Scholar]
- 9.Harata M., Oma Y., Mizuno S., Jiang Y.W., Stillman D.J., Wintersberger U. The nuclear actin-related protein of Saccharomyces cerevisiae, Act3p/Arp4, interacts with core histones. Mol. Biol. Cell. 1999;10:2595–2605. doi: 10.1091/mbc.10.8.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mohrmann L., Verrijzer C.P. Composition and functional specificity of SWI2/SNF2 class chromatin remodeling complexes. Biochim. Biophys. Acta. 2005;1681:59–73. doi: 10.1016/j.bbaexp.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 11.Doyon Y., Selleck W., Lane W.S., Tan S., Côté J. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol. Cell. Biol. 2004;24:1884–1896. doi: 10.1128/MCB.24.5.1884-1896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen X., Mizuguchi G., Hamiche A., Wu C. A chromatin remodelling complex involved in transcription and DNA processing. Nature. 2000;406:541–544. doi: 10.1038/35020123. [DOI] [PubMed] [Google Scholar]
- 13.Shen X., Ranallo R., Choi E., Wu C. Involvement of actin-related proteins in ATP-dependent chromatin remodeling. Mol. Cell. 2003;12:147–155. doi: 10.1016/s1097-2765(03)00264-8. [DOI] [PubMed] [Google Scholar]
- 14.van Attikum H., Fritsch O., Hohn B., Gasser S.M. Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell. 2004;119:777–788. doi: 10.1016/j.cell.2004.11.033. [DOI] [PubMed] [Google Scholar]
- 15.Morrison A.J., Highland J., Krogan N.J., Arbel-Eden A., Greenblatt J.F., Haber J.E., Shen X. INO80 and gamma-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell. 2004;119:767–775. doi: 10.1016/j.cell.2004.11.037. [DOI] [PubMed] [Google Scholar]
- 16.Krogan N.J., Keogh M.C., Datta N., Sawa C., Ryan O.W., Ding H., Haw R.A., Pootoolal J., Tong A., Canadien V., et al. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol. Cell. 2003;12:1565–1576. doi: 10.1016/s1097-2765(03)00497-0. [DOI] [PubMed] [Google Scholar]
- 17.Mizuguchi G., Shen X., Landry J., Wu W.H., Sen S., Wu C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- 18.Kobor M.S., Venkatasubrahmanyam S., Meneghini M.D., Gin J.W., Jennings J.L., Link A.J., Madhani H.D., Rine J. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2004;2:e131. doi: 10.1371/journal.pbio.0020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang H., Richardson D.O., Roberts D.N., Utley R., Erdjument-Bromage H., Tempst P., Côté J., Cairns B.R. The Yaf9 component of the SWR1 and NuA4 complexes is required for proper gene expression, histone H4 acetylation, and Htz1 replacement near telomeres. Mol. Cell Biol. 2004;24:9424–9436. doi: 10.1128/MCB.24.21.9424-9436.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang W., Cote J., Xue Y., Zhou S., Khavari P.A., Biggar S.R., Muchardt C., Kalpana G.V., Goff S.P., Yaniv M., et al. Purification and biochemical heterogeneity of the mammalian SWI/SNF complex. EMBO J. 1996a;15:5370–5382. [PMC free article] [PubMed] [Google Scholar]
- 21.Wang W., Xue Y., Zhou S., Kuo A., Cairns B.R., Crabtree G.R. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 1996b;10:2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- 22.Zhao K., Wang W., Rando O.J., Xue Y., Swiderek K., Kuo A., Crabtree G.R. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell. 1998;95:625–636. doi: 10.1016/s0092-8674(00)81633-5. [DOI] [PubMed] [Google Scholar]
- 23.Ikura T., Ogryzko V.V., Grigoriev M., Groisman R., Wang J., Horikoshi M., Scully R., Qin J., Nakatani Y. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102:463–473. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- 24.Allard S., Utley R.T., Savard J., Clarke A., Grant P., Brandl C.J., Pillus L., Workman J.L., Cote J. NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J. 1999;18:5108–5119. doi: 10.1093/emboj/18.18.5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goldstein A.L., McCusker J.H. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 26.Kitada K., Yamaguchi E., Arisawa M. Cloning of the Candida glabrata TRP1 and HIS3 genes, and construction of their disruptant strains by sequential integrative transformation. Gene. 1995;165:203–206. doi: 10.1016/0378-1119(95)00552-h. [DOI] [PubMed] [Google Scholar]
- 27.Wach A. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast. 1996;12:259–265. doi: 10.1002/(SICI)1097-0061(19960315)12:3%3C259::AID-YEA901%3E3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 28.Gorzer I., Schuller C., Heidenreich E., Krupanska L., Kuchler K., Wintersberger U. The nuclear actin-related protein Act3p/Arp4p of Saccharomyces cerevisiae is involved in transcription regulation of stress genes. Mol. Microbiol. 2003;50:1155–1171. doi: 10.1046/j.1365-2958.2003.03759.x. [DOI] [PubMed] [Google Scholar]
- 29.Liang C., Stillman B. Persistent initiation of DNA replication and chromatin-bound MCM proteins during the cell cycle in cdc6 mutants. Genes Dev. 1997;11:3375–3386. doi: 10.1101/gad.11.24.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Katou Y., Kanoh Y., Bando M., Noguchi H., Tanaka H., Ashikari T., Sugimoto K., Shirahige K. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature. 2003;424:1078–1083. doi: 10.1038/nature01900. [DOI] [PubMed] [Google Scholar]
- 31.Lengronne A., Katou Y., Mori S., Yokobayashi S., Kelly G.P., Itoh T., Watanabe Y., Shirahige K., Uhlmann F. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature. 2004;430:573–578. doi: 10.1038/nature02742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ogiwara H., Ui A., Onoda F., Tada S., Enomoto T., Seki M. Dpb11, the budding yeast homolog of TopBP1, functions with the checkpoint clamp in recombination repair. Nucleic Acids Res. 2006;34:3389–3398. doi: 10.1093/nar/gkl411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kent N.A., Mellor J. Chromatin structure snap-shots: rapid nuclease digestion of chromatin in yeast. Nucleic Acids Res. 1995;23:3786–3787. doi: 10.1093/nar/23.18.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harvey A.C., Jackson S.P., Downs J.A. Saccharomyces cerevisiae histone H2A Ser122 facilitates DNA repair. Genetics. 2005;170:543–553. doi: 10.1534/genetics.104.038570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weinert T. DNA damage and checkpoint pathways: molecular anatomy and interactions with repair. Cell. 1998;94:555–558. doi: 10.1016/s0092-8674(00)81597-4. [DOI] [PubMed] [Google Scholar]
- 36.Skibbens R.V., Hieter P. Kinetochores and the checkpoint mechanism that monitors for defects in the chromosome segregation machinery. Annu. Rev. Genet. 1998;32:307–337. doi: 10.1146/annurev.genet.32.1.307. [DOI] [PubMed] [Google Scholar]
- 37.McAinsh A.D., Tytell J.D., Sorger P.K. Structure, function, and regulation of budding yeast kinetochores. Annu. Rev. Cell Dev. Biol. 2003;19:519–539. doi: 10.1146/annurev.cellbio.19.111301.155607. [DOI] [PubMed] [Google Scholar]
- 38.Measday V., Hieter P. Kinetochore sub-structure comes to MIND. Nat. Cell Biol. 2004;6:94–95. doi: 10.1038/ncb0204-94. [DOI] [PubMed] [Google Scholar]
- 39.Cleveland D.W., Mao Y., Sullivan K.F. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell. 2003;112:407–421. doi: 10.1016/s0092-8674(03)00115-6. [DOI] [PubMed] [Google Scholar]
- 40.Biggins S., Walczak C.E. Captivating capture: how microtubules attach to kinetochores. Curr. Biol. 2003;13:R449–R460. doi: 10.1016/s0960-9822(03)00369-5. [DOI] [PubMed] [Google Scholar]
- 41.Meluh P.B., Yang P., Glowczewski L., Koshland D., Smith M.M. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell. 1998;94:607–613. doi: 10.1016/s0092-8674(00)81602-5. [DOI] [PubMed] [Google Scholar]
- 42.Enquist-Newman M., Cheeseman I.M., Van Goor D., Drubin D.G., Meluh P.B., Barnes G. Dad1p, third component of the Duo1p/Dam1p complex involved in kinetochore function and mitotic spindle integrity. Mol. Biol. Cell. 2001;12:2601–2613. doi: 10.1091/mbc.12.9.2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Janke C., Ortiz J., Tanaka T.U., Lechner J., Schiebel E. Four new subunits of the Dam1-Duo1 complex reveal novel functions in sister kinetochore biorientation. EMBO J. 2002;21:181–193. doi: 10.1093/emboj/21.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Y., Bachant J., Alcasabas A.A., Wang Y., Qin J., Elledge S.J. The mitotic spindle is required for loading of the DASH complex onto the kinetochore. Genes Dev. 2002;16:183–197. doi: 10.1101/gad.959402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tanaka T., Cosma M.P., Wirth K., Nasmyth K. Identification of cohesin association sites at centromeres and along chromosome arms. Cell. 1999;98:847–858. doi: 10.1016/s0092-8674(00)81518-4. [DOI] [PubMed] [Google Scholar]
- 46.Harata M., Zhang Y., Stillman D.J., Matsui D., Oma Y., Nishimori K., Mochizuki R. Correlation between chromatin association and transcriptional regulation for the Act3p/Arp4 nuclear actin-related protein of Saccharomyces cerevisiae. Nucleic Acids Res. 2002;30:1743–1750. doi: 10.1093/nar/30.8.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sato M., Vardy L., Koonrugsa N., Tournier S., Millar J.B.A., Toda T. Deletion of Mia1/Alp7 activates Mad2-dependent spindle assembly checkpoint in fission yeast. Nat. Cell Biol. 2003;5:764–766. doi: 10.1038/ncb0903-764. [DOI] [PubMed] [Google Scholar]
- 48.Minoda A., Saitoh S., Takahashi K., Toda T. BAF53/Arp4 homolog Alp5 in fission yeast is required for histone H4 acetylation, kinetochore-spindle attachment, and gene silencing at centromere. Mol. Biol. Cell. 2005;16:316–327. doi: 10.1091/mbc.E04-06-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Measday V, Hailey D.W., Pot I., Givan S.A., Hyland K.M., Cagney G., Fields S., Davis T.N., Hieter P. Ctf3p, the Mis6 budding yeast homolog, interacts with Mcm22p and Mcm16p at the yeast outer kinetochore. Genes Dev. 2002;16:101–113. doi: 10.1101/gad.949302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Le Masson I., Yu D.Y., Jensen K., Chevalier A., Courbeyrette R., Boulard Y., Smith M.M., Mann C. Yaf9, a novel NuA4 histone acetyltransferase subunit, is required for the cellular response to spindle stress in yeast. Mol. Cell Biol. 2003;23:6086–6102. doi: 10.1128/MCB.23.17.6086-6102.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Krogan N.J., Baetz K., Keogh M.C., Datta N., Sawa C., Kwok T.C., Thompson N.J., Davey M.G., Pootoolal J., Hughes T.R., et al. Regulation of chromosome stability by the histone H2A variant Htz1, the Swr1 chromatin remodeling complex, and the histone acetyltransferase NuA4. Proc. Natl. Acad. Sci. USA. 2004;101:13513–13518. doi: 10.1073/pnas.0405753101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Keogh M.C., Mennella T.A., Sawa C., Berthelet S., Krogan N.J., Wolek A., Podolny V., Carpenter L.R., Greenblatt J.F., Baetz K., et al. The Saccharomyces cerevisiae histone H2A variant Htz1 is acetylated by NuA4. Genes Dev. 2006;20:660–665. doi: 10.1101/gad.1388106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Babiarz J.E., Halley J.E., Rine J. Telomeric heterochromatin boundaries require NuA4-dependent acetylation of histone variant H2A.Z in Saccharomyces cerevisiae. Genes Dev. 2006;20:700–710. doi: 10.1101/gad.1386306. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.