Abstract
In the green alga Chlamydomonas reinhardtii, a Leu290-to-Phe (L290F) substitution in the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco), which is coded by the chloroplast rbcL gene, was previously found to be suppressed by second-site Ala222-to-Thr and Val262-to-Leu substitutions. These substitutions complement the photosynthesis deficiency of the L290F mutant by restoring the decreased thermal stability, catalytic efficiency, and CO2/O2 specificity of the mutant enzyme back to wild-type values. Because residues 222, 262, and 290 interact with the loop between β strands A and B of the Rubisco small subunit, which is coded by RbcS1 and RbcS2 nuclear genes, it seemed possible that substitutions in this loop might also suppress L290F. A mutation in a nuclear gene, Rbc-1, was previously found to suppress the biochemical defects of the L290F enzyme at a posttranslational step, but the nature of this gene and its product remains unknown. In the present study, three nuclear-gene suppressors were found to be linked to each other but not to the Rbc-1 locus. DNA sequencing revealed that the RbcS2 genes of these suppressor strains have mutations that cause either Asn54-to-Ser or Ala57-to-Val substitutions in the small-subunit βA/βB loop. When present in otherwise wild-type cells, with or without the resident RbcS1 gene, the mutant small subunits improve the thermal stability of wild-type Rubisco. These results indicate that the βA/βB loop, which is unique to eukaryotic Rubisco, contributes to holoenzyme thermal stability, catalytic efficiency, and CO2/O2 specificity. The small subunit may be a fruitful target for engineering improved Rubisco.
The green alga Chlamydomonas reinhardtii serves as an excellent model for the study of photosynthesis because photosynthesis-deficient mutants can be maintained with acetate as an alternative source of carbon and energy. Mutations can be assigned to the nuclear or chloroplast genetic compartments by virtue of Mendelian or uniparental inheritance, and both compartments can be transformed with homologous or heterologous DNA (1). These attributes have been particularly useful for examining the structure–function relationships of eukaryotic ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco, Enzyme Commission 4.1.1.39) (2). Because land plants cannot be maintained in the complete absence of photosynthesis, and the subunits of eukaryotic Rubisco cannot be assembled in Escherichia coli (3), it has been difficult to examine the effects of random or directed mutations on the function of land–plant Rubisco in vivo or in vitro (4–6). Tobacco is the only land plant in which the chloroplast genome can be transformed (5, 6).
By screening for photosynthesis-deficient Chlamydomonas mutants and selecting for photosynthesis-competent revertants, several regions of the Rubisco large subunit have been identified that influence the ratio of ribulose 1,5-bisphosphate (RuBP) carboxylase to oxygenase activities (2, 7). Because CO2 and O2 compete at the same large-subunit active site, the CO2/O2 specificity of the enzyme is defined by a kinetic constant, Ω, that equals the catalytic efficiency of carboxylation relative to that of oxygenation (Ω = VcKo/VoKc) (Vc, Vmax for carboxylation; Vo, Vmax for oxygenation; Ko, Km for O2; Kc, Km for CO2) (8). The value of Ω determines the amount of productive CO2 fixation in photosynthesis relative to the loss of CO2 via the nonessential photorespiratory pathway (8, 9). Thus, rbcL mutations and their intragenic suppressors that affect Ω may define potential sites for ultimately engineering an improved Rubisco enzyme (2, 10).
One photosynthesis-competent revertant of a temperature-conditional rbcL mutant has been enigmatic (11–13). It results from a mutation in a nuclear gene, named Rbc-1, the product of which acts posttranslationally to correct decreases in holoenzyme stability and Ω caused by a Leu290-to-Phe (L290F) large-subunit substitution (13). Rbc-1 does not encode the Rubisco small subunit, which is coded by the RbcS1 and RbcS2 genes in Chlamydomonas (14). Because assembly of 8 large (55 kDa) and 8 small (15 kDa) subunits into the Rubisco holoenzyme appears to depend on the action of the chaperonins (15, 16) and perhaps other assembly proteins (17), Rbc-1 may be one of a number of genes required for this process (18, 19).
In a previous study, an attempt was made to discern the nature of the Rbc-1 nuclear gene by selecting and analyzing additional photosynthesis-competent revertants of the rbcL-L290F mutant (20). Two were found to result from intragenic suppressor mutations that cause Ala222-to-Thr (A222T) and Val262-to-Leu (V262L) substitutions. Like the Rbc-1 nuclear-gene mutation (11, 12), these second-site rbcL mutations increase the amount of mutant Rubisco at the 35°C restrictive temperature and also increase Ω of the mutant enzyme back to the wild-type value (20). In the absence of the L290F mutation, the A222T and V262L substitutions improve the thermal stability of otherwise wild-type Rubisco in vitro (7). Based on the x-ray crystal structure of spinach Rubisco (21), residues Leu290, Ala222, and Val262 are far from the active site. They reside at the bottom of the large-subunit α/β-barrel domain where they surround a loop between β strands A and B of the Rubisco small subunit.
Because large-subunit residues 222 and 262 are not in Van der Waals contact with residue 290, they may complement L290F via structural perturbations of the small-subunit βA/βB loop (7, 20). If such is the case, substitutions in the small-subunit loop might complement L290F directly. In the present study, suppressor mutations were found at a second nuclear-gene locus, and this locus does contain the Rubisco RbcS genes.
Materials and Methods
Strains and Culture Conditions.
Chlamydomonas reinhardtii 2137 mt+ is the wild-type strain (22). Mutant rbcL-L290F mt+ lacks photosynthesis and requires acetate for growth at the 35°C restrictive temperature but is indistinguishable from wild type when growth is compared at 25°C (13, 23). Mutant RbcSΔ-T60–3 mt− was used as the host for nuclear transformation. It lacks photosynthesis and requires acetate for growth because of deletion of the 13-kb locus that contains RbcS1 and RbcS2 (24). All Chlamydomonas strains are maintained at 25°C in darkness with 10 mM acetate medium containing 1.5% Bacto-agar (22). For biochemical analysis, cells were grown on a rotary shaker at 220 rpm with 250–500 ml of liquid acetate medium in darkness.
Genetic Analysis.
Crosses were performed as described previously (22, 23). The temperature-conditional acetate-requiring phenotype of rbcL-L290F was scored by replicating dark-grown (25°C) tetrads to minimal medium in the light at 25 and 35°C (11, 23). The pf-2 (paralyzed flagella) centromere marker was included in the crosses to confirm the meiotic basis of the tetrads and to map each nuclear mutation relative to its centromere (22, 23).
RbcS Sequencing and Transformation.
To screen for RbcS mutations, total DNA was extracted from photosynthesis-competent revertants, purified, and digested with EcoRI (25). By using gene-specific oligonucleotides, a 1,303-bp RbcS1 sequence (bases 47–1,349) and a 1,530-bp RbcS2 sequence (bases 47–1,576), which contain much of the coding regions of RbcS1 (bases 1–1,125) and RbcS2 (bases 1–1,267) (14), were PCR amplified (24). The PCR products were then sequenced with Sequenase, deoxyadenosine 5′-[α-[35S]thio]triphosphate, and a set of oligonucleotide primers (11).
To clone mutant RbcS2 genes, gene-specific primers containing engineered XbaI (5′-GCGTCTAGACACCGTGCATTGCTGCCTTAG-3′) or EcoRI (5′-CGCGAATTCGAGGCCAAAATCAACGGAGGATCG-3′) restriction sites were used to PCR amplify a 2,538-bp fragment that contains the entire RbcS2 coding region (bases 1–1,267) between bases −1,063–1,457. The PCR product was digested with XbaI and EcoRI, ligated with XbaI/EcoRI-digested pUC19 (26), and transformed into E. coli XL-1 Blue (27). The plasmids containing the RbcS2-N54S and RbcS2-A57V mutant genes were named pSS2-N54S and pSS2-A57V, respectively.
The RbcSΔ deletion strain (24) was transformed with pSS2-N54S and pSS2-A57V by the glass-bead vortexing method (28), as described previously (24). Photosynthesis-competent transformants were selected on minimal medium in the light (80 μmol photons/m2/sec). After PCR amplification, the coding region of each mutant RbcS2 gene was completely sequenced to ensure that only the expected mutation was present.
Enzyme Kinetics and Thermal Stability.
Rubisco holoenzyme was purified from cell extracts by sucrose gradient centrifugation (29) and quantified (30). Carboxylase and oxygenase kinetic constants were determined by measuring the incorporation of acid-stable 14C from NaH14CO3 as described previously (13). Ω of purified and activated Rubisco (20 μg/reaction) was determined by assaying carboxylase and oxygenase activities simultaneously with [1-3H]RuBP (7.2 Ci/mol) and NaH14CO3 (0.5 Ci/mol) in 30-min reactions at 25°C (31, 32). [1-3H]RuBP and phosphoglycolate phosphatase were synthesized/purified according to standard methods (31, 33).
Rubisco thermal stability was assayed by incubating purified and activated enzymes (10 μg/ml) in 50 mM N,N-bis(2-hydroxyethyl)glycine (Bicine) (pH 8.0)/10 mM NaHCO3/10 mM MgCl2/1 mM DTT at various temperatures for 20 min (7, 12). The samples were then cooled on ice for 5 min, and carboxylase activity was assayed at 25°C by adding the incubated enzymes to 0.5 ml of assay buffer containing 50 mM Bicine (pH 8.0)/0.4 mM RuBP/10 mM NaH 14CO3 (2 Ci/mol)/10 mM MgCl2. After 1 min, the reactions were terminated by adding 0.5 ml of 3 M formic acid in methanol.
Results
Molecular Genetics of Photosynthesis-Competent Revertants.
In previous studies, photosynthesis-competent revertants of mutant rbcL-L290F were selected on minimal medium in the light at the restrictive temperature of 35°C (11, 20). Recent analysis of 11 of these genetically independent revertants revealed that two arose from uniparentally inherited intragenic suppressor mutations that cause A222T and V262L substitutions in the large subunit (7, 20). The remaining nine revertants displayed Mendelian inheritance of the photosynthesis-competent phenotype (20), indicating that they arose from intergenic suppressor mutations in one or more nuclear genes (11). Further genetic analysis in the present study revealed that three of these revertants, named R116–1B, R116–9A, and R116–10C, contained suppressor mutations that assorted independently of the suppressor mutation in the Rbc-1 nuclear gene (11). Reciprocal crosses between mt+ and mt− isolates of revertants R116–1B, R116–9A, and R116–10C indicated that the suppressor mutations were genetically linked (at a locus distinct from Rbc-1).
Because the rbcL-L290F, rbcL-A222T, and rbcL-V262L mutations affect residues at the interface between the large subunit and small-subunit βA/βB loop, and because the new nuclear suppressors were not linked to the Rbc-1 locus, we reasoned that the R116–1B, R116–9A, and R116–10C revertants might result from mutations that affect the small-subunit βA/βB loop (residues 46–73 in Chlamydomonas). When the RbcS genes were PCR amplified and sequenced, no mutation was found in the RbcS1 genes. However, each of the revertants was found to contain a mutation in the RbcS2 gene. In revertant R116–1B, a transition mutation occurred that would change Asn54 (AAC) to Ser (AGC) (N54S). In both of the R116–9A and R116–10C revertants, a transition mutation occurred that would change Ala57 (GCC) to Val (GTC) (A57V). Because this RbcS2-A57V mutation eliminates a HaeIII restriction site (GGCC→GGTC), the mutations in revertants R116–9A and R116–10C were further confirmed by restriction-enzyme analysis. Because R116–9A and R116–10C arose from the same mutation, only one of them (R116–10C) was analyzed further.
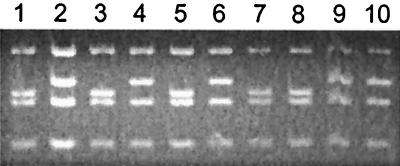
When revertants R116–1B mt+ (rbcL-L290F/RbcS2-N54S) and R116–10C mt+ (rbcL-L290F/RbcS2-A57V) were crossed with the mt− pf-2 centromere marker, the N54S and A57V suppressor mutations were found to be 20 map units from the centromere (Table 1). This map distance provides a point of correlation with the physical map of chromosome II in the Chlamydomonas electronic database (1, 34). R116–1B mt− and R116–10C mt− progeny were recovered from the first crosses and crossed with wild-type mt+. Because the rbcL-L290F chloroplast mutation is not transmitted from the mt− parent, these crosses would allow the phenotypes of the RbcS2-N54S and RbcS2-A57V mutations to be observed in an otherwise wild-type strain. No discernable (deleterious) phenotype could be ascribed to either of the RbcS2 mutations. PCR amplification and DNA sequencing of RbcS2 from complete tetrads of progeny, and also restriction-enzyme analysis in the case of RbcS2-A57V (Fig. 1), confirmed that the N54S and A57V mutations were linked to the suppressor phenotype in the forward crosses and enabled the isolation of RbcS2-N54S and RbcS2-A57V strains (in the absence of the original rbcL-L290F mutation) from the reciprocal crosses.
Table 1.
Genetic analysis of photosynthesis-competent revertants of the temperature-conditional photosynthesis-deficient rbcL-L290F mutant
| Cross | Tetrads (ac-pf)*
|
Map distance† | ||
|---|---|---|---|---|
| PD | NPD | T | ||
| rbcL-L290F, RbcS2-N54S, +, mt+ × +, +, pf-2, mt− | 10 | 13 | 15 | 20 |
| (revertant R116-1B) (wild type) | ||||
| rbcL-L290F, RbcS2-A57V, +, mt+ × +, +, pf-2, mt− | 6 | 7 | 8 | 19 |
| (revertant R116-10C) (wild type) | ||||
All progeny receive the temperature-conditional chloroplast rbcL-L290F mutation, but only two in a tetrad receive the nuclear RbcS2 suppressor mutation. In the absence of the suppressor, progeny lack photosynthesis at 35°C and require acetate for growth (ac phenotype). Segregation of the ac phenotype was scored relative to the centromere-linked paralyzed flagella marker (pf phenotype), which allows parental-ditype (PD), nonparental-ditype (NPD), and tetratype (T) tetrads to be defined.
Distances (% recombination × 100) between the RbcS2 suppressors and their centromeres were calculated as 100 × 0.5T/(PD + NPD + T).
Figure 1.
Segregation of the RbcS2-A57V suppressor mutation in reciprocal crosses. DNA was purified from wild-type (lane 1) revertant R116–10C (rbcL-L290F/RbcS2-A57V) (lane 2), a tetrad of progeny from the cross R116–10C mt+ X wild-type mt− (lanes 3–6), and a tetrad of progeny from the cross wild-type mt+ X R116–10C mt− (lanes 7–10). By using a pair of oligonucleotides specific for the RbcS2 gene, an 818-bp sequence (bases 640-1457 relative to bases 1–1,267 of the coding region of RbcS2) was PCR amplified from each sample, digested with HaeIII, and separated on a 3.5% agarose gel. The RbcS2-A57V mutation eliminates a HaeIII site, increasing the size of a 189-bp fragment to 210 base pairs. Progeny in lanes 3 and 5 had temperature-conditional acetate-requiring phenotypes. Progeny in lanes 4, 6, and 7–10 had wild-type (photosynthesis-competent) phenotypes at 35°C.
Because a wild-type RbcS1 gene is also present and expressed at an appreciable level (14, 24), it would be difficult to conclude from crosses alone that the RbcS2-N54S or RbcS2-A57V mutation had no adverse effect on Rubisco assembly or function in the absence of the original rbcL-L290F mutation. Therefore, the mutant RbcS2-N54S and RbcS2-A57V genes were transformed into the RbcSΔ strain, which lacks both RbcS1 and RbcS2 and requires acetate for growth (24). Photosynthesis-competent colonies were recovered on minimal medium in the light at frequencies (about 3 per 107 cells) comparable to those obtained with the wild-type RbcS2 gene (24). Thus, when the N54S and A57V substitutions are present in all of the small subunits of the holoenzyme, they have no apparent effect on otherwise wild-type Rubisco. These transformant strains are designated RbcS1-Δ/RbcS2-N54S and RbcS1-Δ/RbcS2-A57V.
Small-Subunit Substitutions Improve Rubisco Thermal Stability.
Although the rbcL-L290F/RbcS2-N54S and rbcL-L290F/RbcS2-A57V revertants were indistinguishable from wild type when growth was compared on minimal medium at 35°C, both had less than half the wild-type level of Rubisco holoenzyme when extracts of 35°C-grown cells were fractionated on sucrose gradients (Table 2). However, both revertants had more than twice the level of holoenzyme as the original rbcL-L290F mutant at 35°C (11–13), which would, in part, account for their ability to grow photoautotrophically at 35°C. Extracts of 25°C-grown mutant and revertant cells had nearly equal amounts of holoenzyme, indicating that the N54S and A57V suppressor substitutions may compensate for the altered stability of the L290F enzyme at only the 35°C restrictive temperature (Table 2). In the absence of the rbcL-L290F mutation, the RbcS2-N54S and RbcS2-A57V suppressor strains had wild-type levels of Rubisco holoenzyme, and the RbcS1-Δ/RbcS2-N54S and RbcS1-Δ/RbcS2-A57V transformants had levels of holoenzyme that were not significantly different from those of transformants that contain only the wild-type RbcS2 gene (24) (data not shown).
Table 2.
Amount of Rubisco holoenzyme per total cell protein in wild-type, mutant rbcL-L290F, revertant rbcL-L290F/RbcS2-N54S, and revertant rbcL-L290F/RbcS2-A57V grown at 25 or 35°C in darkness
| Strain | Holoenzyme
|
|||
|---|---|---|---|---|
| 25°C
|
35°C
|
|||
| μg⋅mg−1* | % | μg⋅mg−1* | % | |
| Wild type | 70 ± 9 | 100 | 39 ± 6 | 100 |
| L290F | 44 ± 6 | 63 | 7 ± 1 | 19 |
| L290F/N54S | 46 ± 8 | 66 | 18 ± 2 | 46 |
| L290F/A57V | 40 ± 8 | 57 | 16 ± 1 | 42 |
Values are means ± SD (n − 1) of three or more sucrose-gradient preparations.
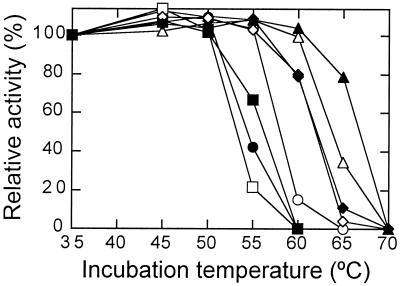
When the thermal stability of Rubisco purified from 25°C-grown cells was assayed in vitro (Fig. 2), the rbcL-L290F/RbcS2-N54S and rbcL-L290F/RbcS2-A57V revertant enzymes were found to be similar to the original rbcL-L290F mutant enzyme. In fact, the N54S substitution may cause a slight decrease in thermal stability relative to that of the rbcL-L290F enzyme (Fig. 2). Thus, increases in the amount of Rubisco in the rbcL-L290F/RbcS2-N54S and rbcL-L290F/RbcS2-A57V revertant strains relative to the amount in the original rbcL-L290F mutant strain (Table 2) may not be explained solely by a direct change in thermal stability. It is more likely that the N54S and A57V suppressor substitutions cause slight improvements in structural stability at 35°C that afford enhanced protection from proteolysis in vivo (11–13).
Figure 2.
Thermal inactivation of purified Rubisco from wild type (○), mutant rbcL-L290F (●), revertant rbcL-L290F/RbcS2-N54S (□), revertant rbcL-L290F/RbcS2-A57V (■), suppressor RbcS2-N54S (⋄), suppressor RbcS2-A57V (♦), transformant RbcS1-Δ/RbcS2-N54S (▵), and transformant RbcS1-Δ/RbcS2-A57V (▴). Purified Rubisco (10 μg/ml) was incubated at each temperature for 20 min. The samples were then cooled on ice, and RuBP carboxylase activity was assayed at 25°C. Activities for each enzyme were normalized against the level of activity measured after the 35°C incubation (wild type, 1.5; L290F, 0.5; L290F/N54S, 1.0; L290F/A57V, 1.0; N54S, 1.4; A57V, 1.4, Δ/N54S, 1.2; Δ/A57V, 1.0 μmol⋅min−1⋅mg−1). Comparing three separate enzyme preparations at 65°C, wild-type Rubisco was completely inactivated, but transformant Δ/N54S and Δ/A57V Rubisco retained 33% ± 2 SD and 76% ± 3 SD of their initial activities, respectively.
In the absence of the L290F large-subunit substitution, the N54S and A57V small-subunit substitutions significantly improve the thermal stability of wild-type Rubisco in vitro (Fig. 2, compare wild type with RbcS2-N54S and RbcS2-A57V). Furthermore, in the absence of the wild-type RbcS1 gene, the Rubisco holoenzymes isolated from the RbcS1-Δ/RbcS2-N54S and RbcS1-Δ/RbcS2-A57V transformants are even more thermally stable. For example, after a 20-min incubation at 65°C, these holoenzymes retained 35 and 80% of their initial carboxylase activities, respectively, but wild-type enzyme was completely inactivated (Fig. 2, compare wild type with RbcS1-Δ/RbcS2-N54S and RbcS1-Δ/RbcS2-A57V). To make sure that this increased thermal stability resulted from the suppressor substitutions and not from a difference in thermal stability between holoenzymes containing only RbcS1 or RbcS2 small subunits, wild-type Rubisco was compared with holoenzymes purified from RbcSΔ cells that had been transformed with either RbcS1 or RbcS2 (24). No difference was found in the thermal stabilities of these “wild-type” holoenzymes in vitro (data not shown).
Small-Subunit Substitutions Increase Ω of Mutant Rubisco.
As shown here (Table 3) and previously (7, 13, 20), the Ω value of rbcL-L290F mutant Rubisco is about 10% lower than that of the wild-type enzyme. This decrease in Ω arises primarily from a decrease in Vc (Table 3). The N54S and A57V small-subunit substitutions increase Ω of the mutant enzyme back to the wild-type value by decreasing Kc and increasing the value of Ko/Kc (Table 3, compare rbcL-L290F with rbcL-L290F/RbcS2-N54S and rbcL-L290F/RbcS2-A57V). The revertant enzymes also have increased Vc and Vc/Kc values relative to those of the mutant enzyme, indicating that they do, in fact, have improved carboxylation efficiency relative to the mutant enzyme (9, 19). However, despite wild-type Ω values, the rbcL-L290F/RbcS2-N54S and rbcL-L290F/RbcS2-A57V revertant enzymes are not as good as the wild-type enzyme (9, 19). They have significant reductions in Vc, Vc/Kc, and Vc/Vo (Table 3, compare wild type with rbcL-L290F/RbcS2-N54S and rbcL-L290F/RbcS2-A57V).
Table 3.
Kinetic properties of Rubisco purified from wild-type, mutant rbcL-L290F, revertants rbcL-L290F/RbcS2-N54S and rbcL-L290F/RbcS2-A57V, and transformants RbcS1-Δ/RbcS2-N54S and RbcS1-Δ/RbcS2-A57V
| Enzymes | Ω* (VcKo/VoKc) | Vc* μmol⋅h−1⋅mg−1 | Kc* μM CO2 | Ko* μM CO2 | Vc/Kc† | Ko/Ko† | Vc/Vo† |
|---|---|---|---|---|---|---|---|
| Wild type | 60 ± 1 | 138 ± 13 | 32 ± 3 | 544 ± 38 | 4.3 | 17 | 3.5 |
| L290F | 54 ± 1 | 47 ± 1 | 43 ± 2 | 763 ± 41 | 1.1 | 18 | 3.0 |
| L290F/N54S | 60 ± 3 | 87 ± 1 | 33 ± 4 | 715 ± 37 | 2.6 | 22 | 2.7 |
| L290F/A57V | 58 ± 1 | 92 ± 3 | 32 ± 1 | 674 ± 79 | 2.9 | 21 | 2.8 |
| Δ/N54S | 61 ± 1 | 99 ± 8 | 26 ± 2 | 485 ± 47 | 3.8 | 19 | 3.2 |
| Δ/A57V | 59 ± 1 | 84 ± 7 | 26 ± 3 | 489 ± 87 | 3.2 | 19 | 3.1 |
The values are the means ± SD (n − 1) of three separate enzyme preparations.
Calculated values.
Rubisco enzymes isolated from the RbcS1-Δ/RbcS2-N54S and RbcS1-Δ/RbcS2-A57V transformants (which have a wild-type rbcL gene, lack RbcS1, and are homogeneous for the N54S and A57V small-subunit substitutions) were found to have wild-type Ω values (Table 3). However, these small-subunit mutant enzymes also have decreases in Vc and Vc/Kc relative to the wild-type enzyme (Table 3, compare wild type with RbcS1-Δ/RbcS2-N54S and RbcS1-Δ/RbcS2-A57V). This negative effect on catalysis was absent from the RbcS2-N54S and RbcS2-A57V suppressor-strain holoenzymes (Table 4), which are heterogeneous for wild-type (coded by RbcS1) and mutant (coded by RbcS2) small subunits. Thus, the RbcS2-N54S and RbcS2-A57V enzymes have improved thermal stability in vitro (Fig. 2) with no apparent decline in Ω or carboxylation catalytic efficiency (Table 4).
Table 4.
Kinetic properties of Rubisco purified from wild-type and suppressor strains RbcS2-N54S and RbcS2-A57V
| Enzymes | Ω (VcKo/VoKc) | Vc μmol⋅h−1⋅mg−1 | Kc μM CO2 | Ko μM O2 |
|---|---|---|---|---|
| Wild type | 61 ± 1 | 152 ± 8 | 34 ± 2 | 537 ± 44 |
| N54S | 60 ± 1 | 149 ± 10 | 30 ± 2 | 521 ± 20 |
| A57V | 62 ± 1 | 137 ± 21 | 29 ± 3 | 497 ± 51 |
The values are the means ± SD (n − 1) of three separate enzyme preparations.
Discussion
The Small-Subunit βA/βB Loop Influences Ω.
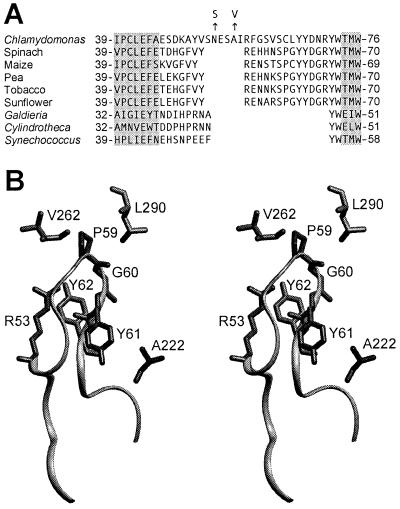
The role of the small subunit in Rubisco holoenzyme structure and function remains poorly defined (9, 19). Because small-subunit primary structures are more divergent than those of large subunits (19), it is interesting to consider to what extent small subunits might contribute to variations in catalytic efficiency and Ω observed between Rubisco enzymes from different species (35, 36). In particular, the small subunits of plants and green algae contain a loop between β strands A and B that is 21 or more residues in length, but this βA/βB loop is only 10 residues long in the small subunits of cyanobacteria and nongreen algae (Fig. 3). Although it has not been possible to exploit directed mutagenesis for studying the eukaryotic small subunit in vivo or in E. coli (19), it has been possible to assess the importance of the βA/βB loop for holoenzyme assembly via transport of small subunits into isolated chloroplasts (42, 43). Synechococcus small subunits, which cannot normally assemble with plant large subunits, were made assembly competent by replacing residues 45 through 53 with the longer pea small-subunit sequence (residues 45 through 65) (Fig. 3A) (42). Replacement of the conserved Arg53 with Glu was also shown to block the assembly of pea small subunits with large subunits in the pea chloroplast (43), further indicating that the longer βA/βB loop of land plants is essential for holoenzyme assembly. However, because only a small amount of holoenzyme assembles in isolated chloroplasts, it is not possible to assess the effect of βA/βB loop replacements on Rubisco function by this method.
Figure 3.
The Rubisco small subunits of plants and green algae contain a long loop between β strands A and B. (A) Sequences of the βA/βB region of small subunits from various species (5, 14, 21, 37–39). The N54S and A57V suppressor substitutions in the small subunit of Chlamydomonas are denoted with arrows. Locations of β strands A and B (gray highlighting) are assigned from the x-ray crystal structures of spinach (21), tobacco (40), Galdieria partita (38), and Synechococcus (41) Rubisco. (B) Stereo view of large- and small-subunit structural features within the x-ray crystal structure of spinach Rubisco (21). Leu290 and Val262 reside in one large subunit, whereas Ala222 is in a neighboring large subunit. These residues are identical to those in Chlamydomonas Rubisco. An L290F substitution in the large subunit of Chlamydomonas is complemented by A222T and V262L large-subunit substitutions (7, 13, 20). Arg53, Pro59, Gly60, Tyr61, and Tyr62 (Arg59, Cys65, Leu66, Tyr67, and Tyr68 in Chlamydomonas Rubisco) reside within the small-subunit βA/βB loop, which is represented as a ribbon. An R53E substitution in the small subunit of pea Rubisco blocks holoenzyme assembly (43).
In the present study, analysis of photosynthesis-competent revertants of the temperature-conditional rbcL-L290F Chlamydomonas mutant identified two mutations in the RbcS2 gene that cause amino acid substitutions in the small-subunit βA/βB loop. The resulting N54S and A57V substitutions occur in an additional 6-residue sequence of the loop that appears to be unique to the small subunits of green algae (Fig. 3A). Either substitution increases the amount of holoenzyme in cells grown at the 35°C restrictive temperature (Table 2), and restores Ω of the rbcL-L290F mutant enzyme to the wild-type value (Table 3). This 10% increase in Ω is apparent even though the revertant enzymes contain some wild-type subunits encoded by the RbcS1 gene. In the absence of the L290F large-subunit substitution, the N54S and A57V substitutions do not increase Ω above the wild-type value regardless of whether some wild-type small subunits are present (Tables 3 and 4). However, they do substantially improve the thermal stability of the otherwise wild-type enzyme in vitro (Fig. 2). Thus, specific residues within the βA/βB loop can, in fact, play a role in holoenzyme stability/assembly as proposed previously (42, 43). Moreover, these small-subunit residues can also contribute to carboxylation efficiency and Ω despite the fact that the large-subunit active site is more than 20 Å distant (21). It is thus apparent that the small-subunit βA/βB loop may be a productive target for ultimately engineering an improved Rubisco (9, 19).
How Does the Small Subunit Influence Catalysis and Thermal Stability?
In previous studies (7, 20), A222T and V262L large-subunit substitutions were also found to complement the original L290F mutant substitution. On the basis of the x-ray crystal structure of spinach Rubisco (21), Ala222, Val262, and Leu290 are in close contact with residues in the small-subunit βA/βB loop (Fig. 3B). Large-subunit Ala222 is in Van der Waals contact with small-subunit Tyr61 (Tyr67 in the Chlamydomonas enzyme), and Val262 is in Van der Waals contact with Pro59 (Cys65 in the Chlamydomonas enzyme). Large-subunit Leu290 may be close to small-subunit residues Pro59, Gly60, and Tyr62 (Cys65, Leu66, and Tyr68 in the Chlamydomonas enzyme). Thus, although Ala222 and Val262 are not in Van der Waals contact with Leu290, all three residues interact with the same region of the small-subunit βA/βB loop. Because the x-ray crystal structure of Chlamydomonas Rubisco has not yet been solved (44), and because the N54S and A57V small-subunit substitutions replace residues that are apparently absent from the spinach Rubisco structure (Fig. 3A), it is difficult to deduce the location of the Asn54 and Ala57 residues within the Chlamydomonas enzyme. However, Arg53 of spinach Rubisco (homologous to the Arg59 of Chlamydomonas that is adjacent to the extra residues) is in Van der Waals contact with small-subunit residues Pro59 and Gly60, and hydrogen bonds with Tyr61 across the hydrophobic core of the loop (Fig. 3B). Thus, all of the small-subunit (N54S and A57V) and large-subunit (A222T and V262L) suppressor substitutions occur at residues that may be in contact with the small-subunit region closest to Leu290 (Cys65 through Tyr68 in Chlamydomonas; Pro59 through Tyr62 in spinach) (Fig. 3).
The L290F large-subunit substitution may affect catalysis by disrupting a series of hydrogen-bonded residues that extends to His327 in the active site (7, 9, 20). It had previously been proposed that the large-subunit A222T and V262L substitutions complemented the original rbcL-L290F mutant enzyme via structural interactions in the hydrophobic core of the large-subunit α/β-barrel, or via structural rearrangements of the small-subunit βA/βB loop (7, 20). Now that small-subunit N54S and A57V substitutions have been identified that can complement the large-subunit L290F substitution, it would seem more likely that the large-subunit A222T and V262L substitutions complement L290F by influencing the structure of the small-subunit βA/βB loop (Fig. 3B).
The large- (A222T and V262L) and small-subunit (N54S and A57V) suppressor substitutions improve the rbcL-L290F mutant enzyme by increasing Ω and Vc (refs. 7, 20; Table 3). However, in the absence of the original L290F substitution, none of the small- or large-subunit suppressor substitutions increases Ω or Vc of the otherwise wild-type enzyme (ref. 7; Table 3). In contrast, all of these large- and small-subunit suppressor substitutions substantially improve the in vitro thermal stability of wild-type Rubisco (ref. 7; Fig. 2). Perhaps each of the suppressor substitutions alters the structure of the βA/βB loop in such a way as to create a similar kind of cavity at the small-/large-subunit interface that accommodates the increased bulk of the Phe side chain. The reduction in steric hindrance at Phe290 would then be responsible for restoring catalysis and improving holoenzyme thermal stability. In the absence of the larger Phe side chain, the cavity produced by each of the suppressor substitutions might permit greater flexibility at the large-/small-subunit interface, and this increased conformational freedom may account for increased thermal stability in vitro (45). If such is the case, one might expect that the introduction of a variety of smaller residues at the subunit interface might improve thermal stability without necessarily affecting catalysis.
A mutation in the Rbc-1 nuclear gene, which is not linked to the RbcS locus, also suppresses the rbcL-L290F mutation in much the same way as the small- (N54S and A57V) and large-subunit (A222T and V262L) suppressor substitutions (11, 12). The Rbc-1 nuclear suppressor increases Ω, Vc and the amount of rbcL-L290F Rubisco (11) and, in the absence of the L290F substitution, increases the thermal stability of wild-type Rubisco (12). Considering that the βA/βB loop is required for the assembly of the eukaryotic Rubisco holoenzyme (42, 43) and that a number of small- (N54S and A57V) and large-subunit (A222T, V262L, and L290F) substitutions in the βA/βB loop region can influence thermal stability, it would seem possible that Rbc-1 encodes a protein that may participate in Rubisco holoenzyme assembly via conformational arrangement of the βA/βB loop. Perhaps the product of the Rbc-1 mutant gene folds the βA/βB loop in a way that also creates a similar kind of cavity at the small-/large-subunit interface.
Additional Regions of the Small Subunit May Control Ω.
In previous studies, coexpression of large subunits from the cyanobacterium Synechococcus (Rubisco Ω = 41) and small subunits from the diatom Cylindrotheca (Rubisco Ω = 107) in E. coli produced a hybrid holoenzyme that had an intermediate Ω value of 65 (46). By exploiting chloroplast transformation of tobacco, large subunits from sunflower (Rubisco Ω = 98) were assembled with the resident small subunits of tobacco (Rubisco Ω = 85) to produce a hybrid holoenzyme with an Ω value of 89 (5). Because the resulting hybrid holoenzymes had greater than 80% decreases in Vc (5, 46), one cannot accurately judge their net carboxylation efficiencies solely from the values of Ω (8, 9, 19). Nonetheless, it is apparent that the Ω values of these hybrid holoenzymes were substantially influenced by the contributed small subunits. The size of the small-subunit βA/βB loop in the hybrid enzymes does not account for this influence on Ω. Synechococcus and Cylindrotheca small subunits lack the larger loop that is characteristic of plants and green algae, and tobacco and sunflower small subunits have larger loops of identical size (Fig. 3A). However, the results of the present study indicate that specific substitutions in the longer βA/βB loop characteristic of plants and green algae can influence Ω (Table 3). Thus, either the nature of the differences between these loops accounts for differences in relative values of Ω (Fig. 3A), or there are additional regions of the small subunit that can also influence Ω. It will be possible to examine these alternatives via directed mutagenesis and transformation of Chlamydomonas (24).
Acknowledgments
This study was supported by the Department of Energy (Grant DE-FG03–00ER15044) and by the Nebraska Agricultural Research Division (Journal Series Paper 13124).
Abbreviations
- Rubisco
ribulose-1,5-bisphosphate carboxylase/oxygenase
- RuBP
ribulose 1,5-bisphosphate
- Ω
CO2/O2 specificity factor
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.260503997.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.260503997
References
- 1.Harris E H. In: The Molecular Biology of Chloroplasts and Mitochondria in Chlamydomonas. Rochaix J D, Goldschmidt-Clermont M, Merchant S, editors. Dordrecht, The Netherlands: Kluwer; 1998. pp. 1–11. [Google Scholar]
- 2.Spreitzer R J. In: The Molecular Biology of Chloroplasts and Mitochondria in Chlamydomonas. Rochaix J D, Goldschmidt-Clermont M, Merchant S, editors. Dordrecht, The Netherlands: Kluwer; 1998. pp. 515–527. [Google Scholar]
- 3.Cloney L P, Bekkaoui D R, Hemmingsen S M. Plant Mol Biol. 1993;23:1285–1290. doi: 10.1007/BF00042362. [DOI] [PubMed] [Google Scholar]
- 4.Getzoff T P, Zhu G, Bohnert H J, Jensen R G. Plant Physiol. 1998;116:695–702. doi: 10.1104/pp.116.2.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kanevski I, Maliga P, Rhoades D F, Gutteridge S. Plant Physiol. 1999;119:133–141. doi: 10.1104/pp.119.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whitney S M, von Cammerer S, Hudson G S, Andrews T J. Plant Physiol. 1999;121:579–588. doi: 10.1104/pp.121.2.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Du Y, Spreitzer R J. J Biol Chem. 2000;275:19844–19847. doi: 10.1074/jbc.M002321200. [DOI] [PubMed] [Google Scholar]
- 8.Laing W A, Ogren W L, Hageman R H. Plant Physiol. 1974;54:678–685. doi: 10.1104/pp.54.5.678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spreitzer R J. Annu Rev Plant Physiol Plant Mol Biol. 1993;44:411–434. [Google Scholar]
- 10.Chen Z, Spreitzer R J. Photosynth Res. 1992;31:157–164. doi: 10.1007/BF00028792. [DOI] [PubMed] [Google Scholar]
- 11.Chen Z, Green D, Westhoff C, Spreitzer R J. Arch Biochem Biophys. 1990;283:60–67. doi: 10.1016/0003-9861(90)90612-3. [DOI] [PubMed] [Google Scholar]
- 12.Chen Z, Hong S, Spreitzer R J. Plant Physiol. 1993;101:1189–1194. doi: 10.1104/pp.101.4.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Z, Chastain C J, Al-Abed S R, Chollet R, Spreitzer R J. Proc Natl Acad Sci USA. 1988;85:4696–4699. doi: 10.1073/pnas.85.13.4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldschmidt-Clermont M, Rahire M. J Mol Biol. 1986;191:421–432. doi: 10.1016/0022-2836(86)90137-3. [DOI] [PubMed] [Google Scholar]
- 15.Hemmingsen S M, Woolford C, van der Vies S M, Tilly K, Dennis D T, Georgopoulos C P, Hendrix R W, Ellis R J. Nature (London) 1988;333:330–334. doi: 10.1038/333330a0. [DOI] [PubMed] [Google Scholar]
- 16.Bertsch U, Soll J, Seetharam R, Viitanen P V. Proc Natl Acad Sci USA. 1992;89:8696–8700. doi: 10.1073/pnas.89.18.8696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roy H, Hubbs A, Gilson M, Chaudhari P. In: From Light to Biosphere. Mathis P, editor. Vol. 5. Dordrecht, The Netherlands: Kluwer; 1995. pp. 53–58. [Google Scholar]
- 18.Gotor C, Hong S, Spreitzer R J. Planta. 1994;193:313–319. [Google Scholar]
- 19.Spreitzer R J. Photosynth Res. 1999;60:29–42. [Google Scholar]
- 20.Hong S, Spreitzer R J. J Biol Chem. 1997;272:11114–11117. doi: 10.1074/jbc.272.17.11114. [DOI] [PubMed] [Google Scholar]
- 21.Andersson I. J Mol Biol. 1996;259:160–174. doi: 10.1006/jmbi.1996.0310. [DOI] [PubMed] [Google Scholar]
- 22.Spreitzer R J, Mets L. Plant Physiol. 1981;67:565–569. doi: 10.1104/pp.67.3.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spreitzer R J, Al-Abed S R, Huether M J. Plant Physiol. 1988;86:773–777. doi: 10.1104/pp.86.3.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khrebtukova I, Spreitzer R J. Proc Natl Acad Sci USA. 1996;93:13689–13693. doi: 10.1073/pnas.93.24.13689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thow G, Zhu G, Spreitzer R J. Biochemistry. 1994;33:5109–5114. doi: 10.1021/bi00183a014. [DOI] [PubMed] [Google Scholar]
- 26.Yanisch-Perron C, Vieira J, Messing J. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 27.Bullock W O, Fernandez J M, Short J M. Biotechniques. 1987;5:376–379. [Google Scholar]
- 28.Kindle K L. Proc Natl Acad Sci USA. 1990;87:1228–1232. doi: 10.1073/pnas.87.3.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spreitzer R J, Chastain C J. Curr Genet. 1987;11:611–616. [Google Scholar]
- 30.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 31.Jordan D B, Ogren W L. Plant Physiol. 1981;67:237–245. doi: 10.1104/pp.67.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spreitzer R J, Jordan D B, Ogren W L. FEBS Lett. 1982;148:117–121. [Google Scholar]
- 33.Kuehn G D, Hsu T C. Biochem J. 1978;175:909–912. doi: 10.1042/bj1750909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Silflow C D. In: The Molecular Biology of Chloroplasts and Mitochondria in Chlamydomonas. Rochaix J D, Goldschmidt-Clermont M, Merchant S, editors. Dordrecht, The Nethlerlands: Kluwer; 1998. pp. 25–40. [Google Scholar]
- 35.Jordan D B, Ogren W L. Nature (London) 1981;291:513–515. [Google Scholar]
- 36.Read B A, Tabita F R. Arch Biochem Biophys. 1994;312:210–218. doi: 10.1006/abbi.1994.1301. [DOI] [PubMed] [Google Scholar]
- 37.McIntosh L, Poulsen C, Bogorad L. Nature (London) 1980;288:556–560. [Google Scholar]
- 38.Sugawara H, Yamamoto H, Shibata N, Inoue T, Okada S, Miyake C, Yokota A, Kai Y. J Biol Chem. 1999;274:15655–15661. doi: 10.1074/jbc.274.22.15655. [DOI] [PubMed] [Google Scholar]
- 39.Hwang S R, Tabita F R. J Biol Chem. 1991;266:6271–6279. [PubMed] [Google Scholar]
- 40.Schreuder H A, Knight S, Curmi P M G, Andersson I, Cascio D, Sweet R M, Branden C I, Eisenberg D. Protein Sci. 1993;2:1136–1146. doi: 10.1002/pro.5560020708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Newman J, Gutteridge S. J Biol Chem. 1993;268:25876–25886. [PubMed] [Google Scholar]
- 42.Wasmann C C, Ramage R T, Bohnert H J, Ostrem J A. Proc Natl Acad Sci USA. 1989;86:1198–1202. doi: 10.1073/pnas.86.4.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Flachmann R, Bohnert H J. J Biol Chem. 1992;267:10576–10582. [PubMed] [Google Scholar]
- 44.Yen A, Haas E J, Selbo K M, Ross II C R, Spreitzer R J, Stezowski J J. Acta Crystallogr. 1998;D54:668–670. doi: 10.1107/s0907444997016211. [DOI] [PubMed] [Google Scholar]
- 45.Fitter J, Heberle J. Biophys J. 2000;79:1629–1636. doi: 10.1016/S0006-3495(00)76413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Read B A, Tabita F R. Biochemistry. 1992;31:5553–5559. doi: 10.1021/bi00139a018. [DOI] [PubMed] [Google Scholar]