Abstract
Conflicting reports have suggested that the silent information regulator 2 (SIR2) protein family employs NAD+ to ADP-ribosylate histones [Tanny, J. C., Dowd, G. J., Huang, J., Hilz, H. & Moazed, D. (1999) Cell 99, 735–745; Frye, R. A. (1999) Biochem. Biophys. Res. Commun. 260, 273–279], deacetylate histones [Landry, J., Sutton, A., Tafrov, S. T., Heller, R. C., Stebbins, J., Pillus, L. & Sternglanz, R. (2000) Proc. Natl. Acad. Sci. USA 97, 5807–5811; Smith, J. S., Brachmann, C. B., Celic, I., Kenna, M. A., Muhammad, S., Starai, V. J., Avalos, J. L., Escalante-Semerena, J. C., Grubmeyer, C., Wolberger, C. & Boeke, J. D. (2000) Proc. Natl. Acad. Sci. USA 97, 6658–6663], or both [Imai, S., Armstrong, C. M., Kaeberlein, M. & Guarente, L. (2000) Nature (London) 403, 795–800]. Uncovering the true enzymatic function of SIR2 is critical to the basic understanding of its cellular function. Therefore, we set out to authenticate the reaction products and to determine the intrinsic catalytic mechanism. We provide direct evidence that the efficient histone/protein deacetylase reaction is tightly coupled to the formation of a previously unidentified acetyl-ADP-ribose product (1-O-acetyl-ADP ribose). One molecule of NAD+ and one molecule of acetyl-lysine are readily catalyzed to one molecule of deacetylated lysine, nicotinamide, and 1-O-acetyl-ADP-ribose. A unique reaction mechanism involving the attack of enzyme-bound acetate or the direct attack of acetyl-lysine on an oxocarbenium ADP-ribose intermediate is proposed. We suggest that the reported histone/protein ADP-ribosyltransferase activity is a low-efficiency side reaction that can be explained through the partial uncoupling of the intrinsic deacetylation and acetate transfer to ADP-ribose.
The silent information regulator 2 (SIR2) gene family of proteins is conserved from bacteria to humans (1). In yeast, SIR2 is required for transcriptional silencing (2) and is also involved in suppressing rDNA recombination and controlling life span (3, 4). Very little is known about the roles of four other SIR2 family members found in yeast (5) or about the function of homologs from other species. The Salmonella typhimurium homolog, CobB, can substitute for CobT as a phosphoribosyltransferase during cobalamin biosynthesis (6). In vitro, CobB has histone/protein deacetylase activity (7, 8). Given the widespread occurrence and extensive conservation of the SIR2-like proteins, understanding their molecular mechanism is essential for identifying the cellular roles played by these proteins. Unfortunately, discrepancies among recent reports (7–11) have added to the general uncertainty as to the true enzymatic function for this family of proteins. These reports have suggested that the SIR2 proteins are histone ADP-ribosyltransferases (9, 11), histone/protein deacetylases (7, 8), or both (10). Therefore, we set out to authenticate the reaction products and to determine the inherent catalytic mechanism. We provide direct evidence that the efficient histone/protein deacetylase reaction is tightly coupled to the formation of a previously unidentified acetyl-ADP-ribose compound (1-O-acetyl-ADP-ribose) as a primary product of the reaction.
Materials and Methods
Materials.
[3H]Acetyl-CoA (1.88 Ci/mmol; 1 Ci = 37 GBq) and sodium [3H]acetate (100,000 cpm/nmol) were from NEN Life Sciences Products. Nicotinamide, ADP-ribose, and NAD were purchased from Sigma. Histone H3 peptide, ARTKQTARKSTGGKAPPKQLC, and the Lys-14-acetylated H3 peptide (AcLys-14), corresponding to the 20 amino-terminal residues of human histone H3, was synthesized by the Protein Chemistry Core Lab at Baylor College of Medicine (Houston). Histidine-tagged full-length HST2 was recombinantly expressed and purified from Escherichia coli BL21DE3 bacteria by using a T7 polymerase-based expression. Harvested cells were lysed by French pressure in 50 mM Tris, pH 8.0/300 mM NaCl/1 mM 2-mercaptoethanol with protease inhibitors (0.1 mM phenylmethylsulfonyl fluoride/10 μg/ml leupeptin/5 μg/ml aprotinin). The clarified extract was rocked batchwise with Ni-NTA-agarose (Qiagen, Chatsworth, CA; 2 ml) for 1 h at 4°C. The Ni-NTA-agarose was then applied to a small column and washed with 50 mM Tris, pH 8.0/300 mM NaCl/1 mM 2-mercaptoethanol. HST2 protein was eluted with a linear 0–500 mM imidazole gradient. HST2 eluted at 200 mM imidazole and was determined to be >95% pure by scanning the densitometry of Coomassie blue-stained SDS/PAGE. Imidazole was removed by extensive dialysis in the Tris buffer mentioned above. CobB and SIR2 proteins were purified as described (7).
Catalytic Analysis by HPLC.
Standards of AcLys-14 H3 peptide and NAD+ (HST2 substrates) and potential products of the HST2 reaction (H3 peptide, nicotinamide, [3H]acetate, and ADP-ribose) were resolved by reverse-phase HPLC on a Beckman Ultrasphere column (4.6 mm × 15 cm). Samples were injected in 0.05% trifluoroacetic acid (TFA)/H2O for 1 min before eluting with a linear gradient of 0–40% acetonitrile in 1–41 min. The peaks of absorbance at 214/260 nm were collected and subjected to mass spectral analysis. [3H]Acetate was detected by liquid scintillation counting. To authenticate the reaction products, HST2 (3.6 μM) was mixed with substrates AcLys-14 H3 peptide (525 μM) and NAD+ (175 μM) and allowed to react at 37°C for 5 min before being quenched with TFA to 1%. Substrates and products were resolved on reverse-phase HPLC and subjected to mass spectral analysis.
Typical HST2 steady-state reactions to monitor deacetylation of AcLys-14 H3 peptide or nicotinamide formation were carried out at 37°C in 50 mM Tris (pH 7.5) buffer. Rates of product formation were determined at various concentrations of AcLys-14 H3 peptide (5–300 μM) and NAD+ (5–600 μM). During the linear portion of the initial velocity, the reactions were quenched with TFA to 1%, and product formation was monitored by the HPLC analysis described above. Standard curves were generated to quantify H3 peptide and nicotinamide product formation by injecting known amounts of authentic standards and monitoring the corresponding peak height at 214 nm. The standard curves were linear in the range of 0–2 nmol and were reproducible from day to day.
P/CAF [3H]Monoacetylation of H3 Peptide.
The histone acetyltransferase P/CAF was used to monoacetylate H3 peptide on lysine 14 (12). P/CAF (0.175 μM) was mixed with [3H]acetyl-CoA (33.3 μM/1.88 Ci/mmol) and H3 peptide (175 μM) for 20 min at 25°C before being quenched with TFA to 1%. Mono-[3H]acetylated H3 peptide was separated from unmodified H3 by HPLC analysis as described above.
Matrix-Assisted Laser Desorption Ionization (MALDI)/Electrospray Ionization (ESI) Mass Spectrometry Analyses.
MALDI mass spectrometry was performed (Oregon State University, Environmental Health Sciences Center) on a custom-built reflector time-of-flight mass spectrometer equipped with a two-stage delayed extraction source. Approximately 1 μl of sample solution was mixed with 2 μl of 2,4,6-THAP [2,4,6-trihydroxy-acetophenone; 10 mg/ml in 70:30 water/acetonitrile (vol/vol)/50 mg/ml diammonium citrate in water]; a 1.0-μl droplet of this analyte–matrix solution was deposited onto a matrix precrystallized sample probe and allowed to air dry. Mass spectra were produced by irradiating the sample with an Nd:YAG laser (355 nm; Spectra-Physics) and operating the ion source at 23 kV with a 150-ns/1.0-kV delay. Every mass spectrum was recorded as the sum of 20 consecutive spectra, each produced by a single pulse of photons. Ions from an added standard were used for mass calibration. ESI mass spectrometry was carried out at Emory University School of Medicine Microchemical Facility (Atlanta). The precursor ion (phosphate anion) scanning procedure was performed as described (13). Chemical database searches (Available Chemicals Directory database, MDL Information Systems; CAS online, American Chemical Society; SciFinder Scholar, American Chemical Society) revealed that 1-O-acetyl-ADP-ribose is a previously unidentified compound.
Rapid-Reaction Kinetic Analysis.
For the quench-flow analysis under single turnover conditions, HST2 (10 μM) and NAD+ (300 μM) were rapidly mixed with 2.5 μM [3H]AcLys-14 H3 peptide at 22 ± 3°C (pH 7.5) in a Hi-Tech rapid quench-flow device, RQF-63 (Hi-Tech Scientific, Salisbury, U. K.). After various reaction times between 31 ms and 8 s, the reactions were quenched with TFA to a final concentration of 1%. The amount of [3H]AcLys-14 H3 peptide and 1-O-[3H]acetyl-ADP-ribose was determined by liquid scintillation counting of these species separated on reverse-phase HPLC.
Results and Discussion
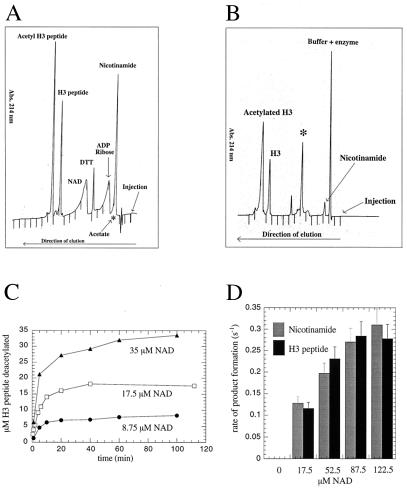
Because of the high yield and activity of recombinant enzyme, the yeast SIR2 homolog HST2 was used as the prototypical SIR2 member for the extensive study described here. Where noted, yeast SIR2 and the Escherichia coli homolog CobB were used to demonstrate the conservation in catalytic function among the family. Although the requirement for NAD+ has been demonstrated, surprisingly, the true identity and stoichiometry of the products has not been investigated. The extent of coupling between NAD+ consumption and histone deacetylation was examined initially by using the monoacetylated histone H3 peptide ARTKQTARKSTGG(AcK)APRKQL (corresponding to the first 20 amino-terminal residues in histone H3), subsequently referred to as AcLys-14 H3 peptide. To identify the authentic products of the NAD-dependent deacetylation reaction unambiguously, HST2-catalyzed products were resolved and quantified by reverse-phase HPLC and verified by mass spectrometry. First, standards of substrates (NAD+ and AcLys-14 H3 peptide) and of the potential products (H3 peptide, acetate, ADP-ribose, and nicotinamide) were separated by reverse-phase HPLC (Fig. 1A). The components eluted in the following order: nicotinamide, acetate, ADP-ribose, NAD+, H3 peptide, and AcLys-14 H3 peptide. The elution position of acetate was determined by using [3H]acetate and detection by liquid scintillation counting, whereas all others were detected by UV absorption at 214 nm or 260 nm (Fig. 1A).
Figure 1.
HPLC analysis of the HST2 NAD-dependent deacetylation reaction. (A) Reverse-phase HPLC elution of substrate standards (NAD+ and AcLys-14 H3 peptide) and of potential products (H3 peptide, acetate, ADP-ribose, and nicotinamide). Approximately 1.5 nmol AcLys-14 H3 peptide, 1.0 nmol H3 peptide, 10 nmol NAD, 15 nmol ADP-ribose, and 15 nmol nicotinamide were mixed and subjected to reverse-phase chromatography. In a separate HPLC run, 1.0 nmol sodium [3H]acetate (100,000 cpm/nmol) was subjected to chromatography. Order of elution: nicotinamide, acetate, ADP-ribose, NAD+, H3 peptide, and AcLys-14 H3 peptide. The elution position of acetate (∗) was determined by using authentic [3H]acetate and detection by liquid scintillation counting. All others were detected by UV absorption at 214 nm. (B) Elution of products from the HST2 NAD-dependent deacetylation reaction, detected by UV absorbance at 214 nm. ∗, Previously unidentified product. Conditions: 3.6 μM HST2/175 μM NAD+/525 μM Lys-14 AcH3/1 mM DTT for 30 min at 37°C before quenching with TFA to final concentration of 1%. (C) Amount of deacetylation correlates exactly with the consumption of NAD+. This graph displays the progress curves of deacetylation at fixed but limiting [NAD+]. HST2 reaction was allowed to proceed to completion under various limiting [NAD+], and the amount of deacetylated H3 peptide was determined. Conditions: 375 nM HST2/175 μM Lys-14 AcH3/8.75, 17.5, or 35 μM NAD+/1 mM DTT for 20 min at 37°C before quenching with TFA to 1%. (D) Steady-state rate of nicotinamide and deacetylated H3 peptide formation at various fixed [NAD+]. Conditions: 375 nM HST2/175 μM Lys-14 AcH3/8.75 μM–280 μM NAD+/1 mM DTT for 1 min at 37°C before quenching with TFA to a final concentration of 1%.
HST2 deacetylation reactions in the absence of NAD+ produced no detectable H3 peptide deacetylation or nicotinamide formation, consistent with the described NAD dependence (7, 8, 10). When NAD+ was included, robust deacetylation and nicotinamide formation were observed (Fig. 1B). However, to our great surprise, no significant amounts of ADP-ribose were detected (Fig. 1B). Nicotinamide and ADP-ribose are the predicted products from the hydrolysis of the NAD+ glycosidic bond. To explore this finding and establish the degree of coupling between NAD+ consumption and AcLys-14 H3 peptide deacetylation, the amount of deacetylated H3 formed and the amount of NAD+ consumed were determined (Fig. 1C). Moles of product formed in the enzyme-catalyzed reaction were calculated from standard curves generated with known amounts of authentic H3 peptide and nicotinamide. In reactions with several different initial concentrations of NAD+, every mole of NAD+ consumed by the enzyme produced a mole of deacetylated H3 peptide, suggesting that these two chemical events are tightly coupled. Mass spectrometry confirmed the identity of nicotinamide and deacetylated H3 peptide as the HPLC product peaks coeluting with authentic standards (Fig. 1 A and B). The previously described NAD+–nicotinamide exchange reaction was consistent with nicotinamide being a product of the reaction (7). The tight coupling of nicotinamide formation and H3 deacetylation was established further by comparing the steady-state rate of nicotinamide formation with the rate of AcLys-14 H3 peptide deacetylation (Fig. 1D). The NAD-concentration dependence of the steady-state rate of AcLys-14 H3 deacetylation matches exactly the rate of nicotinamide formation (Fig. 1D). Substrate saturation curves indicated that the Km for NAD+ is 70 μM and the Km for AcLys-14 H3 is estimated to be less than 500 nM. The fact that NAD+ is consumed in the reaction indicates that it is not an allosteric regulator but rather a bona fide substrate. Together, these results (Fig. 1 B and C) suggested an exquisitely linked enzymatic reaction in which NAD+ cleavage and deacetylation are directly coupled.
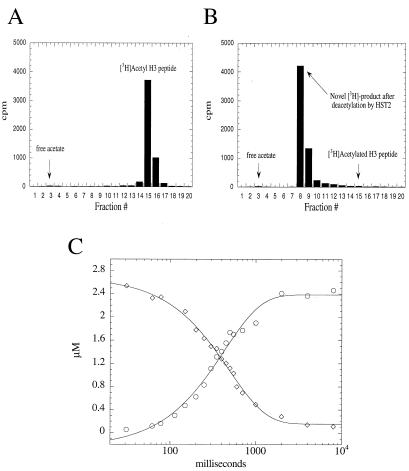
Next, we attempted to verify and quantify acetate as one of the obligate products. To accomplish this step, H3 peptide was stoichiometrically acetylated on Lys-14 by the histone acetyltransferase P/CAF by using [3H]AcCoA. Monoacetylated [3H]AcLys-14 H3 peptide was then purified by HPLC (Fig. 2A) and used as a substrate in the HST2 deacetylation reaction (Fig. 2B). To our surprise, the product profiles clearly indicated that acetate was not the primary product in the reaction. The HPLC elution position of the radioactive [3H]acetate product (Fig. 2B) did not correspond to the position of authentic acetate (Fig. 1A). Instead, the 3H-labeled product eluted at a position that was significantly more hydrophobic than ADP-ribose. On complete consumption of the [3H]AcLys-14 H3 peptide, only a trace amount of acetate (<2% of the total product formed) was detected in the reactions, whereas >98% of the original radioactivity from [3H]AcLys-14 H3 was transferred to a previously unidentified acetate adduct (Fig. 2B). Control experiments in the absence of enzyme did not result in transfer of acetate to this more hydrophobic form. Also, the acetate–adduct was not formed from a nonenzymatic solution reaction between the putative products [3H]acetate, ADP-ribose, nicotinamide, and H3 peptide.
Figure 2.
Acetate is not a primary product of HST2-catalyzed histone/protein deacetylation. H3 peptide was stoichiometrically acetylated on Lys-14 by the histone acetyltransferase P/CAF and [3H]AcCoA. Monoacetylated [3H]AcLys-14 H3 peptide was then purified by HPLC (A) and used as a substrate in the HST2 deacetylation reactions (B). On complete consumption of [3H]AcLys-14 H3 by HST2, all of the original 3H from H3 peptide was converted to a labeled product that eluted much later than authentic acetate. Conditions: 375 nM HST2/175 μM NAD+/5 μM [3H]Lys-14 AcH3/1 mM DTT for 1 min at 37°C before quenching with TFA to a final concentration of 1%. (C) Single turnover rapid quench-flow analysis. HST2 (10 μM) and NAD+ (300 μM) were mixed rapidly with 2.5 μM [3H]AcLys-14 H3 peptide at 22 ± 3°C (pH 7.5) in a Hi-Tech rapid quench-flow device, RQF-63. Between 31 ms and 8 s, reactions were quenched with TFA (1%). Quantification of [3H]AcLys-14 H3 peptide (⋄) and the [3H]acetate adduct (○) was accomplished by liquid scintillation counting of these species separated by using reverse-phase HPLC. Data were fitted to a single exponential, with yielding rate constants of 2.0 ± 0.1 s−1 for [3H]AcLys-14 H3 peptide deacetylation and 2.3 ± 0.2 s−1 for [3H]acetate adduct formation.
To explore the idea that the acetate–adduct was a direct product of enzymatic deacetylation and not the product of a slower side reaction, a rapid-reaction single turnover experiment was performed. By using a quench-flow apparatus, HST2 was reacted rapidly with excess NAD+ and substoichiometric levels of [3H]AcLys-14 H3. Under these conditions, the enzyme will perform only a single round of catalysis, allowing us to quantify the time-dependent loss of [3H]AcLys-14 H3 and the generation of the [3H]acetate adduct. At various times between 30 ms and 8 s, the reactions were quenched and were analyzed by HPLC and liquid scintillation counting. The progress curves (Fig. 2C) for the rapid-reaction single turnover clearly demonstrated that AcLys-14 H3 substrate consumption (rate constant of 2.0 ± 0.1 s−1) and acetate adduct formation (rate constant of 2.3 ± 0.2 s−1) were concomitantly linked, providing strong evidence that the acetate adduct is a primary product of HST2-catalyzed deacetylation.
UV-visible spectral analysis revealed that the acetate adduct absorbed strongly at 260 nm, which is consistent with an analog containing an adenine ring. During catalytic turnover, the appearance of a new UV-absorbing species (Fig. 1B) correlated exactly with the fractions containing the 3H-labeled product (Fig. 2B). Because ADP-ribose was not detected as a bona fide product of the reaction (Fig. 1B), it was logical to suggest that the previously unknown product was an adduct between ADP-ribose and acetate. In similar experiments with SIR2 and CobB, the identical-eluting radioactive product was observed, and again, no acetate or ADP-ribose was detected as a primary product in these reactions.
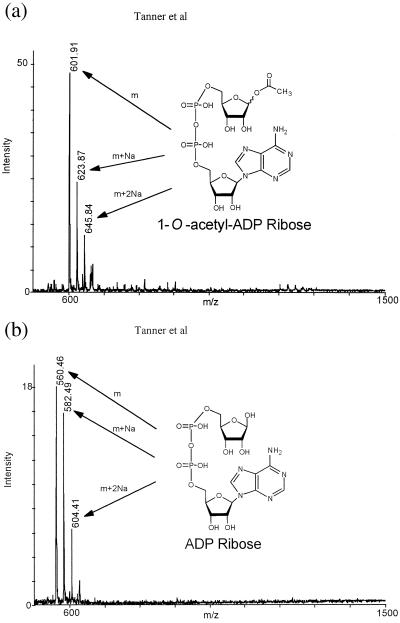
To confirm our hypothesis that an adduct between ADP-ribose and acetate is the authentic product, HPLC fractions containing this product were subjected to both MALDI and ESI mass spectral analysis for mass determination. The previously unknown adduct yielded a mass (positive molecular ion in MALDI) of 602 m/z (Fig. 3A), consistent with the enzymatic formation of acetyl-ADP-ribose. For comparison, the mass results of an ADP-ribose standard are displayed in Fig. 3B, indicating the predicted mass of 560 m/z. The difference of 42 m/z between the two masses is equivalent to the difference between replacing a hydrogen on an ADP-ribose hydroxyl with an acetyl group. Confirming these results, the identical mass was also observed by separate ESI/MALDI analyses from samples submitted to two independent facilities. Consistent with the formation of an O-acetyl bond to ribose, hydroxylamine treatment of the acetyl-ADP-ribose compound liberated free ADP-ribose. Similarly, high glycine concentrations also liberated ADP-ribose from the HST2, SIR2, and CobB-catalyzed formation of acetyl-ADP-ribose. Thus, we provide direct evidence that the efficient histone/protein deacetylase reaction is tightly coupled to the formation of a previously unidentified acetyl-ADP-ribose product. One molecule of NAD+ and one molecule of acetyl-lysine (histone) are readily catalyzed to one molecule of deacetylated lysine, nicotinamide, and acetyl-ADP-ribose. Although mass spectrometry cannot provide direct information on the position of the acetyl group on ADP-ribose, the chemical evidence discussed below strongly suggests that the C1 position of ribose (1-O-acetyl-ADP-ribose) is the site of attachment. In NAD+, C1 forms the glycosidic bond to nicotinamide. Searches of chemical databases for 1-O-acetyl-ADP-ribose produced no matches, indicating that it is a previously unknown compound produced by a previously unidentified enzymatic reaction.
Figure 3.
Mass spectral analysis of the previously unknown product generated by HST2-catalyzed histone/protein deacetylation. MALDI mass spectrometry was used to identity a mass of 602 m/z, consistent with the formation of acetyl-ADP- ribose (1-O-acetyl-ADP-ribose). For comparison, authentic ADP-ribose yielded a predicted mass of 560 m/z. With both ADP-ribose and acetyl-ADP-ribose, masses corresponding to the association of one and two sodium ions also were observed.
How do the SIR2 enzymes accomplish this unique and tightly coupled reaction? It is well established that many NAD-dependent enzymes (NAD+ glycohydrolases, ribosyltransferases, and ADP-ribosyl cyclases) form a putative oxocarbenium ADP-ribose cation as the direct product of nicotinamide elimination (14–16). Given this precedent for oxocarbenium cation formation and the previously observed NAD+–nicotinamide exchange reaction (7), SIR2 enzymes will likely form a similar intermediate. Interestingly, in the case of the SIR2 enzymes, oxocarbenium cation formation seems to require acetyl-lysine binding (and/or deacetylation). Only in the presence of acetyl-lysine and NAD+ can exogenously added nicotinamide exchange with the enzyme intermediate to reform NAD+ (7).
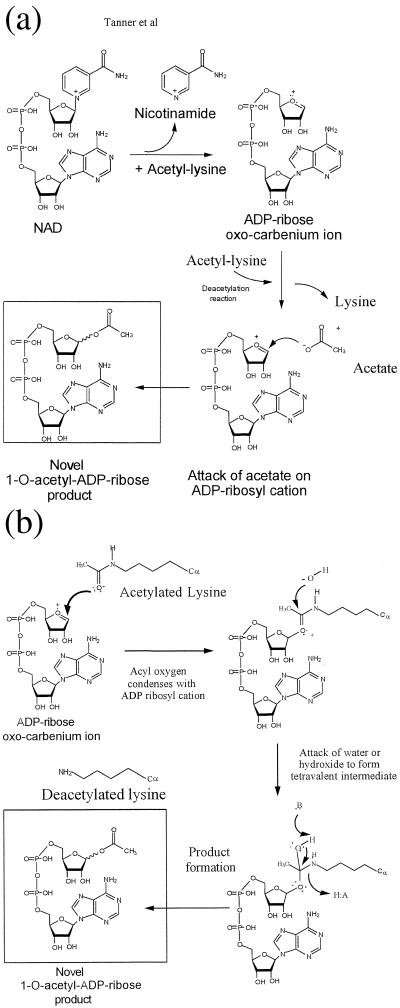
We propose two possible chemical mechanisms for catalysis by the SIR2 family (Fig. 4). In both cases, a putative oxocarbenium ADP-ribose intermediate is formed by the elimination of nicotinamide from NAD+. In the first mechanism (Fig. 4A), formation of the oxocarbenium is coupled to acetyl-lysine binding or hydrolysis. On acetyl-lysine hydrolysis, enzyme-bound acetate attacks the oxocarbenium cation to produce 1-O-acetyl-ADP-ribose. Alternatively, acetyl-lysine condenses directly with the oxocarbenium cation (Fig. 4B). This mechanism would imply that acetyl-lysine binding induces the elimination of nicotinamide to form initially the oxocarbenium intermediate. The acyl oxygen of acetyl-lysine condenses with the oxocarbenium cation. A hydroxide ion then attacks this intermediate to form a tetravalent intermediate, which can collapse to produce 1-O-acetyl-ADP-ribose through the use of enzyme general acid/base catalysis. Although unlikely based on the above arguments, a different isomer of 1-O-acetyl-ADP-ribose may be formed. For clarity, we have drawn the chemical events as stepwise events (Fig. 4 A and B); however, the chemistry could occur in either stepwise or concerted fashion.
Figure 4.
Proposed catalytic mechanisms for the production of 1-O-acetyl-ADP-ribose. (A) Attack of transiently formed acetate on an oxocarbenium ADP-ribose intermediate. Nicotinamide elimination from NAD+ to produce an oxocarbenium ADP-ribose intermediate is coupled to acetyl-lysine binding or hydrolysis. The enzyme-bound acetate generated in the deacetylation reaction attacks the oxocarbenium cation to produce 1-O-acetyl-ADP-ribose. (B) Acetyl-lysine condenses directly with the oxocarbenium cation. After the formation of the oxocarbenium cation as in A, the acyl oxygen of acetyl-lysine condenses with the oxocarbenium cation. A hydroxide ion then attacks this intermediate to form a tetravalent intermediate, which can collapse to produce 1-O-acetyl-ADP-ribose through the use of enzyme general acid/base catalysis. With either mechanism (A or B), the chemistry could occur in either stepwise or concerted fashion. For clarity, we have drawn the chemical events as stepwise events.
Given our findings, we suggest that the reported histone/protein ADP-ribosyltransferase activity (9–11) is a low-efficiency side reaction that can be explained through the partial uncoupling of the intrinsic deacetylation/acetate ADP-ribosylation reactions. The fact that these enzymes are capable of an NAD+–nicotinamide exchange reaction suggests that the oxocarbenium cation of ADP-ribose is at least partially susceptible to attack by the base nucleophile. However, the proposed oxocarbenium cation of SIR2 enzymes seems to be exquisitely constructed to limit other possible side reactions, such as attack by H2O or by nucleophilic amino acid side chains, which would result in ADP-ribose or protein ADP-ribosylation, respectively. We observed, at most, protein ADP-ribosylation that is ≈0.1% of the authentic reaction described in this study. Also, ADP-ribose was not detected as a primary enzymatic product. It may be possible that some uncoupling of this reaction to yield protein ADP-ribosylation could result from perturbations in native protein structure (partially unfolded protein, mutagenesis, inappropriate reaction conditions) and from extremely high concentrations of an alternate acceptor, such as reactive protein side chains.
Why do the SIR2 enzymes go to great lengths to couple histone/protein deacetylation to the formation of 1-O-acetyl-ADP-ribose? It is attractive to propose that this previously unknown molecule has a unique cellular function or functions that may be linked to SIR2's gene-silencing effects, raising the possibility that 1-O-acetyl-ADP-ribose has an important signaling role in which other enzymes/proteins may use 1-O-acetyl-ADP-ribose to elicit the proper cellular response. Such targets might include ATP-dependent chromatin remodeling enzymes, histone/protein acetyltransferases, or poly(ADP-ribosyl)transferases. It is interesting to note that poly(ADP-ribosyl)transferases use NAD+ to poly(ADP-ribosyl)ate proteins involved in the metabolism of nucleic acids and in the maintenance of chromatin architecture (15, 17). One intriguing possibility is that 1-O-acetyl-ADP-ribose could bind poly(ADP-ribosyl)transferases and block poly(ADP ribosyl)ation. Moreover, NAD+ levels in cells are inversely affected by the level of protein poly(ADP-ribosyl)ation (15, 17). Because poly(ADP-ribosyl)transferases and SIR2 enzymes exhibit similar Km values (≈50–70 μM) for NAD+, they may compete for the available NAD+ and oppose each other's function. Recently, the observation has been made that caloric restriction leading to increased life span seems to be linked through an NAD+- and SIR2-dependent pathway (18), which raises the possibility that 1-O-acetyl-ADP-ribose may play a role in this phenomenon. It is important to note that bacteria do not have histones, and yet they do have SIR2-like proteins with similar activity, as has been shown here for HST2/SIR2/CobB. Thus, histones need not be the only substrates for deacetylation by these enzymes. Identification of the authentic products and the catalytic mechanism of the SIR2-like enzymes has provided the initial basis for understanding the cellular role(s) played by this important family of proteins.
Acknowledgments
We thank Jack Kyte (University of California, San Diego) and Dick Goodman (Vollum Institute, Portland, OR) for helpful discussions, Lilo Barofsky for the MALDI mass spectral analysis, Jan Pohl for the ESI analysis, Mike Langer for quench-flow analysis, and Youngjoo Kim and Margie Borra for assistance with HPLC. This work was supported by National Institutes of Health Grants GM59785–02 (to J.M.D.) and GM55641 (to R.S.) and American Cancer Society Post-Doctoral Fellowship PF-00-162-01-GMC (to K.T.).
Abbreviations
- TFA
trifluoroacetic acid
- MALDI
matrix-assisted laser desorption ionization
- ESI
electrospray ionization
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
See commentary on page 14030.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.250422697.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.250422697
References
- 1.Frye R A. Biochem Biophys Res Commun. 2000;273:793–798. doi: 10.1006/bbrc.2000.3000. [DOI] [PubMed] [Google Scholar]
- 2.Loo S, Rine J. Annu Rev Cell Dev Biol. 1995;11:519–548. doi: 10.1146/annurev.cb.11.110195.002511. [DOI] [PubMed] [Google Scholar]
- 3.Gottlieb S, Esposito R E. Cell. 1989;56:771–776. doi: 10.1016/0092-8674(89)90681-8. [DOI] [PubMed] [Google Scholar]
- 4.Guarente L. Nat Genet. 1999;23:281–285. doi: 10.1038/15458. [DOI] [PubMed] [Google Scholar]
- 5.Brachmann C B, Sherman J M, Devine S E, Cameron E E, Pillus L, Boeke J D. Genes Dev. 1995;9:2888–2902. doi: 10.1101/gad.9.23.2888. [DOI] [PubMed] [Google Scholar]
- 6.Tsang A W, Escalante-Semerena J C. J Biol Chem. 1998;273:31788–31794. doi: 10.1074/jbc.273.48.31788. [DOI] [PubMed] [Google Scholar]
- 7.Landry J, Sutton A, Tafrov S T, Heller R C, Stebbins J, Pillus L, Sternglanz R. Proc Natl Acad Sci USA. 2000;97:5807–5811. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith J S, Brachmann C B, Celic I, Kenna M A, Muhammad S, Starai V J, Avalos J L, Escalante-Semerena J C, Grubmeyer C, Wolberger C, Boeke J D. Proc Natl Acad Sci USA. 2000;97:6658–6663. doi: 10.1073/pnas.97.12.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tanny J C, Dowd G J, Huang J, Hilz H, Moazed D. Cell. 1999;99:735–745. doi: 10.1016/s0092-8674(00)81671-2. [DOI] [PubMed] [Google Scholar]
- 10.Imai S, Armstrong C M, Kaeberlein M, Guarente L. Nature (London) 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 11.Frye R A. Biochem Biophys Res Commun. 1999;260:273–279. doi: 10.1006/bbrc.1999.0897. [DOI] [PubMed] [Google Scholar]
- 12.Tanner K G, Langer M R, Denu J M. Biochemistry. 2000;39:11961–11969. doi: 10.1021/bi001272h. [DOI] [PubMed] [Google Scholar]
- 13.Annan R S, Carr S A. Anal Chem. 1996;68:3413–3421. doi: 10.1021/ac960221g. [DOI] [PubMed] [Google Scholar]
- 14.Scheuring J, Schramm V L. Biochemistry. 1997;36:8215–8223. doi: 10.1021/bi970379a. [DOI] [PubMed] [Google Scholar]
- 15.Ziegler M. Eur J Biochem. 2000;267:1550–1564. doi: 10.1046/j.1432-1327.2000.01187.x. [DOI] [PubMed] [Google Scholar]
- 16.Cakir K-S C, Muller-Steffner H, Schuber F. Biochem J. 2000;349:203–210. doi: 10.1042/0264-6021:3490203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.D'Amours D, Desnoyers S, D'Silva I, Poirier G G. Biochem J. 1999;342:249–268. [PMC free article] [PubMed] [Google Scholar]
- 18.Lin S J, Defossez P A, Guarente L. Science. 2000;289:2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]