Abstract
Human respiratory syncytial virus (HRSV) is a major cause of a number of severe respiratory diseases, including bronchiolitis and pneumonia, in infants and young children. The HRSV F protein, a glycoprotein essential for viral entry, is a primary target for vaccine and drug development. Two heptad-repeat regions within the HRSV F sequence were predicted by the computer program learncoil-vmf. These regions are thought to form trimer-of-hairpins–like structures, similar to those found in the fusion proteins of several enveloped viruses. The hairpin structure likely brings the viral and cellular membranes into close apposition, thereby facilitating membrane fusion and subsequent viral entry. Here, we show that peptides, denoted HR-N and HR-C, corresponding to the heptad-repeat regions from the N-terminal and C-terminal segments of the HRSV F protein, respectively, form a stable α-helical trimer of heterodimers. The HRSV N/C complex was crystallized and its x-ray structure was determined at 2.3-Å resolution. As anticipated, the complex is a six-helix bundle in which the HR-N peptides form a three-stranded, central coiled coil, and the HR-C peptides pack in an antiparallel manner into hydrophobic grooves on the coiled-coil surface. There is remarkable structural similarity between the HRSV N/C complex and the fusion protein core of other viruses, including HIV-1 gp41. In addition, earlier work has shown that HRSV HR-C peptides, like the HIV-1 gp41 C peptides, inhibit viral infection. Thus, drug discovery and vaccine development strategies aimed at inhibiting viral entry by blocking hairpin formation may be applied to the inhibition of HRSV.
Keywords: x-ray crystallography, membrane fusion, pneumovirus
Human respiratory syncytial virus (HRSV), a member of the pneumovirus subfamily of the Paramyxoviridae family (1), is a leading cause of severe respiratory infections in infants and young children worldwide (2). A safe and effective HRSV vaccine is not yet available, so it is important to pursue potential therapeutic measures against HRSV infection.
HRSV carries three surface glycoproteins: F, G, and SH (1). The HRSV F protein is an attractive target for drug and vaccine development as it is essential for viral entry, is highly conserved, and is the major virus neutralization antigen (1). In contrast, the G protein is highly variable among different strains of HRSV (3), and the SH protein has been found to be dispensable for virus growth (4).
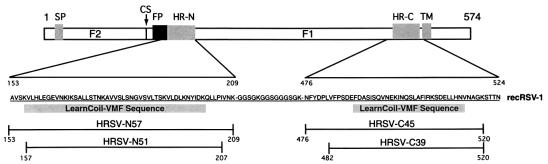
The F protein is synthesized as a 67-kDa precursor (denoted F0) that is processed by proteolytic cleavage to yield two disulfide-linked subunits: F1 and F2 (Fig. 1). The fusion peptide region, a hydrophobic/glycine-rich segment that inserts into the target cellular membrane during the fusion process, is located at the N terminus of the F1 subunit (1). The transmembrane segment is close to the C terminus of the F1 subunit. Adjacent to both the fusion peptide and transmembrane segments are two regions containing 4,3-hydrophobic heptad repeats (HR), a sequence motif suggestive of coiled-coil structures (5–7). These regions are denoted HR-N and HR-C, respectively, and are separated by an intervening domain of ≈270 residues. In general, the arrangement of these structural elements in the F1 protein is highly conserved among the Paramyxoviridae family (8).
Figure 1.
Schematic diagram of the HRSV F protein sequence. F1 and F2 are formed after proteolytic cleavage of the precursor F0. The signal peptide (SP), the cleavage site (CS), the putative fusion peptide (FP), the N-terminal HR (HR-N) region, the C-terminal HR (HR-C) region, and the transmembrane segment (TM) within the amino acid sequence of the recombinant recRSV-1 construct are indicated. The HR-N and HR-C regions predicted by learncoil-vmf are represented by shaded boxes. Protease-resistant fragments from trypsin (HRSV-N57 and HRSV-C45) and proteinase K (HRSV-N51 and HRSV-C39) cleavage are indicated.
Previous studies with viral fusion proteins (ref. 7 and references therein), including hemagglutinin of influenza (9, 10), TM subunit of Moloney murine leukemia virus (11), gp41 of HIV-1 and simian immunodeficiency virus (12–17), GP2 of Ebola (18, 19), gp21 of human T cell leukemia virus type 1 (20), and F1 of simian parainfluenza virus 5 (SV5) (21), suggest that HR-N and HR-C form trimeric hairpin-like structures, with the HR-C regions packing in an antiparallel manner against the inner coiled coil formed by the HR-N regions. The likely role of the hairpin structure is to facilitate apposition of the viral and cellular membranes by bringing the fusion peptide, which inserts into the cellular membrane, close to the transmembrane segment, which is anchored in the viral membrane. Hence, the trimer-of-hairpins structure presumably corresponds to the fusion-active state, distinct from the native (nonfusogenic) state. But whether hairpin formation precedes the actual membrane-fusion event or occurs simultaneously with fusion is unknown.
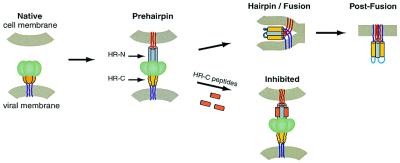
There is extensive biochemical evidence to suggest that viral fusion proteins undergo conformational changes when activated (22, 23). Earlier work on the HIV-1 fusion protein gp41 led to a model for viral membrane fusion (Fig. 2) in which gp41 exists in at least three different conformations: (i) the native (nonfusogenic) form, where the fusion peptide and HR-N region are inaccessible, (ii) the prehairpin intermediate, where the HR-N region is exposed and the fusion peptide is projected into the target cellular membrane, and (iii) the fusogenic hairpin form, where the HR-N and HR-C regions associate and bring the viral and the cellular membranes together to facilitate fusion (24).
Figure 2.
A model for virus–cell membrane fusion (24). The model shown is based primarily on studies of the HIV-1 gp41-mediated membrane fusion process. The HRSV F protein likely undergoes similar conformational changes. In the native state, the fusion peptide (not shown) is inaccessible. Upon activation, the fusion protein undergoes a conformational change to the prehairpin intermediate, in which the fusion peptide (red lines) is inserted into the target-cell membrane, and the HR-N peptide region (gray) is a trimeric coiled coil. The HR-C peptide region (yellow) has not yet associated with the N peptide coiled coil. This intermediate is vulnerable to HR-C peptide inhibition (Lower, inhibitory peptides shown in orange). The prehairpin intermediate resolves to the fusion-active hairpin structure when the HR-C peptide region binds to the HR-N peptide coiled coil and adopts a helical conformation. This rearrangement results in membrane apposition. Whether hairpin formation precedes the actual membrane-fusion event or occurs simultaneously with fusion is unknown. Figure was adapted from Chan and Kim (24).
Agents that prevent these conformational changes by stabilizing the native or intermediate states are expected to prevent fusion activation and thus block viral entry. In the case of HIV-1 gp41, peptides corresponding to the HR-C region of gp41, referred to as C peptides, can effectively inhibit infection (24). One such C peptide is in clinical trials and shows antiviral activity in humans (25). C peptides function in a dominant-negative manner by binding to the transiently exposed coiled coil in the prehairpin intermediate and consequently blocking the formation of the fusion-active hairpin structures (24).
A similar approach may be effective in identifying inhibitors of HRSV infection. HR-C regions from HRSV have been shown to inhibit viral infection and thus provide evidence that the F protein also may display the prehairpin intermediate conformation (26). Analogous to the HIV-1 C peptides, these HRSV HR-C peptides likely act in a dominant-negative manner to prevent the formation of final fusion-active structures, thereby inhibiting viral entry.
To verify that the HRSV F1 protein core indeed forms a trimer-of-hairpins structure and to provide a structural basis for the development of fusion inhibitors, we characterized the interaction of the two HR regions of HRSV F1 protein. We identified two peptides, HRSV-N57 and HRSV-C45, which correspond to the HR-N and HR-C regions of HRSV F1 protein, respectively, that form a stable trimer of heterodimers. This result is consistent with recent reports that indicate the HR-N and HR-C regions from HRSV associate to form an α-helical trimer of heterodimers (27, 28). The HRSV-N57/C45 complex characterized here was crystallized, and its x-ray structure was determined to 2.3-Å resolution. The structure confirms the similarity between the HRSV F1 protein core and several other viral fusion proteins, including HIV-1 gp41, suggesting that methods for discovery of potential therapeutics and prophylactics developed against HIV-1 gp41 also might apply to the HRSV F1 protein.
Materials and Methods
Gene Construction and Purification of recRSV-1.
Using optimal codons for Escherichia coli expression (29), a synthetic gene sequence denoted recRSV-1 was constructed that encodes residues 153–209 and 476–524 of HRSV (strain RSS-2; Swiss-Prot accession no. P11209), connected by a glycine-rich linker (Fig. 1). A factor Xa cleavage site was incorporated upstream of the HRSV coding sequence. The constructed gene was inserted into the BamHI–HindIII restriction site of the hexahistidine expression vector pQE9 (Qiagen, Chatsworth, CA). The resulting plasmid, denoted pRSV-1, was transformed into E. coli XL1-Blue competent cells for protein expression. Cells were grown in Luria–Bertani medium to an optical density of 0.6 at 600 nm. Protein expression then was induced with 1 mM isopropyl-β-D-thiogalactopyranoside, and cells were harvested after 3 h.
Cells were lysed in 6 M guanidine-HCl, and the lysate was clarified by centrifugation. The recombinant protein was purified by nickel-nitrilotriacetic acid metal-affinity chromatography, followed by reverse-phase HPLC (Waters) using a Vydac C18 preparative column (Vydac, Hesperia, CA) with a water/acetonitrile gradient of 0.1%/min in the presence of 0.1% trifluoroacetic acid. The mass of the purified protein was verified by mass spectrometry on a Voyager Elite matrix-assisted laser desorption ionization-time of flight mass spectrometer (PerSeptive Biosystems, Framingham, MA). The protein was lyophilized and resuspended in ultrapure water and dialyzed against factor Xa cleavage buffer (50 mM Tris⋅HCl, pH 8.0/100 mM NaCl/2 mM CaCl2). To remove the His tag, factor Xa was added at a 1:500 wt/wt ratio of protease to tagged protein, and the reaction was incubated for 2 days at room temperature. The cleavage mixture then was purified by reverse-phase HPLC on a Vydac C18 preparative column. Peak fractions containing recRSV-1 were verified by mass spectrometry and lyophilized.
Proteolysis.
All proteolysis reactions were performed with 1 mg/ml protein and 0.1 mg/ml protease in PBS, pH 7.4, at room temperature and quenched with 2 mM PMSF (final concentration). Proteolysis samples were analyzed by reverse-phase HPLC connected to an LCQ electrospray mass spectrometer (Finnigan-MAT, San Jose, CA). Fragments were assigned by matching observed masses with a list of possible fragment masses predicted by the computer program fragment mass (E. Wolf and P. S. Kim, http://www.wi.mit.edu/kim/computing.html). Assigned fragments were within 1 Da of their predicted values.
CD Spectroscopy.
CD spectra were measured at 10 μM protein concentration in PBS buffer with an AVIV 62 DS spectrometer (Aviv Associates, Lakewood, NJ) as described (30). Protein concentrations were determined by absorbance at 280 nm in 20 mM phosphate-buffered 6 M guanidine-HCl (pH 6.5) (31).
Sedimentation Equilibrium Analysis.
Sedimentation equilibrium analysis was performed on a Beckman XLA-90 analytical ultracentrifuge (Beckman Instruments) at 15,000 rpm and 20,000 rpm, and data were collected after spinning for 18 h at 20°C. Three protein samples at concentrations of 10, 50, and 100 μM were spun, following dialysis against PBS buffer overnight. Data analyses were performed as described (32).
Purification, Crystallization, and Structure Determination of the HRSV-N57/C45 Trimer.
The HRSV-N57 and HRSV-C45 peptides were generated by trypsin digestion of recRSV-1 protein and purified to homogeneity by reverse-phase HPLC on a Vydac C18 preparative column. The purified HRSV peptides were lyophilized and dissolved in water and 10 mM Tris⋅HCl (pH 8.5), respectively. Equimolar amounts of HRSV-N57 and HRSV-C45 were mixed, and the complex was separated from the free peptide and aggregated species by gel filtration on a Sephacryl S-100 HR column (Amersham Pharmacia) in buffer (10 mM Tris⋅HCl, pH 8.5/50 mM NaCl). The purified HRSV-N57/C45 complex was concentrated to 10 mg/ml.
Crystals were obtained by using the hanging drop method by equilibrating 2-μl drops (protein solution mixed 1:1 with reservoir solution) against a reservoir solution containing 24–26% polyethylene glycol 4000/200 mM Tris⋅HCl, pH 8.5/300 mM Li2SO4. The crystals belong to the space group P1 (a = 67.9 Å, b = 71.5 Å, c = 76.5 Å, α = 81.3°, β = 73.8°, γ = 60.7°). There are four trimers per asymmetric unit that give rise to a solvent content of ≈46%. For data collection, crystals were directly mounted in nylon loops (Hampton Research, Riverside, CA) from the drop and flash-frozen in liquid nitrogen. Diffraction data were collected at 100 K at the Howard Hughes Medical Institute Beamline X4A at Brookhaven National Laboratory, Brookhaven, NY, using a Quantum-4 CCD detector. Diffraction intensities were integrated by using denzo and scalepack software (33) and reduced to structure factors with the program truncate from the CCP4 program suite (34). The structure of the HRSV-N57/C45 complex was solved by molecular replacement with the program amore (47). A polyserine model derived from the SV5 N1/C1 trimer (21) was used as a search model. Initially, only three molecules per asymmetric unit were found. After applying solvent flattening, histogram matching, and 9-fold noncrystallographic averaging with the program dm (35), the electron density map was interpretable in most regions. Three HRSV-N57/C45 trimer molecules were built into the density using the program o (36). After a few cycles of refinement, the Rfree value remained high, and the existence of unassigned continuous density was suggestive of a fourth molecule. Molecular replacement with a partially refined HRSV-N57/C45 trimer molecule readily located the fourth trimer in the unit cell. Crystallographic refinement of the structure (Table 1) was done with the cns programs (37) (reflections beyond 2.3 Å were not used due to poor quality). Noncrystallographic symmetry restraints were applied during the first few cycles of refinement but then were removed and not used in the final refinement. The quality of coordinates was examined by procheck (38). No residues were in disallowed regions, and 94.5% were in the most favored regions of the Ramachandran plot.
Table 1.
X-ray data collection and refinement statistics
| Data collection | |
| Resolution range, Å | 20.0–2.20 |
| Observed reflections | 119,905 |
| Unique reflections | 56,973 |
| Completeness, % | 91.2 (64.7)* |
| Rmerge† | 0.041 (0.269)* |
| Refinement | |
| Resolution range, Å | 10.0–2.30 |
| Protein nonhydrogen atoms | 8073 |
| Water molecules | 888 |
| Rcryst‡ | 0.233 |
| Rfree‡ | 0.286 |
| rms difference from ideal geometry | |
| Bond lengths, Å | 0.008 |
| Bond angles, ° | 1.2 |
| Average B-factor, Å2 | 45.4 |
Values in parentheses correspond to highest resolution shell 2.28 to 2.20 Å.
Rmerge = ∑∑j | Ij (hkl) − 〈I(hkl)〉 |/∑∑j | 〈I(hkl)〉 |, where Ij is the intensity measurement for reflection hkl and 〈I〉 is the mean intensity over j reflections.
Rcryst (Rfree) = ∑ | |Fobs(hkl) | − | Fcalc(hkl) | |/∑ | Fobs (hkl) | , where Fobs and Fcalc are observed and calculated structure factors, respectively. No σ-cutoff was applied. 10% of the reflections were excluded from refinement and used to calculate Rfree.
Results
HRSV-N57 and HRSV-C45 Peptides Form a Stable Trimeric α-Helical Complex.
learncoil-vmf, a program for identifying coiled-coil-like regions in viral fusion proteins, predicts such regions in the HRSV F1 protein (7). The first region (residues 156–201) is near the putative fusion peptide, and the second region (residues 488–516) is adjacent to the transmembrane segment (Fig. 1).
To study the regions predicted by learncoil-vmf, we constructed a recombinant protein, recRSV-1, corresponding to residues 153–209 and residues 476–524 of HRSV F protein connected by a glycine-rich linker (see Materials and Methods) (Fig. 1). CD spectroscopy indicates that recRSV-1 is highly helical (≈70% helix content) and extremely thermostable (the midpoint of thermal denaturation [Tm] ≈90°C in the presence of 4 M guanidine-HCl, pH 7.4); recRSV-1 forms a trimer in PBS buffer as judged by sedimentation equilibrium analysis (X.Z. and P.S.K., unpublished results).
Proteolysis of the recRSV-1 protein reveals a protease-resistant core containing the entire HR-N and HR-C regions predicted by learncoil-vmf. Digestion of recRSV-1 by trypsin generates two major fragments readily separated by reverse-phase HPLC. Mass spectrometry analysis unambiguously assigns these as an N-terminal fragment corresponding to residues 153–209, denoted HRSV-N57 [observed molecular weight (MW) = expected MW = 6,130], and a C-terminal fragment corresponding to residues 476–520, denoted HRSV-C45 (observed MW = expected MW = 5,111) (Fig. 1).
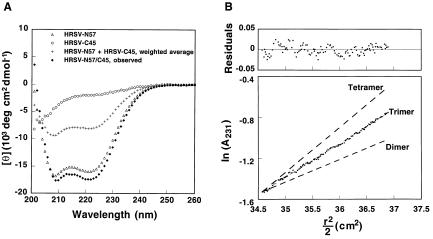
The isolated HRSV-N57 peptide folds into an α-helical structure in PBS (>40% helix content) as determined by CD but does not form a monodisperse species according to sedimentation equilibrium analysis (X.Z. and P.S.K., unpublished results). The apparent Tm of HRSV-N57 varies (≈40–50°C at 10 μM in PBS), presumably due to aggregation. In contrast, the isolated HRSV-C45 peptide is unfolded, and the CD signal does not show a thermal unfolding transition (X.Z. and P.S.K., unpublished results). The CD spectrum of an equimolar mixture of the HRSV-N57 and HRSV-C45 peptides shows a substantially higher α-helical signal than the weighted average of the spectra of the individual peptides, indicating a major conformational change upon association of the two peptides (Fig. 3A). The HRSV-N57/C45 complex has an apparent Tm of 88°C at 10 μM in PBS (X.Z. and P.S.K., unpublished results). Sedimentation equilibrium centrifugation clearly indicates that in the HRSV-N57/C45 complex the peptides are present in a 3:3 ratio (Fig. 3B). These results are in agreement with those of Lambert and colleagues (28).
Figure 3.
Biophysical characterization of HRSV peptides. (A) CD spectra of HRSV-N57, HRSV-C45, and HRSV-N57/C45 in PBS at 25°C. The predicted spectrum for noninteracting HRSV-N57 plus HRSV-C45 peptides is shown for comparison. (B) Equilibrium sedimentation of HRSV-N57/C45 at 20°C and 15,000 rpm in PBS. The data fit closely to a trimeric model. Lines expected for dimeric and tetrameric models are indicated for comparison. The deviation in the data from the linear fit for a trimeric model is plotted (Upper). No systematic deviation in the residuals is observed.
HRSV-N57/C45 can be further trimmed by proteinase K to generate smaller fragments denoted HRSV-N51 (residues 157–207) and HRSV-C39 (residues 482–520) (Fig. 1). Proteinase K is not sequence specific; therefore, its proteolytic fragments are expected to better represent the boundaries of a well-folded domain. The HRSV-N51/C39 complex has an apparent Tm of 88°C at 10 μM in PBS (X.Z. and P.S.K., unpublished results), the same as that of HRSV-N57/C45. Thus, the extra-terminal residues in HRSV-N57 and HRSV-C45 do not seem to contribute to the stability of the core complex.
Crystal Structure of HRSV-N57/C45.
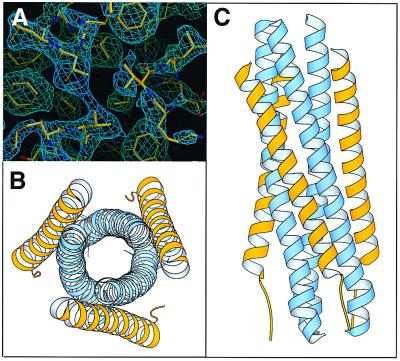
Crystals of HRSV-N57/C45 contain four trimers of HRSV-N57/C45 heterodimers per unit cell, giving rise to 12 independently refined heterodimers. Both the final 2Fo–Fc map and simulated annealing omit map are readily interpretable (Fig. 4A). However, some terminal residues, including 153–159, 208–209, 476–479, and 517–520, are disordered to a different degree among different chains. Although it is possible that this variation is due to the differences in local lattice contact environments, these terminal residues are more likely to be intrinsically flexible, especially in solution, as they are removed in the proteinase K-digested HRSV-N57/C45.
Figure 4.
Crystal structure of HRSV-N57/C45 complex. (A) A region of a simulated annealing omit map of HRSV-N57/C45, calculated with 2Fo–Fc coefficients in the absence of the fragments shown. The map is displayed as a map cover contoured at 1σ. The view is down the 3-fold axis of the trimer. Figure was generated with the program o (36). (B and C) Ribbon diagram of the HRSV-N57/C45 trimer. N helices are shown in blue and C helices in yellow. (B) A top view of the HRSV-N57/C45 trimer looking down the 3-fold axis of the trimer. (C) A side view with the amino termini of the N helices pointing toward the top of the page and those of the C helices pointing toward the bottom. Figure was prepared with molscript (45).
We focused our structural analysis on the well-defined regions, including residues 160–207 in HRSV-N57 and residues 480–516 in HRSV-C45. For these regions, the structural differences between the 12 copies of the HRSV-N57/C45 heterodimer are small (e.g., the average rms difference in Cα positions between any pair of heterodimers is 0.34 Å).
HRSV-N57/C45 forms a rod-shaped, six-helix bundle of approximately 68 Å in length and 27 Å in diameter (Fig. 4 B and C). The HRSV-N57 peptides form a three-stranded coiled coil. Three HRSV-C45 peptides pack in an antiparallel manner against long, hydrophobic grooves formed on the surface of the HRSV-N57 core. This packing arrangement would put the fusion peptide segment, located immediately before HRSV-N57, and transmembrane segment, located immediately after HRSV-C45, in close proximity.
The Interface Between the HRSV-N57 Core and HRSV-C45.
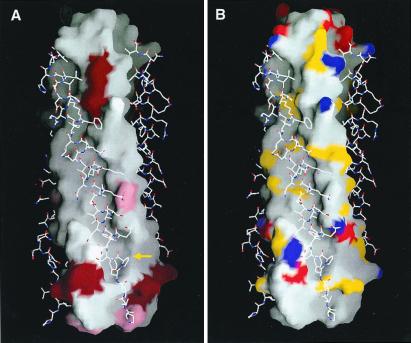
Approximately 3,000 Å2 of solvent-accessible surface area are buried at the interface between the HRSV-N57 core and each HRSV-C45 peptide. Twenty amino acid residues from each HRSV-C45 peptide and 26 residues from two adjacent HRSV-N57 peptides contribute to the interface interaction. These residues are highly conserved among different HRSV strains, including 20 HRSV isolates and seven bovine respiratory syncytial virus isolates (analysis based on sequences available in GenBank and Swiss-Prot as of September 2000) (Fig. 5A). Most of the amino acid differences between strains are limited to conservative mutations.
Figure 5.
The surface properties of the HRSV-N57 trimer with the HRSV-C45 peptides displayed in a stick-style representation. View same as Fig. 4C. (A) Surface variability of the HRSV-N57 trimer (analysis based on RSV sequences available in GenBank and Swiss-Prot). The residues shown in dark red vary among different human virus strains. The residues shown in pink are identical among 20 human strains of HRSV but are different in bovine respiratory syncytial virus. The cavity region is indicated by a yellow arrow. (B) Surface mapping of groups with the potential to form electrostatic and polar interactions. Nitrogen and oxygen atoms from charged amino acid side chains are shown in blue and red, respectively. Nitrogen and oxygen atoms from polar amino acid side chains are shown in yellow. Figure was drawn with the program grasp (46).
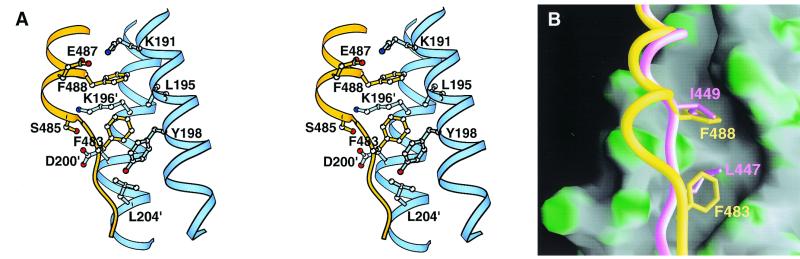
Various features of the deep groove on the surface of the HRSV-N57 trimer (Fig. 5B) suggest that this groove is a potentially attractive drug target. The carboxyl-terminal end of the groove features a hydrophobic cavity. Within the cavity region, the majority of contacts are between six cavity-lining residues from the HRSV-N57 peptides (Lys-191, Leu-195, and Tyr-198 from one chain and Lys-196, Asp-200, and Leu-204 from another) and two aromatic residues, Phe-483 and Phe-488, from the HRSV-C45 peptide (Fig. 6). A patch in the middle of the hydrophobic groove (Fig. 5B) is enriched with polar residues. Multiple hydrogen bonds are observed in this area. For example, Ser-509 Oγ of the HRSV-C45 peptide forms an elaborate hydrogen-bond network with Thr-174 Oγ1, Ala-170 O, and Asn-175 Nδ2 from the HRSV-N57 helices. The abundance of hydrogen-bond donor/acceptors in the groove is likely to greatly enhance binding specificity due to the requirement of electrostatic complementarity. Thus, this cavity and groove are potentially good binding sites for small molecule inhibitors (cf. refs. 39–41).
Figure 6.
The cavity on the surface of the HRSV-N57 coiled coil. (A) A stereo view of interactions in the HRSV-N57 cavity. Two phenylalanine residues of HRSV-C45 (yellow) fit into the cavity formed by two neighboring HRSV-N57 peptides (blue). A prime symbol is used to distinguish residues from the two HRSV-N57 chains. Figure was prepared with molscript (45). (B) Comparison of cavity interactions of the HRSV and SV5 structures. The HRSV-N57 coiled coils, which superimpose closely with the SV5 N peptide coiled coils (21), are represented as a molecular surface (the most convex part shown in green and the most concave in gray). The HRSV-C45 (yellow) and SV5 C peptide (pink) helices are shown as ribbons with selected side chains that pack into the cavity. The relative shift of the C peptides is clearly visible. Figure was drawn with the program grasp (46).
Comparison with SV5 F1 Trimer Core and Other Viral Fusion Proteins.
Among paramyxovirus fusion proteins, only the structure of the SV5 F1 trimer core has been determined (21). In general, the F1 core from HRSV and SV5 adopt a similar fold, despite a low sequence identity between the two proteins (18.4% for the entire protein sequence and 19.1% for the HR regions). These structures can be superimposed with an rms difference of 2.1 Å between all Cα atoms. The central N peptide cores superimpose closely, with an rms difference of 0.65 Å, whereas the C peptides are more divergent, with an rms difference of 2.9 Å.
A stutter or increase in spacing pattern (3-4-4-4-3) for HRs, instead of the regular 3-4-3-4-3, has been observed in the SV5 F1 trimer core (21). A similar stutter is observed within the last two HRs in the HRSV-N57 coiled-coil core. This supports the prediction by Baker et al. that the 3-4-4-4-3 pattern will be conserved among paramyxovirus F proteins because it can maintain hydrophobic residues within the coiled-coil core (21). However, because the stutter in HRSV-N57 occurs at the end of the peptide helix, structural irregularities resulting from helix end effects cannot be ruled out.
It has been suggested that the C peptides of HRSV and other paramyxoviruses will have a similar main-chain conformation as observed in the SV5 crystal structure (21). However, our results show that the HRSV C peptide has one additional helical turn not observed in the SV5 C peptide. As a consequence, the position of the C peptide residues that pack into the hydrophobic cavity is shifted in HRSV compared with SV5 (Fig. 6B), and such a packing arrangement is not readily predicted from primary sequence alignment of the two proteins.
Discussion
The structure of HRSV-N57/C45 adds to the repertoire of viral fusion proteins that have been shown to form a trimer-of-hairpins motif. The remarkable similarity between the HRSV F1 and HIV-1 gp41 core structures, as well as similar C peptide inhibition phenomena, suggest a conserved mechanism of fusion. Similar to HIV-1 gp41, HRSV F1 likely undergoes a series of conformational changes to become fusion active. The distinct conformational states proposed for HIV-1 gp41 (24), including the native state, the prehairpin intermediate, and fusion-active hairpin state, also may apply to HRSV F1. Electron microscopy studies of the full-length F protein reveal two distinct conformations, one cone-shaped and the other lollipop-shaped, which may represent the native and fusion-active states, respectively (42). The HRSV-N57/C45 structure presented here likely corresponds to the fusion-active hairpin conformation, as the hairpin formation is structurally coupled to the apposition of viral and cellular membranes.
The existence of the prehairpin intermediate conformation for the HRSV F protein is strongly supported by the observation that peptides corresponding to the HRSV HR-C regions can efficiently inhibit viral fusion (26). Presumably, these HR-C peptides function by binding to the exposed HR-N regions, thereby blocking the conformational transition to the fusion-active form.
If small, oral, bioavailable molecules that disrupt hairpin formation are identified, they may be effective drugs against viral infection. In the case of HIV-1, a strategy to block hairpin formation has been developed to find D peptide inhibitors of viral entry that bind to a hydrophobic pocket on the surface of the central coiled coil consisting of HIV-1 gp41 N peptides (39). The cavity and well-defined groove on the surface of the central coiled coil of the HRSV-N57/C45 complex identified here may serve as useful drug targets.
Finally, the HRSV-N57/C45 structure may provide a new direction in vaccine development against HRSV infection. Antibodies from vaccine trials using purified, native, fulllength F protein have very low neutralizing activities compared with those generated by live HRSV (1). One suggested strategy for eliciting neutralizing HIV-1 antibodies is to target transient intermediates or fusion-competent conformations (39, 43). The N-terminal coiled coil observed in the current structure might be formed in a prehairpin intermediate analogous to that found in HIV-1 and thus may be a viable target for antifusion antibodies. A neutralizing antibody against HRSV has been mapped to the N-terminal coiled-coil region (44); it will be interesting to see whether it targets a prehairpin intermediate.
Acknowledgments
We thank James Pang for mass spectrometric analysis; Alexandra Evindar and Leslie Gaffney for assistance in preparation of the manuscript and figures; Christopher Liu for valuable comments on the manuscript; Michael Burgess, David Akey, and Brian Schneider for technical assistance and advice; and members of the Kim lab for helpful discussions. We also thank Dr. Craig Ogata and the staff of the Howard Hughes Medical Institute beamline (X4A) at Brookhaven National Laboratory for help in data collection. X.Z. was supported by a postdoctoral fellowship from the Anna Fuller Fund. This work was supported by National Institutes of Health Grants GM44162 and GM56552 and used the W.M. Keck Foundation x-ray Crystallography Facility at the Whitehead Institute.
Abbreviations
- HRSV
human respiratory syncytial virus
- HR
heptad repeat
- SV5
simian parainfluenza virus 5
Footnotes
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1G2C).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.260499197.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.260499197
References
- 1.Collins P L, McIntosh K, Chanock R M. In: Fields Virology. Fields B N, Knipe D M, Howley P M, editors. Philadelphia: Lippincott; 1996. pp. 1313–1351. [Google Scholar]
- 2.Heilman C A. J Infect Dis. 1990;161:402–406. doi: 10.1093/infdis/161.3.402. [DOI] [PubMed] [Google Scholar]
- 3.Johnson P R, Spriggs M K, Olmsted R A, Collins P L. Proc Natl Acad Sci USA. 1987;84:5625–5629. doi: 10.1073/pnas.84.16.5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bukreyev A, Whitehead S S, Murphy B R, Collins P L. J Virol. 1997;71:8973–8982. doi: 10.1128/jvi.71.12.8973-8982.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chambers P, Pringle C R, Easton A J. J Gen Virol. 1990;71:3075–3080. doi: 10.1099/0022-1317-71-12-3075. [DOI] [PubMed] [Google Scholar]
- 6.Buckland R, Wild F. Nature (London) 1989;338:547. doi: 10.1038/338547a0. [DOI] [PubMed] [Google Scholar]
- 7.Singh M, Berger B, Kim P S. J Mol Biol. 1999;290:1031–1041. doi: 10.1006/jmbi.1999.2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lamb R A. Virology. 1993;197:1–11. doi: 10.1006/viro.1993.1561. [DOI] [PubMed] [Google Scholar]
- 9.Bullough P A, Hughson F M, Treharne A C, Ruigrok R W, Skehel J J, Wiley D C. J Mol Biol. 1994;236:1262–1265. doi: 10.1016/0022-2836(94)90027-2. [DOI] [PubMed] [Google Scholar]
- 10.Chen J, Skehel J J, Wiley D C. Proc Natl Acad Sci USA. 1999;96:8967–8972. doi: 10.1073/pnas.96.16.8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fass D, Harrison S C, Kim P S. Nat Struct Biol. 1996;3:465–469. doi: 10.1038/nsb0596-465. [DOI] [PubMed] [Google Scholar]
- 12.Chan D C, Fass D, Berger J M, Kim P S. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 13.Weissenhorn W, Dessen A, Harrison S C, Skehel J J, Wiley D C. Nature (London) 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 14.Tan K, Liu J, Wang J-H, Shen S, Lu M. Proc Natl Acad Sci USA. 1997;94:12303–12308. doi: 10.1073/pnas.94.23.12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malashkevich V N, Chan D C, Chutkowski C T, Kim P S. Proc Natl Acad Sci USA. 1998;95:9134–9139. doi: 10.1073/pnas.95.16.9134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caffrey M, Cai M, Kaufman J, Stahl S J, Wingfield P T, Covell D G, Gronenborn A M, Clore G M. EMBO J. 1998;17:4572–4584. doi: 10.1093/emboj/17.16.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang Z N, Mueser T C, Kaufman J, Stahl S J, Wingfield P T, Hyde C C. J Struct Biol. 1999;126:131–144. doi: 10.1006/jsbi.1999.4116. [DOI] [PubMed] [Google Scholar]
- 18.Malashkevich V N, Schneider B J, McNally M L, Milhollen M A, Pang J X, Kim P S. Proc Natl Acad Sci USA. 1999;96:2662–2667. doi: 10.1073/pnas.96.6.2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weissenhorn W, Carfi A, Lee K H, Skehel J J, Wiley D C. Mol Cell. 1998;2:605–616. doi: 10.1016/s1097-2765(00)80159-8. [DOI] [PubMed] [Google Scholar]
- 20.Kobe B, Center R J, Kemp B E, Poumbourios P. Proc Natl Acad Sci USA. 1999;96:4319–4324. doi: 10.1073/pnas.96.8.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baker K A, Dutch R E, Lamb R A, Jardetzky T S. Mol Cell. 1999;3:309–319. doi: 10.1016/s1097-2765(00)80458-x. [DOI] [PubMed] [Google Scholar]
- 22.Hughson F M. Curr Biol. 1997;7:R565–R569. doi: 10.1016/s0960-9822(06)00283-1. [DOI] [PubMed] [Google Scholar]
- 23.Hernandez L D, Hoffman L R, Wolfsberg T G, White J M. Annu Rev Cell Dev Biol. 1996;12:627–661. doi: 10.1146/annurev.cellbio.12.1.627. [DOI] [PubMed] [Google Scholar]
- 24.Chan D C, Kim P S. Cell. 1998;93:681–684. doi: 10.1016/s0092-8674(00)81430-0. [DOI] [PubMed] [Google Scholar]
- 25.Kilby J M, Hopkins S, Venetta T M, DiMassimo B, Cloud G A, Lee J Y, Alldredge L, Hunter E, Lambert D, Bolognesi D, et al. Nat Med. 1998;4:1302–1307. doi: 10.1038/3293. [DOI] [PubMed] [Google Scholar]
- 26.Lambert D M, Barney S, Lambert A L, Guthrie K, Medinas R, Davis D E, Bucy T, Erickson J, Merutka G, Petteway S R., Jr Proc Natl Acad Sci USA. 1996;93:2186–2191. doi: 10.1073/pnas.93.5.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matthews J M, Young T F, Tucker S P, Mackay J P. J Virol. 2000;74:5911–5920. doi: 10.1128/jvi.74.13.5911-5920.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lawless-Delmedico M K, Sista P, Sen R, Moore N C, Antczak J B, White J M, Greene R J, Leanza K C, Matthews T J, Lambert D M. Biochemistry. 2000;39:11684–11695. doi: 10.1021/bi000471y. [DOI] [PubMed] [Google Scholar]
- 29.Makrides S C. Microbiol Rev. 1996;60:512–538. doi: 10.1128/mr.60.3.512-538.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fass D, Kim P S. Curr Biol. 1995;5:1377–1383. doi: 10.1016/s0960-9822(95)00275-2. [DOI] [PubMed] [Google Scholar]
- 31.Edelhoch H. Biochemistry. 1967;6:1948–1954. doi: 10.1021/bi00859a010. [DOI] [PubMed] [Google Scholar]
- 32.Laue T M, Shah B D, Ridgeway T M, Pelletier S L. In: Analytical Ultracentrifugation in Biochemistry and Polymer Science. Harding S E, Rowe A J, Horton J C, editors. Cambridge: Royal Society of Chemistry; 1992. pp. 90–125. [Google Scholar]
- 33.Otwinowski Z. In: Data Collection and Processing. Sawer L, Isaacs N, Bailey S, editors. Daresbury Laboratory, Warrington, U.K.: SERC; 1993. pp. 55–62. [Google Scholar]
- 34.CCP4. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 35.Kowtan K. Proteins. 1994;31:34–38. [Google Scholar]
- 36.Jones T A, Zou J W, Cowan S, Kjeldgaard M. Acta Crystallogr D. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 37.Brünger A T, Adams P D, Clore G M, DeLano W L, Gros P, Grosse-Kunstleve R W, Jiang J-S, Kuszewski J, Nilges M, Pannu N S, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 38.Laskowski R A, MacArthur M W, Moss D S, Thornton J M. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 39.Eckert D M, Malashkevich V N, Hong L H, Carr P A, Kim P S. Cell. 1999;99:103–115. doi: 10.1016/s0092-8674(00)80066-5. [DOI] [PubMed] [Google Scholar]
- 40.Chan D C, Chutkowski C T, Kim P S. Proc Natl Acad Sci USA. 1998;95:15613–15617. doi: 10.1073/pnas.95.26.15613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ferrer M, Kapoor T M, Strassmaier T, Weissenhorn W, Skehel J J, Oprian D, Schreiber S L, Wiley D C, Harrison S C. Nat Struct Biol. 1999;6:953–960. doi: 10.1038/13324. [DOI] [PubMed] [Google Scholar]
- 42.Calder L J, Gonzalez-Reyes L, Garcia-Barreno B, Wharton S A, Skehel J J, Wiley D C, Melero J A. Virology. 2000;271:122–131. doi: 10.1006/viro.2000.0279. [DOI] [PubMed] [Google Scholar]
- 43.LaCasse R A, Follis K E, Trahey M, Scarborough J D, Littman D R, Nunberg J H. Science. 1999;283:357–362. doi: 10.1126/science.283.5400.357. [DOI] [PubMed] [Google Scholar]
- 44.Langedijk J P, Meloen R H, van Oirschot J T. Arch Virol. 1998;143:313–320. doi: 10.1007/s007050050288. [DOI] [PubMed] [Google Scholar]
- 45.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 46.Nicholls A, Sharp K A, Honig B. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 47.Navaza J. Acta Crystallogr A. 1994;50:157–163. [Google Scholar]