Summary
σ factors are transcriptional regulatory proteins that bind to the RNA polymerase and dictate gene expression. The extracytoplasmic function σ factors (ECF) govern the environment dependent regulation of transcription. ECF σ factors have two domains σ2 and σ4 that recognize the −10 and −35 promoter elements. However, unlike the primary σ factor σA, the ECF σ factors lack σ3, a region that helps in the recognition of the extended −10 element and σ1.1, a domain involved in the auto-inhibition of σA in the absence of core RNA polymerase. Mycobacterium tuberculosis σC is an ECF σ factor that is essential for the pathogenesis and virulence of M. tuberculosis in the mouse and guinea pig models of infection. However, unlike other ECF σ factors, σC does not appear to have a regulatory anti-σ factor located in the same operon. We also note that Mycobacterium tuberculosis σC differs from the canonical ECF σ factors as it has an N-terminal domain comprising of 126 amino acids that precedes the σC2 and σC4 domains. In an effort to understand the regulatory mechanism of this protein, the crystal structures of the σC2 and σC4 domains of σC were determined. These promoter recognition domains are structurally similar to the corresponding domains of σA despite the low sequence similarity. Fluorescence experiments using the intrinsic tryptophan residues of σC2 as well as surface plasmon resonance measurements reveal that the σC2 and σC4 domains interact with each other. Mutational analysis suggests that the Pribnow box binding region of σC2 is involved in this inter-domain interaction. Interaction between the promoter recognition domains in M. tuberculosis σC are thus likely to regulate the activity of this protein even in the absence of an anti-σ factor.
Introduction
The slow growing pathogen, Mycobacterium tuberculosis, has only one ribosomal RNA operon. An efficient transcription mechanism is thus essential for the survival and pathogenecity of this bacillus. Although a third of the human population are carriers of this pathogen, only a few contract this disease. This bacillus can lie latent in the host for prolonged periods of time till a mycobacterial infection gets triggered, most often in the setting of decreased host immunity. Studies that help further our understanding of the latent phase of this bacillus are thus vital to understand the mechanisms of pathogenesis and virulence in this organism. This ability of M. tuberculosis to survive in the hostile environmental conditions in the carrier is brought about by adaptability to rapidly changing environmental conditions. The survival mechanism is thus crucially dependent on efficient communication between the mechanism that senses the environment and the molecules that regulate transcription.
The DNA dependent RNA polymerase (RNAP) that controls gene expression consists of five domains- two α subunits, one ω subunit and the β and β′ domains. The σ-factor reversibly associates with the RNAP and provides the RNAP enzyme with the ability to recognize promoter regions on the DNA template. M. tuberculosis has a total of 13 σ-factors: 3 primary σ factors and 10 extra-cytoplasmic function (ECF) σ factors. Differences in the promoter specificity of these σ factors lead to differential gene expression (1, 2). The task of environment-dependent transcription modulation is performed by the ECF σ factors. Regulatory mechanisms, in turn, dictate which σ factors get activated to bind to the apo-RNAP and initiate transcription and which ones are not. This regulatory mechanism is coupled to a signal transduction system that senses the environment. The interplay between signal transduction and the transcriptional regulatory mechanisms allows the bacillus to respond to changes in the environment by synthesizing new proteins or down-regulating others.
Studies on the transcription mechanism in E. coli have shown that a σ factor directs promoter-specific transcription initiation by specific interactions between two hexamers of a consensus DNA sequence- the Pribnow box (−10 element) and the −35 element. The crystal structure and limited proteolysis experiments on the principal σ factor σA revealed a structural arrangement comprising of a series of domains connected by flexible linkers. The utility of this structural arrangement is borne out by the observation that substantial structural rearrangements take place in the σ-factor during transcription initiation. Exposed surfaces of each of these domains were found to be important for RNAP binding (3). Interaction with the core RNAP has been demonstrated to structurally rearrange the σ factor into an active conformation in which the DNA binding regions of σ2 and σ4 domains are exposed and appropriately positioned to recognize the −10 and −35 elements (3,4,5,6).
Two distinct types of mechanisms have been proposed to describe the regulation of the activity of the σ factors that belong to the σ70 family. Thus while auto-inhibition by the N-terminal domain mediates the activity of the principal σ factor, σA, protein-protein interactions mediated by the anti- and anti-anti-σ factors control σ factor concentrations in the case of the ECF σ factors. Other regulatory mechanisms have been proposed wherein σ factor activity is controlled by stimulatory signals (such as the phosphorylation and dephosphorylation processes controlled by the kinases and phosphatases) or mechanisms that can control the intracellular σ factor concentration by localizing it on the inner surface of the cell membrane. In the case of the E. coli σ factor σE (7) for example, many of the key regulators of σE activities (products of the rse genes) are co-transcribed with RpoE to form an operon : rpoE, rseA, rseB and rseC. The protein RseA is an anti-σ factor for σE whereas the periplasmic RseB protein enhances its activity. The activity of σE can also be regulated by cytoplasmic factors like guanosine 3′,5′-bispyrophosphate (ppGpp). The signaling pathway employed in this process is distinct from the pathway that is activated upon extra-cytoplasmic stress (8). Very little is known of these processes in mycobacteria although recent reports indicate that work to delineate the regulatory mechanism of σE and another σE-like factor σH are presently in progress (9,10,11). The anti-σ factor, as seen in the cases of the SpoIIAB (12) and the σE-RseA complex (4), appears to bind the free σ factor, thereby preventing its interaction with the core RNAP. The σ28/FlgM complex (6) on the other hand is different from the others in that FlgM can also form a ternary complex with the σ28 holoenzyme, thereby destabilizing the σ28/RNAP interaction. The modes of σ factor anti-σ factor interactions also differ between the σ70 and σ28 families. The anti-σ factor RseA is sandwiched between σE2 and σE4 domains of σE whereas the FlgM protein wraps around the outside of σ28 and occludes the core RNAP binding determinants on σ282 and σ284
M. tuberculosis has about 190 transcription regulators (13) which include 13 σ factors, 11 two-component systems, 5 unpaired response regulators, 11 protein kinases and 140 other proteins. The resulting interaction amongst these proteins suggests a complex system of overlapping functions and redundancies (9). Four σ factors, σA, σB, σC and σE of the 13 in M. tuberculosis are conserved in all the pathogenic mycobacteria. Only four σ factors in M. tuberculosis σA, σC, σD and σL have been examined in detail for their roles in virulence and pathogenecity. The ECF σ factor σC is essential for lethality in mice but is not required for bacterial survival in this species (14). Based on genomic microarray data, σC was demonstrated to modulate the expression of several key virulence associated genes including hspX, senX3 and mtrA, a two component sensor kinase and a two component response regulator. The phenotype of the Δ σC mutant of M. tuberculosis can thus persist in tissues but is attenuated in its ability to elicit lethal immunopathology (14). A recent report also suggests σC to be a key regulator of pathogenesis and adaptive survival in the lung and spleen of guinea pigs (15). These in vivo studies in two different animal models thus demonstrate the role of σC in the pathogenesis and virulence of M. tuberculosis. Although σC does not appear to have an anti-σ factor located in the same operon, in silico analysis for the identification of potential interacting partners for σC suggested two proteins SirR and the unannotated protein Rv0093c. However, the interaction between these proteins and σC could not be substantiated by experimental evidence.
In this manuscript, we report the biochemical and structural analysis of the domain organization of σC. Despite the low sequence similarity between the promoter recognition domains of σC with the primary σ-factor σA, we observe that the crystal structure of the two promoter recognition domains are conserved. Spectroscopic studies using the intrinsic tryptophan fluorescence as well as surface plasmon resonance experiments suggest that the σC2 and σC4 domains interact in vitro. Interactions between the two promoter recognition domains of σC which involves the occlusion of the Pribnow box recognition region of σC2 suggests that substantial inter- domain rearrangements would be needed to activate σC even in the absence of an anti σ-factor.
Methods
Cloning, Expression and Purification of σC, and the σC2 and σC4 domains
The details of the expression constructs used in this study are compiled in Table 1. After transforming the plasmid into BL21 (DE3) or Rosetta origami cells (Novagen, Inc.), the cells were grown in Luria broth with an antibiotic selection (ampicillin 100 μg/ml and chloramphenicol 30 μg/ml) to an OD600 nm of 0.5-0.6. The cells were induced with 0.2 mM IPTG (final concentration). Subsequently, the growth temperature was lowered to 290 K and cells were grown for further 12-18 hrs before they were spun down and stored at 193 K. Full length σC (311 amino acids) was purified in denaturing conditions using 8M urea. Refolding of the denatured protein was carried out in a step-wise fashion with five buffer changes (each lasting about 3-4 hours) with decreasing concentration of urea. The recombinant proteins from the other three constructs were purified under native conditions as described earlier (16). The proteins were further purified by size exclusion chromatography using a Superdex S-200 column (Amersham-Pharmacia, Inc.) after the affinity chromatography step. The proteins were concentrated using membrane based centrifugal ultra filtration (Amicon). L-Arginine and L-glutamic acid were added to the final concentration of 50mM each during the concentration step (17). The purity of the samples was analyzed using SDS-PAGE followed by Coomassie blue staining. Both full length σC as well as the σC4 domain have poor solubility and are very unstable. These proteins were freshly purified prior to each biochemical experiment. The molecular weights of the recombinant proteins were also verified by Mass spectrometry on a MALDI-TOF (Bruker Daltonics, Inc) mass spectrometer.
Table 1.
List of Expression constructs used in the analysis of the domain organization of M. tuberculosis σC
| S.No | Construct | Restriction sites used 5′ , 3′ |
Construct size |
|---|---|---|---|
| 1 | pET15b-σC127-311aa | NdeI, BamHI | 127-311aa |
| 2 | pET15b-σC2 | NheI, XhoI | 127-238aa |
| 3 | pET15b-σC4 | NdeI, BamHI | 243-311aa |
| 5 | pETduet1-σC | EcoRI, HindIII | 1-311aa |
| 6 | pETduet-σC-Rv0093c | EcoRI, HindIII; Nde1-Xho1 | 127-311; 1-95 |
| 7 | pETduet-σC-SirR | EcoRI, HindIII; Nde1-Kpn1 | 127-311; 88-149 |
The western blot analysis to determine the molecular weight of σC in situ was performed according to the standard ocedure outlined in Molecular Cloning (18). Cell free lysate of H37Rv was obtained from the TB vaccine testing and research material contract at the Colorado state University. The cell lysate and purified protein samples were resolved in 10% SDS-PAGE, and transferred to a nitrocellulose membrane. A polyclonal antibody raised in rabbit against σC127-311 (a smaller construct containing residues 127-311aa based on the annotated sequence of H37Rv) was used as the primary antibody for σC detection. Western blots were developed using the AEC (Sigma-Aldrich, Inc.) substrate with HRP-labeled sheep anti-rabbit IgG (Bangalore Genei Co.)
Site directed mutagenesis experiments were performed using a PCR based method (19). The primers GATAGGCGACGAACCGCGCCACGTCTTGCTGGG (for W164A) and CGATGGCCAGCAACGCAGTTCGGGCGCTGG (for the W203A point variant) were used. These mutations were confirmed by DNA sequencing.
Fluorescence Spectroscopy
All fluorescence studies were performed on a JOBIN YVON FlouroMax-3 fluorimeter at room temperature. The fluorescence excitation was set at 280 nm and emission spectra were recorded from 300 to 400 nm at a band-width of 1 nm. The excitation and emission slit widths were set to 3 and 5 nm, respectively. Each spectrum is an average of five scans. The spectra were obtained at a protein concentration of ∼ 3 μM in 10 mM Tris-HCl, 50 mM NaCl pH 7.5. Interactions between the σC2 and σC4 domains were monitored after the two protein samples were mixed and incubated at room temperature for 2 minutes prior to data acquisition.
Surface Plasmon Resonance Experiments
Surface plasmon resonance (SPR) assays were performed on a BIAcore 2000 instrument (BIAcore AB). All experiments were conducted at 25°C. SPR experiments were performed with either the σC2 and σC4 domains immobilized onto carboxylated dextran chips (sensor chip CM5 from BIAcore AB) using the standard amine coupling procedure as recommended by the manufacturer. Immobilization resulted in a change in the refractive index corresponding to 700 and 640 resonance units (RU) for σC2 and σC4 respectively. Binding and kinetic assays were performed in 50 mM sodium phosphate (pH 7.4), 100 mM NaCl at flow rate of 20 μL/min. The proteins were diluted in this buffer and experiments were carried out at concentrations ranging from 0.8–200 μM. Freshly purified protein samples were used for these binding assays. Dissociation was initiated by replacing the analyte with buffer. The association and dissociation curves were monitored for 120s. Sensograms were analyzed with BIAevaluation software version 2 (BIAcore AB). A 1:1 langmuir binding model was used to fit the curves.
Model building and Refinement of the two domains of σC
Crystallization conditions and data collection strategies have been reported previously (16). The data collection and refinement statistics for both the domains are reported in Table 2. The N- and C-terminal domains of E.coli σE (PDB code 1OR7) were used as starting model(s) for Molecular Replacement (MR). The sequence identity between σC2 and N-term σE is 23% whereas that between the C-term σE and σC4 domain is 45%. MR calculations were performed using the program PHASER(20). The solution for σC2 and σC4 had log likelihood gains of 71.52 and 489.97 with corresponding Z-scores of 5.21 and 15.53 respectively. There were two molecules in the asymmetric unit in the case of σC2 and three in the asymmetric unit for the σC4 domain. Both structures were refined using tight non-crystallographic symmetry (NCS) restraints. Iterated cycles of model building using Coot (21) and refinement using Refmac(22) led to the final model for σC2 with an Rcryst of 18.9% and Rfree of 24.3%. The final model σC4 had Rcryst of 21.0 % and Rfree of 26.7% The final model of σC2 consists of 88 residues, 4 sulfate ions and 20 water molecules whereas that for the σC4 domain has 60 residues, 8 sulfate ions and 17 water molecules.
Table 2.
Summary of data collection, processing and refinement statistics
| σC2 | σC4 | |
|---|---|---|
| Wavelength (Å) | 1.54 | 1.54 |
| Resolution (Å) | 35.01-2.70 (2.89-2.70) | 40.29-3.0 (3.16-3.0) |
| Number of reflections | 45969 (6060) | 23915 (2920) |
| Number of unique reflections | 17709 (2612) | 7040 (1002) |
| Completeness | 99.8 (99.2) | 99.4 (97.9) |
| Redundancy | 2.6 (2.3) | 3.4 (2.9) |
| Rsym | 0.087 (0.387) | 0.105 (0.546) |
| I/σ | 10.9 (2.3) | 11.1 (2.0) |
| Rcryst | 20.1 (26.0) | 20.8 (33.8) |
| Rfree | 24.7 (24.7) | 26.4 (33.2) |
Values for outer shells are given in paranthesis.
Rsym=Σj|<I>-Ij|/Σ<I> where Ij is the intensity of the jth reflection and <I> is the average intensity.
Rcryst=Σhkl|Fo – Fc|/Σhkl|Fo|
Rfree was calculated as for Rcryst but on 5 % of the data excluded from the refinement calculation.
Computational strategies to identify potential σC interacting proteins
In M. tuberculosis the following σ-anti σ pairs have been reported: σD-Rv3413c, σE-Rv1222, σF-Rv3287c, σH-Rv3222c and σL-Rv0736 (23,24,25,26,27). σD, σF, σH and their respective anti-σ factors are canonical in the sense that these pairs occur as a part of one operon. σC the only member in an operon in both the M. tuberculosis strains H37Rv and CDC1551. Assuming that interacting proteins would show correlated expression levels under certain conditions, the correlation coefficients and t values (Student's t test) between a given σ factor and the corresponding anti-σ pair were calculated in different conditions (7H9 medium(N=4), Balb mice(N=4), Scid mice(N=4), stationary state(N=6), Low oxygen dormancy(N=9) using the equations given below:
To predict all the interacting proteins/anti-σ factor(s) for σC, we examined all the transcription regulatory genes that are present in M. leprae (M. Leprae has only 4 σ factors σA, σB, σC, and σE). An amino-acid sequence based search was also performed to locate putative anti-σ factors on the basis of the sequence signature (HXXXCXXC) (27). Transmembrane protein prediction was done using the program Conpred (28). These results are compiled in Supplementary Table 1.
Results and Discussion
Characterization of the domain organization of M. tuberculosis σC
Differences in the annotation of σC in the two M. tuberculosis strains H37Rv (13) and CDC1551 (29) led to the identification of an N-terminal 126 residue long polypeptide preceding the structured domain σC2. The σC2 and σC4 domains are connected by a flexible ∼25 amino acids long linker. Sequence based prediction of low complexity regions (30) in this protein (Figure 1a) suggests that the N-terminal domain preceding σC2 is largely unfolded. M. tuberculosis extra-cytoplasmic σC thus appears to be more primary σ factor-like in terms of the domain organization except that it lacks the region 3.2 that lies between the σC2 and σC4 domains and is involved in extended −10 promoter recognition. The sequence based structure alignment of M. tuberculosis σC is shown in Figure 2 (adapted from reference 2). Interestingly, Myobacterium leprae σC does not have this N-terminal region. Based on the sequence alignment and patterns of protease sensitivity, four expression constructs of σC were examined: the full length σC, the smaller construct containing only the σC2 and σC4 domains and the two independent domains σC2 and σC4 (Table 1 and Figure 3). In a western blot experiment using cell free lysate of M. tuberculosis H37Rv (obtained from the TB research center, Colorado State University), the polyconal antibody raised against the shorter length construct (lacking the 126 amino acid polypeptide at the N-terminus) recognized the full-length σC. σC is thus a 311 amino acids long protein in both the laboratory (H37Rv) as well as the clinical (CDC1551) strains of M. tuberculosis (Figure 3).
Figure 1.
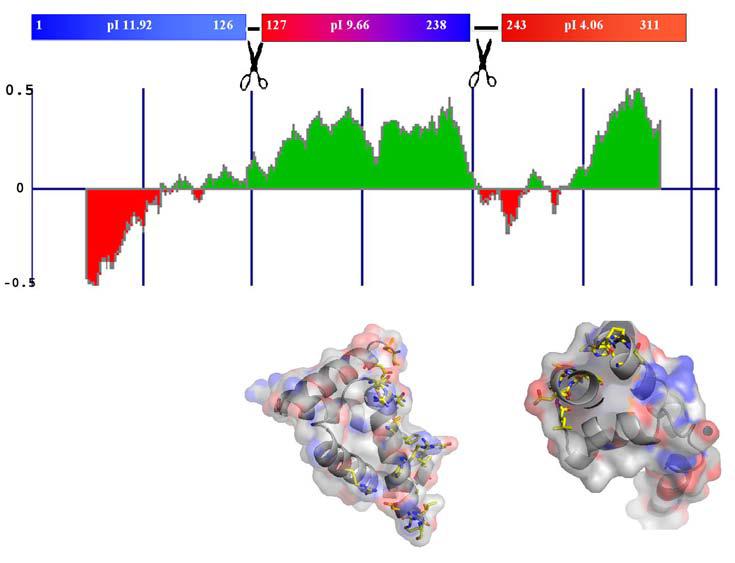
Domain Organization of M. tuberculosis σC. (A) Estimate of the folded nature of the three domains of σC calculated using the program FoldIndex (30). These estimates are consistent with presently available biochemical data on the principal σ factor σA. The top panel shows the pI of different domains in σC. (B) Molecular surface representation of the two domains of σC. Residues that interact with the −10 promoter element (σC2) and the −35 element (σC2) are highlighted in a ball-and-stick representation. These two domains are connected by a flexible linker.
Figure 2.
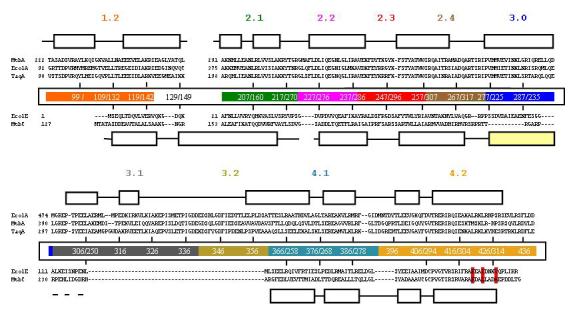
Structure based sequence alignment of σ70 families of σfactors. The top three sequences correspond to those of the principal σ factors with the secondary elements that correspond to the T. aquaticus σA (3). The two sequences below those of the principal σ factors correspond to ECF σ factors. The secondary structure representation in this case is based on M. tuberculosis σC. Regions of the σ factor that have been implicated in promoter recognition are highlighted. The residues crucial for the interactions with RseA in case of E. coli σE are shown in red. The abbreviations are: MtbC: M. tuberculosis σC, MtbA: M. tuberculosis σA, EcolA: E. coli σA, EcolE: E. coli σE, TaqA: T. aquaticus σA.
Figure 3.
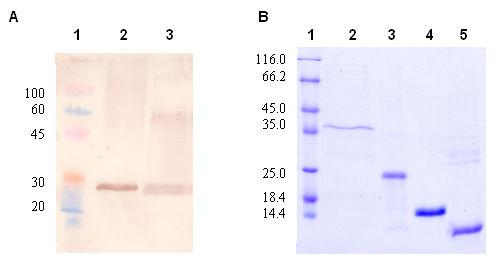
M. tuberculosis σC has an N-terminal polypeptide preceding the σC2 and σC4 domains. (A) Western blot analysis using the cell free lysate of M. tuberculosis H37Rv. The size of the protein corresponds to that of the full-length 311 residue protein of σC. Lane 1:prestained protein marker, lane 2: M. tuberculosis H37RV cell free lysate, lane 3:purified σC1-311aa (B) 12% SDS PAGE gel showing different constructs used in the present study. Lane 1: protein marker, lane 2: σC1-311aa (35.3KDa), lane 3: σC127-311aa (22.5KDa), lane 4: σC 2 (14.7KDa), lane 5: σC 4 (9.5KDa). The details of these expression constructs are given in Table 1.
Structure determination of the σC2 and the σC4 domains
The σE2 and σE4 domains of E. coli σE were used as search models to solve the structures of M. tuberculosis σC2 and σC4 domains by Molecular Replacement. Overall, these two proteins have 32 % identity in the amino acid sequence although the C-terminal region is more conserved than the N-terminal polypeptide. Thus while M. tuberculosis σC2 and E. coli σE2 have 23 % identity, M. tuberculosis σC4 and E. coli σE4 are more similar with ∼ 44 % sequence identity. The crystallization of the σC2 and the σC4 domains has been reported earlier (16). The data collection and refinement statistics are compiled in Table 2 (more details in Supplementary Figure 1). Despite the low sequence similarity, the overall topology of the σC2 and the σC4 domains is similar to that of the known σ factors reported till date (3,4,5,6). The backbone Cα superposition between σC2 with E. coli σA2 shows a root-mean-squared-deviation (rmsd) of 2.97 Å (∼8 % identity over 77 amino acid residues) whereas σC2 superposes better with E. coli E2 with an rmsd of 1.4 Å (24 % identity over 88 residues). The σC4 domain which interacts with the −35 promoter element is structurally more conserved with a backbone rmsd of 1.6 Å with the corresponding domain of the primary σ-factor σA4. Molecular modeling and docking based on the structure of the Thermus aquaticus RNAP suggests that the σC2 and σC4 domains would be spatially separated by a distance of ∼50 Å (data not shown) when bound to the RNAP. This distance is compatible with the ca 25 residue linker joining the σC2 and σC4 domains. Studies on the primary σ-factor σA have shown that aromatic and charged residues in region 2 are involved in recognizing the −10 element and subsequent strand separation (31). On the basis of multiple sequence alignment, W203 was found to be universally conserved and is likely to be involved in interactions with the Pribnow box element. Based on a model of E. coli σE-holoenzyme and the σE-RseA structure (4), Campbell et al proposed that the E. coli anti-σ factor RseA functions by sterically occluding the two primary RNAP-binding regions on σE. Activation of the σE regulon occurs when σE is released from RseA upon degradation by the Hho (DegS) protease. In the σE-RseA structure, the anti-σ factor was found to be sandwiched between the σE2 and the σE4 domains. Several substitutions in E. coli σE4, R178G, I181A and V185A were identified as those that led to defective RseA binding (4). In the case of M. tuberculosis, the equivalent Arg residue is conserved whereas the other two positions (E. coli/M. tub) are replaced by (Ile/Leu) and (Val/Ala).
Identification and verification of potential σC interacting proteins in M. tuberculosis
M. tuberculosis σC is different from other ECF σ factors as there is no anti-σ factor located downstream of σC in the same operon. To understand the mechanism of regulation of transcription by σC, attempts were made to identify the interacting partners that could activate/inactivate this protein. Coimmunoprecipitation using the purified σC antibody using the cell free lysate (obtained from the TB research center, Colorado State University) was used to isolate proteins that interact with σC. These proteins were identified by in situ digestion using trypsin followed by peptide mass-fingerprinting using a MALDI-TOF mass spectrometer and analyzed using MASCOT (Bruker Daltonics, Inc). Few components of RNAP could be detected at high confidence levels (data not shown). However, no potential anti-σ factor could be identified in this experiment. Several computational tools were also used to identify proteins that could potentially interact with σC. These results including relevant entries from the database STRING (32) are compiled in Table 3. The application of this tool for the prediction of protein–protein interactions suffers from the problem that functional interaction does not necessarily imply direct physical interaction. Based on the co-occurrence of genes in related species, proteins exhibiting similar phylogenetic profiles can be predicted to be functionally linked i.e. they occur as a structural complex or are involved in a common metabolic pathway. Although 29 potential interacting proteins could be identified using this approach, none showed a significant correlation in their mRNA expression levels with that of σC. The Rosetta tool for gene fusion analysis (33) led to the identification of the gene Rv0093c. In this method the existence of a fusion protein in one genome allows the prediction of the interaction between the single domain proteins in other genomes. Rv0093c also has an amino-acid sequence signature proposed for anti-σ factors that regulate oxidative stress (27) and a conditional correlation was observed between the expression level of this protein versus σC in one DNA microarray data-set. A pertinent observation in this regard is that notwithstanding the large errors involved in the gene expression levels obtained using DNA microarray technology, well characterized σ-anti σ pairs exhibit significant correlation (α=0.05) under specific environmental or growth conditions (Table 3b).
Table 3.
Computational analysis of proteins that can interact with σC. (A) Results compiled from the program STRING (32) and (B). Analysis of co-expression levels of known σ / anti σ pairs in M. tuberculosis.
| (A) | ||||||
|---|---|---|---|---|---|---|
| Gene- name |
Neighborhood | Gene fusion |
Co-occurence | Text mining |
Homology | Score |
| SigM | − | − | + | + | − | 0.726 |
| Rv0093c | − | + | − | − | − | 0.693 |
| RpoE | − | − | + | + | + | 0.390 |
| SigE | − | − | + | + | + | 0.352 |
| Upk | − | − | − | + | − | 0.267 |
| LepA | + | − | − | − | − | 0.256 |
| DapF | − | − | + | − | − | 0.226 |
| Rv3676 | − | − | + | − | − | 0.223 |
| SigG | − | − | + | − | + | 0.222 |
| Rv1318c | − | − | + | − | − | 0.222 |
| (B) | |||||
|---|---|---|---|---|---|
| Name | Stationary | Low oxygen | 7h9 | Balb | Scid |
| Rv3416(whiB3) | −0.69 (−1.9) | 0.64 (1.7) | −0.96 (−6.9) |
−0.87 (−3.9) |
0.89 (3.9) |
| Rv3413(anti-σD) | 0.92 (4.7) | −0.67(−1.8) | 0.55 (1.3) | −0.38 (−0.8) | −0.34 (−0.7) |
| Rv3287(anti-σF) | 0.50 (1.2) |
0.41 (0.9) |
0.99 (14.0) |
−0.63 (−1.6) |
−0.66 (−1.8) |
| Rv3229(anti-σH) | 0.28 (0.6) |
0.99 (14.0) |
0.43 (1.0) |
0.34 (0.7) |
−0.28 (−0.6) |
| Rv0736(anti-σL) |
0.98 (9.8) |
0.96 (8.9) |
−0.98 (9.8) |
−0.67 (−1.8) |
0.96 (6.9) |
| Rv0093c | −0.76 (−2.3) |
−0.81 (−2.8) |
0.92 (4.7) |
−0.42 (−0.9) |
−0.64 (−1.6) |
Legend: Expression levels are correlated under different conditions. Stationary: stationary phase of growth; Low oxygen:anaerobic conditions; 7h9: growth in 7H9 medium; Balb: M. tuberculosis isolated from immune competent Balb/c mice; scid: M. tuberculosis isolated from severe combined immune-defecient (SCID) mice.
A sequence based search for RseA-like proteins in the M. tuberculosis genome led to the identification of a protein SirR (32 % identity between the E. coli RseA sequence that interacts with σE and M. tuberculosis SirR). The annotation of SirR in the M. tuberculosis genome describes it as an Iron dependent transcription repressor. Two strategies, one involving co-expression using pET-Duet1 vector and the other by in vitro studies using purified recombinant proteins were adopted to examine interactions between σC and the predicted anti-σ factors SirR and Rv0093c. In the co-expression approach, σC was cloned with a poly-histidine tag at the N-terminus to enable the complex to be purified by affinity chromatography. No interaction could be detected between either of these proteins and σC in vitro (data not shown). This finding could probably be rationalized on the basis of the variations seen in residues that are involved in RseA recognition.
Interaction between the two promoter recognition domains σC2 and σC4
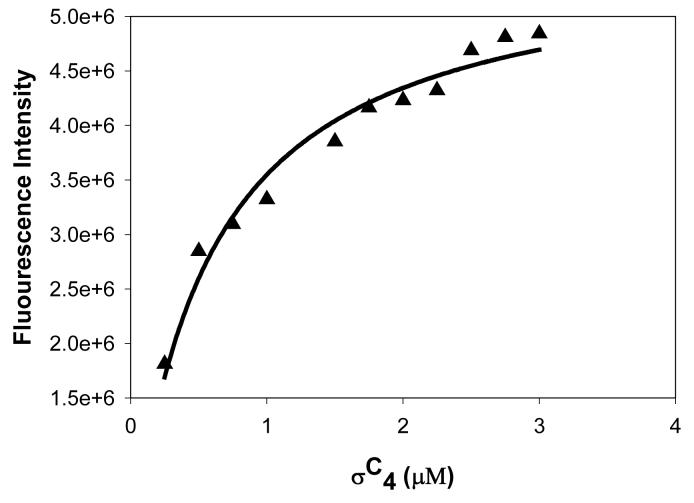
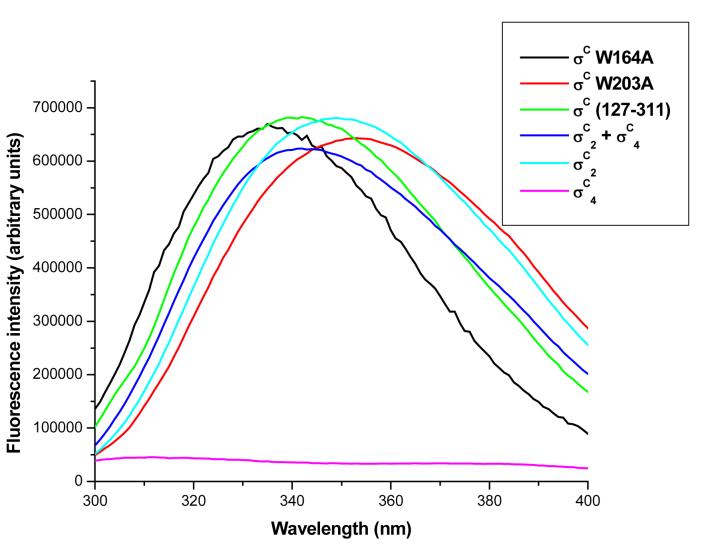
Fluorescence measurements using the intrinsic tryptophan residue in σC2 were used to monitor the interactions between the two promoter recognizing domains of σC. The fluorescence spectra are shown in Figure 4. There are two tryptophans in σC2 (region 2) and none in σC4 (region 4) (Figure 4b). This fortuitous distribution of tryptophan residues helped us examine the interactions between the σC2 and σC4 domains. A fluorescence spectrum of native σC (127-311 residues) showed an emission maximum at 342 nm while σC2 showed an emission maximum at 349 nm. Upon the addition of σC4 in stoichiometric ratio, a blue shift (∼7nm) in the fluorescence spectrum was observed with a reduction in the emission intensity. A representative binding reaction wherein the σC4 domain is titrated into the σC2 domain is shown in Figure 4a. This fluorescence titration suggests a weak interaction between the σC4 and σC2 domains (Kd ∼ 2 μM).
Figure 4.
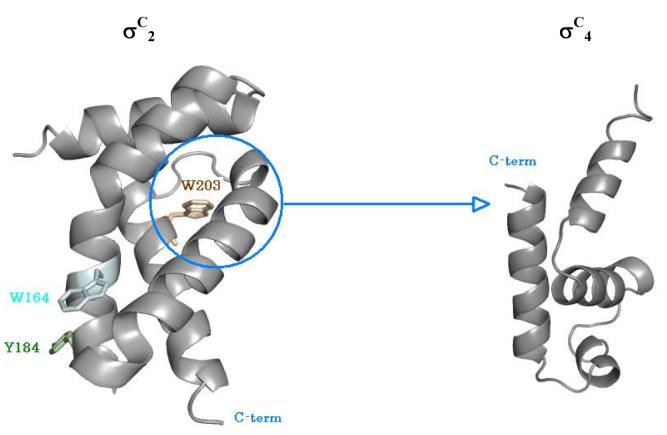
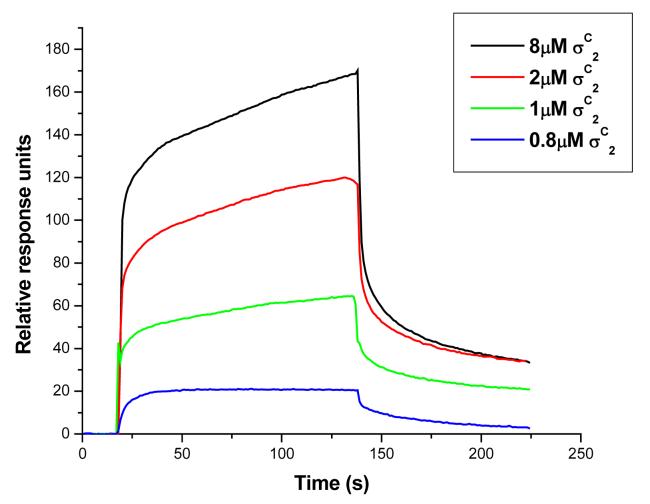
The σC2 and σC4 domains of σC interact with each other in the absence of core RNA polymerase. A. Interaction between the σC2 and σC4 domains monitored by the Fluorescence spectroscopy. In this experiment the quenching of the intrinsic tryptophan flurescence was monitored as a titration with increasing concentrations of the σC4 domain (that has no tryptophan residues). This data was analyzed using the SigmaPlot software suite (Systat, Inc.). B. Changes in the intrinsic tryptophan fluorescence suggests an interaction between the σC2 and σC4 domains. C. Spatial orientations of the two tryptophan and one tyrosine residues on σC2. The probable site for interaction is shown with the circle. D. A representative SPR sensogram that shows interactions between the σC2 and σC4 domains. In this experiment, σC4 was immobilized on the CM-5 chip (Amersham-Pharmacia, Inc.) while the σC2 domain was used as the analyte. The experiments were performed with different concentrations of σC2.
Of the two tryptophans in σC2, Trp-203 is involved in DNA melting (Pribnow Box element). Based on molecular modeling analysis (data not shown) Trp-164 is likely to interact with the RNAP. These observations led us to investigate the specificity of interactions (given the fact that the two domains are connected by a flexible linker of ca 25 amino acids long) and identify the region where σC4 binds σC2. To address these questions, two point variants of σC viz W164A and W203A were constructed in σC127-311 (medium length σC that lacks the positively charged N terminal domain which contains three tryptophan residues). The W164A mutant protein had a fluorescence emission maximum at 335 nm whereas the W203A variant had its emission maximum at 353 nm (Figure 4b). The blue shift seen in the W164A variant suggests that σC4 interacts σC2 in the region that would lead to the partial burial of W203 (Figure 4c). Interactions between the σC2 and σC4 domains thus result in the occlusion of the Pribnow box recognition element.
The interaction between the σC2 and σC4 domains was also monitored by Surface Plasmon Resonance experiments (Figure 4d). SPR studies of the interaction between the σC2 and σC4 domains suggest a low affinity binding between these two domains (Kd=1.81±0.03μM). The binding affinity and kinetic parameters were comparable when either the σC2 and σC4 domain was immobilized. The highly polar nature of this interdomain interaction was apparent from a size exclusion chromatography experiment performed at medium salt concentrations (250mM NaCl) where the two domains migrate separately. This was also supported by fluorescence quenching experiments where increasing salt concentrations not only reduced the fluorescence quenching but also resulted in a red shift in the emission maximum (Supplementary Figure 3).
In summary, the structural and biophysical studies on the two promoter recognition domains of σC shows an interaction between the σC2 and σC4 domains that is mostly governed by polar residues. This interaction involves the occlusion of the region of σC that participates in the recognition of the −10 promoter element. The binding of M. tuberculosis σC to the core RNAP would thus involve substantial structural re-arrangement to release σC from its auto-inhibited state. These observations thus provide an alternate mechanism for the regulation of the ECF σ-factor, σC.
Supplementary Material
References
- 1.Helmann JD. Advances in Microbial Physiol. 2002;46:47–109. doi: 10.1016/s0065-2911(02)46002-x. [DOI] [PubMed] [Google Scholar]
- 2.Waagmeester A, Thompson J, Reyrat JM. Trends Microbiol. 2005;13:505–509. doi: 10.1016/j.tim.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 3.Campbell EA, Muzzin O, Chlenov M, Sun JL, Olson CA, Weinman O, Trester-Zedlitz ML, Darst SA. Mol Cell. 2002;9:527–39. doi: 10.1016/s1097-2765(02)00470-7. [DOI] [PubMed] [Google Scholar]
- 4.Campbell EA, Tupy JL, Gruber TM, Wang S, Sharp MM, Gross CA, Darst SA. Mol. Cell. 2003;11:1067–1078. doi: 10.1016/s1097-2765(03)00148-5. [DOI] [PubMed] [Google Scholar]
- 5.Murakami K, Darst SA. Curr. Opin. struct. Biol. 2003;13:131–139. doi: 10.1016/s0959-440x(02)00005-2. [DOI] [PubMed] [Google Scholar]
- 6.Sorenson MK, Ray SS, Darst SA. Mol. Cell. 2004;14:127–138. doi: 10.1016/s1097-2765(04)00150-9. [DOI] [PubMed] [Google Scholar]
- 7.Lonetto M, Gribskov M, Gross CA. J. Baceriol. 1992;174:3843–3849. doi: 10.1128/jb.174.12.3843-3849.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Costanzo A, Ades SE. J. Bacteriol. 2006;188:4627–4634. doi: 10.1128/JB.01981-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manganelli R, Dubnau E, Tyagi S, Kramer FR, Smith I. Mol. Microbiol. 1999;31:715–724. doi: 10.1046/j.1365-2958.1999.01212.x. [DOI] [PubMed] [Google Scholar]
- 10.Manganelli R, Voskuil MI, Schoolnik GK, Dubnau E, Gomez M, Smith I. Mol. Microbiol. 2002;45:365–374. doi: 10.1046/j.1365-2958.2002.03005.x. [DOI] [PubMed] [Google Scholar]
- 11.Manganelli R, Proveddi R, Rodrigue S, Beaucher J, Gaudreau L, Smith I. J. Bact. 2004;186:895–902. doi: 10.1128/JB.186.4.895-902.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Campbell EA, Masuda S, Sun JL, Muzzin O, Olson CA, Wang S, Darst SA. Cell. 2002;108:795–807. doi: 10.1016/s0092-8674(02)00662-1. [DOI] [PubMed] [Google Scholar]
- 13.Camus JC, Pryor MJ, Medigue C, Cole ST. Microbiology. 2002;148:2967–2973. doi: 10.1099/00221287-148-10-2967. [DOI] [PubMed] [Google Scholar]
- 14.Sun R, Converse PJ, Ko C, Tyagi S, Morrison NE, Bishai WR. Mol. Microbiol. 2004;52:25–38. doi: 10.1111/j.1365-2958.2003.03958.x. [DOI] [PubMed] [Google Scholar]
- 15.Karls RK, Guarner J, McMurray DN, Birkness KA, Quinn FD. Microbiology. 2006;152:1591–1600. doi: 10.1099/mic.0.28591-0. [DOI] [PubMed] [Google Scholar]
- 15.Costanzo DA, Ades SE. J. Bacteriol. 2006;188:4627–4634. doi: 10.1128/JB.01981-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thakur KG, Gopal B. Acta Cryst. 2005;F61:779–781. doi: 10.1107/S1744309105022669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Golovanov AP, Hautbergue GM, Wilson SA, Lian LY. J. Am. Chem. Soc. 2004;126:8933–8939. doi: 10.1021/ja049297h. [DOI] [PubMed] [Google Scholar]
- 18.Sambrook J, Russell DW. Molecular Cloning: A laboratory manual. Third edition Cold Spring Harbour Press; 2001. [Google Scholar]
- 19.Shenoy AR, Visweswariah SS. Anal. Biochem. 2003;319:335–336. doi: 10.1016/s0003-2697(03)00286-0. [DOI] [PubMed] [Google Scholar]
- 20.Storoni LC, McCoy AJ, Read RJ. Acta Cryst. 2004;D60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- 21.Emsley P, Cowtan K. Acta Cryst. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 22.Murshudov GN, Vagin AA, Dodson EJ. Acta Cryst. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 23.Raman S, Hazra R, Dascher CC, Husson RN. J. Bacteriol. 2004;186:6605–6616. doi: 10.1128/JB.186.19.6605-6616.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manganelli R, Voskuil MI, Schoolnik GK, Smith I. Mol. Microbiol. 2001;41:423–437. doi: 10.1046/j.1365-2958.2001.02525.x. [DOI] [PubMed] [Google Scholar]
- 25.Beaucher J, Rodrigue S, Jacques P-E, Smith I, Brzezinski R, Gaudreau L. Mol Microbiol. 2002;45:1527–1540. doi: 10.1046/j.1365-2958.2002.03135.x. [DOI] [PubMed] [Google Scholar]
- 26.Song T, Dove SL, Lee KH, Husson RN. Mol. Microbiol. 2003;50:949–959. doi: 10.1046/j.1365-2958.2003.03739.x. [DOI] [PubMed] [Google Scholar]
- 27.Hahn MY, Raman S, Ananya M, Husson RN. J. Bacteriol. 2005;187:7062–7071. doi: 10.1128/JB.187.20.7062-7071.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arai M, Mitsuke H, Ikeda M, Xia J-X, Kikuchi T, Satake M, Shimizu T. Nuc Acids Res. 2004;32:W390–W393. doi: 10.1093/nar/gkh380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fleischmann RD, Alland D, Eisen JA, Carpenter L, White O, Peterson J, DeBoy R, Dodson R, Gwinn M, Haft D, Hickey E, Kolonay JF, Nelson WC, Umayam LA, Ermolaeva M, Salzberg SL, Delcher A, Utterback T, Weidman J, Khouri H, Gill J, Mikula A, Bishai W, Jacobs WR, Jr, Venter JC, Fraser CM. J Bacteriol. 2002;184:5479–90. doi: 10.1128/JB.184.19.5479-5490.2002. Jr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Priluski J, Felder CE, Zeev-Ben-Mordehai T, Rydberg EH, Man O, Beckmann JS, Silman I, Sussman JL. Bioinformatics. 2005;21:3435–3438. doi: 10.1093/bioinformatics/bti537. [DOI] [PubMed] [Google Scholar]
- 31.Tomsic M, Tsujikawa L, Panaghie G, Wang Y, Azok J, deHaseth PL. J. Biol. Chem. 2001;276:31891–31896. doi: 10.1074/jbc.M105027200. [DOI] [PubMed] [Google Scholar]
- 32.Von Mering C, Jensen LJ, Snel B, Hooper SD, Krupp M, Foglierini M, Jouffre N, Huynen MA, Bork P. Nuc. Acid. Res. 2005;33:D433–D437. doi: 10.1093/nar/gki005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strong M, Graeber TG, Beeby M, Pellegrini M, Thomson MJ, Yeates TO, Eisenberg D. Nuc. Acid. Res. 2003;31:7099–7109. doi: 10.1093/nar/gkg924. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.