Abstract
Polo kinases play crucial conserved roles in controlling the eukaryotic cell cycle through orchestrating several events during mitosis1,2. An essential element of cell cycle control is exerted by altering the expression of key regulators3. Here, we demonstrate an important role for the polo kinase, Cdc5p, in controlling cell cycle-dependent gene expression which is critical for the execution of mitosis in the model eukaryote Saccharomyces cerevisiae. In particular, we find that Cdc5p is temporally recruited to promoters of the cell cycle-regulated CLB2 gene cluster, where it targets the Mcm1p-Fkh2p-Ndd1p transcription factor complex, through direct phosphorylation of the coactivator protein Ndd1p. This phosphorylation event is required for the normal temporal expression of cell cycle-regulated genes such as CLB2 and SWI5 in G2/M phases. Furthermore, severe defects in cell division occur in the absence of Cdc5p-mediated phosphorylation of Ndd1p. Thus, polo kinase is required for the production of key mitotic regulators, in addition to previously defined roles in controlling other mitotic events.
In Saccharomyces cerevisiae, the polo kinase Cdc5p has been implicated in a number of processes associated with mitosis, including mitotic entry, spindle assembly, cytokinesis and mitotic exit2. Although several direct targets for Cdc5p have been identified including Swe1p, Scc1p and Bfa1p4,5,6,7, no direct links have been made between polo kinases and the transcription factor complexes which control cell cycle-dependent transcription.
In S. cerevisiae transcriptional regulation of the CLB2 gene cluster in the G2 and M phases represents a key cell cycle control point. This gene cluster encodes several factors required for mitotic progression including the B-type cyclin Clb2p, transcriptional regulators like Swi5p, and the Cdc5p polo kinase3,8. A major determinant of this coordinated gene expression is a transcription factor complex that consists of Mcm1p, the forkhead transcription factor, Fkh2p, and the coactivator protein, Ndd1p3,9,10,11,12,13. The regulation of the Mcm1p-Fkh2p-Ndd1p complex is linked to the cell cycle through sequential phosphorylation of Fkh2p by Clb5pCdc28p14 and Ndd1p, by Clb2p-Cdc28p15,16 kinase complexes. Although these regulatory phosphorylation events promote Ndd1p recruitment and target gene activation it is less clear whether they alone account for the complete regulation of the Mcm1p-Fkh2p-Ndd1p complex. We therefore investigated whether the polo kinase Cdc5p has a role in controlling the activity of the Mcm1p-Fkh2p-Ndd1p complex.
First, we investigated whether we could detect Cdc5p at the promoters of genes within the CLB2 cluster using chromatin immunopreciptation (ChIP) analysis. We used a strain overexpressing Cdc5p to permit analysis of Cdc5p recruitment at times where endogenous Cdc5p is difficult to detect due to its cyclical expression17. Little Cdc5p could be detected at CLB2 cluster promoters in cells arrested in S phase by hydroxyurea (HU) treatment (Fig. 1a, lane 1). However, upon release from the HU block, recruitment of Cdc5p to the promoters of all of the CLB2 cluster genes tested was observed, which was further enhanced when cells were arrested in M phase with nocodazole (Noc) (Fig. 1a, compare lanes 2 and 3). We also tested whether Cdc5p recruitment could be observed in synchronised cells following release from α factor arrest in G1 phase. In agreement with the hydroxurea and nocodazole treated cultures, recruitment of Cdc5p was induced in a cell-cycle dependent manner at both the CLB2 and SWI5 promoters, at a time consistent with the onset of transcription of these genes (Fig. 1b). However, in contrast to CLB2 and SWI5, Cdc5p recruitment to the promoter of a G1 phase-expressed cyclin-encoding gene, CLN213, could not be observed at any time point following release from α factor (Fig. 1c). Importantly, GST-Cdc5p was expressed at times when promoter recruitment was not observed, demonstrating that recruitment is a temporal event that is not dependent on the levels of Cdc5p (Supplementary Fig. S1). Moreover, ChIP analysis of a control strain expressing GST alone, showed little enrichment of the CLB2 promoter (Supplementary Fig. S2), further emphasising the specificity of Cdc5p recruitment. Importantly, recruitment of Cdc5p expressed at endogenous levels could also be specifically detected at CLB2 cluster promoters (Fig. 1d). Thus despite the catalogued localisation of Cdc5p to specific subcellular structures during mitosis such as the spindle pole bodies2, a portion of the cellular pool is associated with promoters.
Fig. 1.
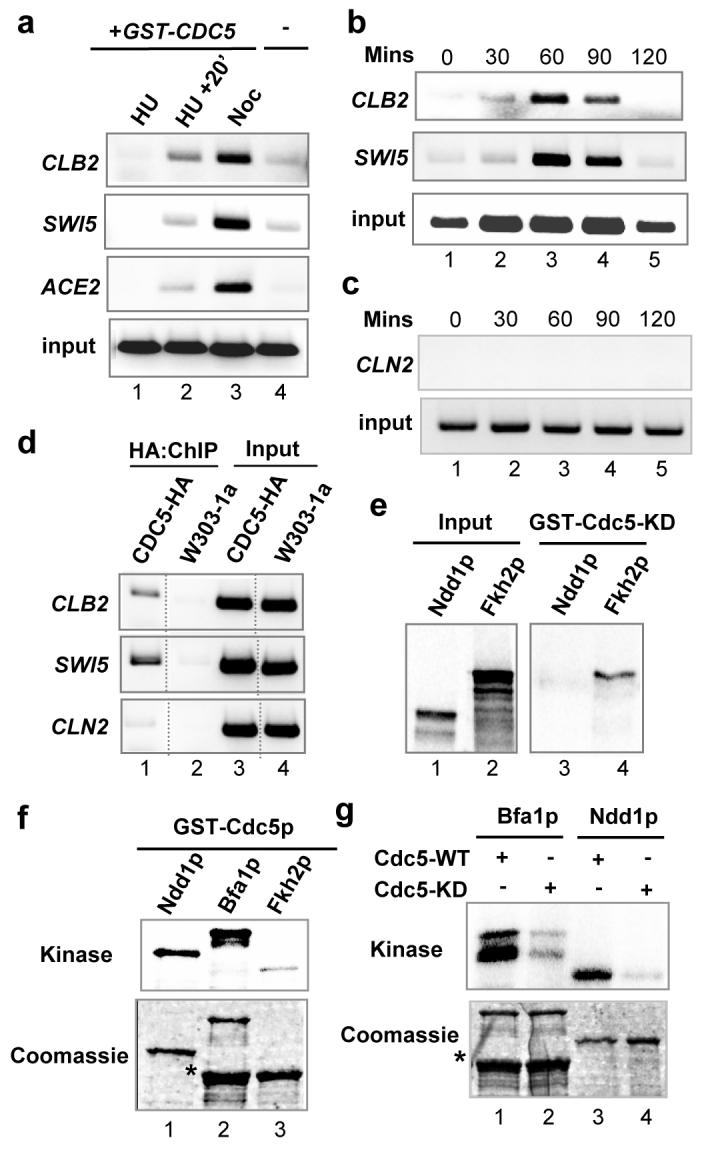
Cdc5p is recruited to CLB2 gene cluster promoters and phosphorylates Ndd1p. (a) ChIP analysis using an anti-GST antibody and extracts isolated from mid-log-phase growing W303-1a cells (lane 4) or from MGY96 cells (lanes 1-3) treated with hydroxyurea (HU), released from hydroxyurea for 20 mins (HU+20'), or treated with nocodazole (Noc). Input represents precipitated DNA amplified using CLB2-specific primers. (b and c) ChIP analysis using an anti-GST antibody and extracts of MGY96 cells treated with α factor and released for the indicated times. Input is shown for the CLB2 (b) and CLN2 (c) promoters. (d) ChIP analysis using anti-HA antibody and extracts from nocodazole-treated cells from either RB15 (expressing HA-tagged Cdc5p) or W303-1a strains. (e) GST pulldown analysis of GST-Cdc5-KD and in vitro-translated full-length Ndd1p and Fkh2p. 25% input is shown. (f and g) Protein kinase assays with wild-type (WT) or catalytically-inactive (KD) GST-Cdc5p and full-length GST-Ndd1p, full-length MBP-Bfa1p or GST-Fkh2(458-862). The asterisk on the coomassie gels represents a degradation product.
As cell cycle-dependent transcription of the CLB2 cluster is controlled by the Mcm1p-Fkh2p-Ndd1p transcription factor complex9,10,11,12, we tested whether the Fkh2p and/or Ndd1p components of this complex could bind directly to Cdc5p. GST pulldown assays revealed that Fkh2p, but not Ndd1p, formed a complex with Cdc5p (Fig. 1e). Similarly, GST-Cdc5p could co-precipitate Fkh2p when co-expressed in vivo (data not shown). The observed recruitment of Cdc5p to CLB2 cluster promoters suggested that Cdc5p may directly phosphorylate one or more components of the Mcm1p-Fkh2p-Ndd1p complex. In contrast to Fkh2p, which was only poorly phosphorylated by Cdc5p, Ndd1p was phosphorylated as efficiently as the known Cdc5p substrate Bfa1p (Fig. 1f, and Supplementary Fig. S3)4,6. A catalytically-inactive version of Cdc5p, Cdc5-KD exhibited much reduced ability to phosphorylate Ndd1p, demonstrating that phosphorylation of Ndd1p was dependent on the kinase activity of Cdc5p rather than a co-precipitating kinase (Fig. 1g).
Ndd1p contains a perfect match at serine 85 (Ser85) to the consensus phosphoacceptor motif for the mammalian polo kinase, Plk118 (Fig. 2a). We therefore substituted Ser85 of Ndd1p with an alanine residue (S85A), and analysed the phosphorylation of the mutant protein by Cdc5p. In vitro kinase assays revealed a reduction in total phosphorylation of Ndd1p(S85A) by Cdc5p compared to wild-type Ndd1p (Fig. 2b, lanes 3 and 4, top panel), and moreover, Western blotting using an anti-phosphoSer-85 antibody showed that phosphorylation at Ser85 was abolished in Ndd1p(S85A) (Fig. 2b, lane 4, middle panel). In contrast, the S85A mutation had little effect on Ndd1p phosphorylation by Clb2p-Cdc28p complexes and phosphorylation at Ser85 was not observed (Fig. 2b, lanes 1-2).
Fig. 2.
Ndd1p is phosphorylated at Ser85 by Cdc5p. (a) The Cdc5p phosphorylation site, Ser85, in Ndd1p and the consensus Plk site18. (b) In vitro phosphorylation of wild-type (WT) and S85A mutant GST-Ndd1p by Clb2p-Cdc28p or Cdc5p was detected by either 32P phosphate incorporation (top panel) or Western blot analysis (WB) with an anti-phospho-Ser85 antibody (middle panel). Input protein was detected using an anti-GST-antibody (bottom panel). (c) In vivo phosphorylation of Ser85 in Ndd1p. HA-tagged wild-type Ndd1p (WT) or Ndd1p-S85A proteins were expressed in cdc5ts cells (KKY021) grown at the indicated temperatures. (d) ChIP analysis using anti-HA antibody and extracts isolated from either RB15 (FKH2) or RB16 (fkh2Δ) strains expressing HA-tagged Cdc5p. Cells were taken at the indicated times after release from α factor block. (e) Fkh2p is required for phosphorylation of Ser85 in Ndd1p. Wild-type HA-tagged Ndd1p was expressed in W303-1a (FKH1 FKH2), fkh1Δ or fkh2Δ cells (lanes 1-3). Lane 4 represents W303-1a (FKH1 FKH2) cells containing no HA-Ndd1p. (c and e) Ndd1p proteins were immunoprecipitated (IP) with anti-HA antibodies. Phosphorylation was detected by Western blot (WB) analysis with an anti-phospho-Ser85 antibody (top panels) and total precipitated protein was detected using an anti-HA antibody (bottom panels).
To probe the potential role of Cdc5p in the phosphorylation of Ndd1p in vivo, and also the importance of Ser85 in this process, we analysed the phosphorylation of wild-type and mutant (S85A) versions of Ndd1p in yeast strains containing a temperature-sensitive allele of CDC5 (cdc5ts). Significantly, phosphorylation of Ser85 could be detected in the wild-type, but not the mutant Ndd1p at the permissive temperature (Fig. 2c, lanes 1 and 3). Furthermore, phosphorylation of Ser85 in wild-type Ndd1p was attenuated upon inactivation of Cdc5p at 37°C (Fig. 2c, lane 2).
Clb2p-Cdc28p-mediated phosphorylation of Thr319 in Ndd1p has previously been shown to play an important role in Ndd1p-mediated activation of the CLB2 cluster16. Hence, we next investigated whether Thr319 phosphorylation was required for Cdc5p-dependent phosphorylation of Ndd1p. However, Ndd1p phosphorylation at Ser85 in vivo was unaffected by mutation of Thr319 suggesting that phosphorylation of Thr319 is not required for this Cdc5p-dependent phosphorylation (Supplementary Fig. S4).
Fkh2p is important for the recruitment of Ndd1p to promoters of genes in the CLB2 cluster9. As Cdc5p is also recruited to these promoters, and binds to Fkh2p in vitro, we tested whether Fkh2p is required for Cdc5p recruitment in vivo. HA epitope-tagged Cdc5p was expressed in FKH2 (wild-type control) and fkh2Δ strains and promoter recruitment monitored by ChIP analysis. Cell cycle-dependent temporal recruitment of Cdc5p to CLB2 cluster promoters was observed in wild-type cells, but was largely lost in fkh2Δ cells (Fig. 2d and Supplementary Fig.S5). Using the same approach, we also determined whether Fkh2p was required for Ndd1p phosphorylation in vivo. Ndd1p was phosphorylated at Ser85 in wild-type cells whereas phosphorylation of Ser85 was severely diminished in fkh2Δ cells (Fig. 2e, lane 3). In contrast, phosphorylation of Ser85 was only slightly reduced in mutants lacking Fkh1p (Fig. 2e, lane 2). Thus, these results suggest that Fkh2p plays a role in nucleating the formation of a Fkh2p-Ndd1p-Cdc5p complex on CLB2 cluster promoters, and the subsequent Cdc5p-dependent phosphorylation of Ndd1p at Ser85.
To understand the functional consequences of Cdc5p-mediated phosphorylation of Ndd1p at Ser85, we analysed the cellular phenotypes associated with mutations of this site. In contrast to wild-type control cells (>90% normal morphology), over 90% of the ndd1Δ cells expressing Ndd1p(S85A) displayed aberrant morphologies, such as elongated daughter cells, hyphal-like growth and cells with problems separating (Fig. 3a, Supplementary Fig. S6). Moreover, these cells exhibited multiple, different severe defects in nuclear morphology and mitotic spindle structure (Fig. 3a- see supplementary information for details). In addition, cytokinesis was defective, as evidenced by the lack of formation of normal septa (Fig. 3b). Many of the defects observed in Ndd1p(S85A) expressing cells resemble those observed in cdc5 mutant cells, including defects in septum formation and cytokinesis19. This suggests that at least some of the cdc5-dependent defects are primarily due to deficiencies in Ndd1p-mediated gene regulation rather than problems in other functions of Cdc5p.
Fig. 3.
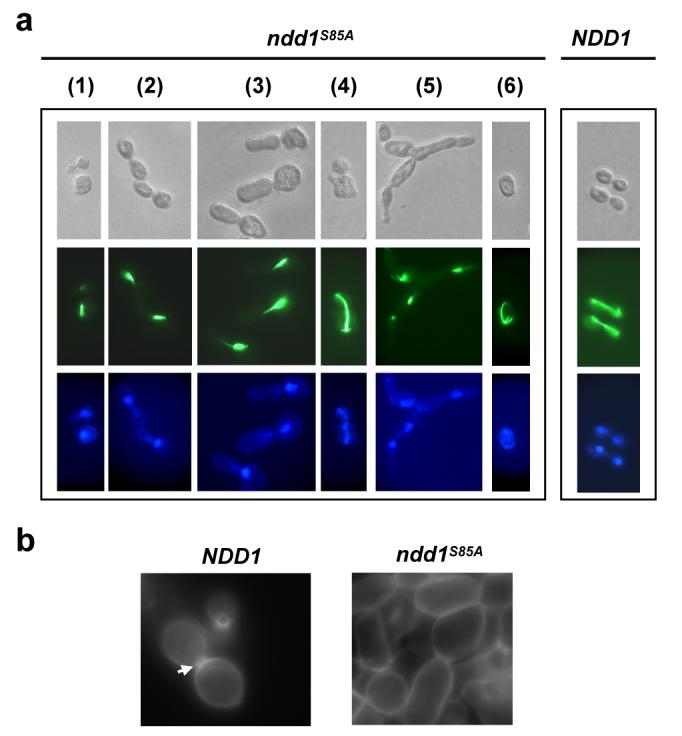
Ser85 of Ndd1p is required for several cell cycle-dependent processes. Wild-type Ndd1p in AP179 was replaced by FOA selection with CEN-based plasmids, expressing the indicated wild-type and mutant Ndd1p proteins. (a) Cellular morphologies were analysed by DIC microscopy (top panel). Mitotic spindles (middle panel) and nuclei (bottom panel) were analysed by fluorescence microscopy (FITC, spindles; DAPI, nuclei). 1-6 refer to different cell morphologies (see Supplementary Material for details). (b) Septa were visualised by fluorescence microscopy following treatment with calcofluor. White arrow indicates normal septa.
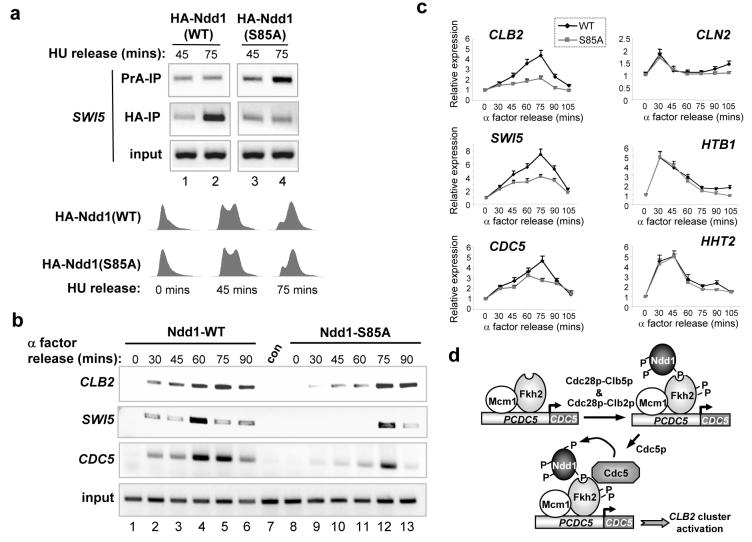
As Ndd1p is an important co-activator, controlling genes in the CLB2 cluster3,9,13,15,16, we examined whether phosphorylation of Ser85 is involved in the recruitment of Ndd1p to promoters in the CLB2 cluster. HA epitope-tagged versions of wild-type Ndd1p and Ndd1p(S85A) were expressed from plasmids in a strain expressing TAP-tagged wild-type Ndd1p from the normal chromosomal locus, to allow the cell cycle to proceed normally. Under these conditions, plasmid-expressed HA-tagged wild-type Ndd1p is more efficiently recruited to the SWI5 promoter than HA-tagged Ndd1p(S85A) in the presence of TAP-tagged wild-type Ndd1p (Fig. 4a, lanes 2 and 4). In contrast, TAP-tagged wild-type Ndd1p is preferentially recruited only in the presence of plasmid-expressed HA-tagged Ndd1p(S85A) (Fig. 4a, lanes 2 and 4). These data suggested that wild-type Ndd1p is more efficiently recruited to CLB2 gene cluster promoters than Ndd1p(S85A). Indeed, consistent with this hypothesis, delayed recruitment of mutant Ndd1p(S85A) to several CLB2 gene cluster promoters was observed in synchronised cells co-expressing wild-type Ndd1p (Fig. 4b and Supplementary Fig. S7). Moreover, Gal4p-DBD fusions to Ndd1p demonstrated equal recruitment of wild-type and mutant Ndd1p to a Gal4p-driven promoter but reduced recruitment of Ndd1p mutated at Ser85 to the CLB2 promoter (Supplementary Fig. S8b). Thus, taken together, these data suggest that Cdc5p-mediated phosphorylation of Ndd1p at Ser85 is important in controlling the temporal recruitment of Ndd1p at promoters of CLB2 cluster genes.
Fig. 4.
Ser85 of Ndd1p is required for normal promoter recruitment and CLB2 gene cluster expression. (a) ChIP analysis on the SWI5 promoter was performed using either protein A (to bind TAP-tagged Ndd1p) or anti-HA antibody (to bind HA-tagged Ndd1p) and extracts isolated from S288C cells expressing TAP-tagged wild-type Ndd1p from the normal chromosomal locus and one of the indicated plasmid-borne HA epitope-tagged Ndd1p proteins. Cells were synchronised with HU and released for the indicated times. DNA content analysis of propidium iodide-stained cells expressing the indicated wild-type (WT) and mutant (S85A) Ndd1p proteins is shown. (b) ChIP analysis on the CLB2, SWI5 and CDC5 promoters was performed using anti-HA antibody and extracts isolated from α factor-synchronised W303-1a cells expressing the indicated HA epitope-tagged Ndd1p proteins. Times after release from α factor arrest are indicated. Input is from the CLB2 promoter. Control (con) represents extracts from cells which do not express HA-Ndd1p. (c) Real time RT-PCR analysis of the indicated genes in the cells expressing only wild-type (WT) or mutant (S85A) Ndd1p proteins (see Supplementary Figs. S9 and S10). Data are presented relative to the expression of each gene at the zero timepoint (taken as 1). (d) Model of the impact of cell cycle-dependent kinases on the regulation of the Mcm1p-Fkh2p-Ndd1p complex and the regulation of CLB2 cluster genes like CDC5.
Next we asked whether the Cdc5p phosphorylation site in Ndd1p has a role in the regulation of CLB2 cluster gene expression. Due to the severe defects observed in strains expressing only Ndd1p(S85A) (see Fig. 3), we were unable to analyse cell cycle-dependent expression of the CLB2 gene cluster in these cells. Therefore, to examine the potential role of Ser85 on CLB2 cluster gene expression we developed a system whereby we could rapidly switch between endogenous and plasmid-borne expression of Ndd1p (Supplementary Fig. S9). To establish whether there were any defects in periodic gene expression during the cell cycle, we compared cells expressing either wild-type Ndd1p or Ndd1p(S85A) following release from an α factor block (Supplementary Fig. S10). Significantly, periodic gene expression that occurs early in the cell cycle in G1 phase (CLN2) and S phase (HHT2 and HTB1) was similar in cells expressing either wild-type Ndd1p or Ndd1p(S85A) (Fig. 4c). However, in contrast, cells expressing Ndd1p(S85A) exhibited reduced expression of the CLB2 cluster genes CLB2, SWI5 and CDC5 (Fig. 4c). These data suggested that Cdc5p has a role in specifically controlling late cell cycle-dependent gene expression. To investigate this further cdc5ts cells were arrested in G1 phase with α factor, then released at the permissive (25°C) or non-permissive temperatures (37°C). Consistent with the effects observed in Fig. 4c, cell cycle-dependent induction of CLB2 was found to be severely attenuated upon inactivation of Cdc5p (Supplementary Fig. S11). As Ndd1p acts as a co-activator in the Mcm1p-Fkh2p-Ndd1p complex, it was possible that phosphorylation of Ser85 regulates the transactivation function of Ndd1p. A GAL1pr-LacZ reporter gene assay was used to monitor the intrinsic transactivation capacity of Gal4p-DBD fusions to wild-type and mutant (S85A and S85E) versions of Ndd1p. Gal4p-DBD-Ndd1p(S85A) exhibited reduced ability to activate transcription of this reporter which could be rescued by introducing a phosphomimetic substitution (S85E) (Supplementary Fig. S8a).
Collectively, our data demonstrate that Cdc5p-mediated phosphorylation of Ndd1p plays an important role in controlling CLB2 cluster gene expression and normal cell cycle progression. Importantly, as CDC5 itself is part of the CLB2 gene cluster, the phenotypic defects associated with mutation of Ser85 of Ndd1p are likely to be due in part to downstream defects caused by reduced Cdc5p levels. Moreover, our data extend a previous model in which Clb2p-mediated phosphorylation of Ndd1p helps establish a positive feedback loop that in turn leads to further increases in CLB2 transcription and M phase entry3,15,16. Here, Cdc5p is identified as another cell cycle regulator that can generate a positive feedback loop, as the CDC5 gene is also part of the CLB2 gene cluster that is a target of the Mcm1p-Fkh2p-Ndd1p complex10,12,20,21,22. Hence, Cdc5p and Clb2p-Cdc28p together converge on Ndd1p to activate one of the key switches that controls cell cycle progression (Fig. 4d). This is reminiscent of the convergence of Cdc5p and Clb2p-Cdc28p on the regulation of Swe1p kinase and subsequent promotion of mitotic entry23. Thus, the combinatorial action of these kinases appears to be a common mechanism to precisely control late cell cycle events.
Forkhead transcription factors are also important for late cell cycle-dependent gene expression in the evolutionary divergent yeast Schizosaccharomycespombe24,25,26,27 and in mammalian cells28,29. Similarly, some of the previously described functions of polo kinases are also conserved between yeast and higher eukaryotes2 and genetic studies link the Cdc5p homologue Plo1p to the regulation of M/G1 phase-specific transcription in S. pombe30. Given these parallels, it is quite possible that a role for polo kinases in regulating cell cycle-dependent gene expression in conjunction with forkhead transcription complexes will eventually be uncovered in mammalian systems.
MATERIALS AND METHODS
More detailed methods are supplied in the Supplementary Information.
Plasmid constructs and yeast strains
Details of plasmid construction and yeast strains used in this study can be found in the Supplementary Information.
RNA analysis and chromatin immunoprecipitations
RNA levels were analysed by Northern blotting11 or by real time RT-PCR (for further details see Supplementary Material). Chromatin immunoprecipitation (ChIP) assays were performed as described previously14.
Protein production, pulldown assays and Western blotting
GST pulldown assays with in vitro-translated proteins were carried out, as described previously14. MBP fusion proteins were prepared by maltose affinity chromatography (New England Biolabs). Western blots were performed with antibodies recognising epitope tags or with an antibody raised against a phosphorylated peptide (NSSSNESS[P]LVENS; Eurogentec).
Protein kinase production and kinase assays
Protein kinase assays using Flag-tagged Clb2p-kinase complexes purified from yeast cells, bacterially-expressed and purified Clb2p and Cdc28p15 or GST-Cdc5p(WT) and GST-Cdc5p(KD)6 with bacterially-expressed and purified substrates were performed as described previously15.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Anne Clancy for excellent technical assistance. We are grateful to Iain Hagan, Peter March, Mike Jackson and Gislene Pereira for advice, Alan Whitmarsh, Shen-Hsi Yang and members of our laboratories for comments on the manuscript and helpful discussions and to Mark Walberg, Uttam Surana, Gislene Pereira and David Lydall for reagents.
This work was supported by Cancer Research UK, the BBSRC, the MRC and the Wellcome Trust.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
REFERENCES
- 1.Barr FA, Silljé HHW, Nigg EA. Polo-like kinases and the orchestration of cell division. Nature Reviews Molecular Cell Biology. 2004;5:429–441. doi: 10.1038/nrm1401. [DOI] [PubMed] [Google Scholar]
- 2.Lee KS, Park JE, Asano S, Park CJ. Yeast polo-like kinases: functionally conserved multitask mitotic regulators. Oncogene. 2005;24:217–229. doi: 10.1038/sj.onc.1208271. [DOI] [PubMed] [Google Scholar]
- 3.Wittenberg C, Reed SI. Cell cycle-dependent transcription in yeast: promoters, transcription factors, and transcriptomes. Oncogene. 2005;24:2746–2755. doi: 10.1038/sj.onc.1208606. [DOI] [PubMed] [Google Scholar]
- 4.Alexandru G, Uhlmann F, Mechtler K, Poupart M, Nasmyth K. Phosphorylation of the Cohesin Subunit Scc1 by Polo/Cdc5 Kinase Regulates Sister Chromatid Separation in Yeast. Cell. 2001;105:459–472. doi: 10.1016/s0092-8674(01)00362-2. [DOI] [PubMed] [Google Scholar]
- 5.Hu F, et al. Regulation of the Bub2/Bfa1 GAP Complex by Cdc5 and Cell Cycle Checkpoints. Cell. 2001;107:655–665. doi: 10.1016/s0092-8674(01)00580-3. [DOI] [PubMed] [Google Scholar]
- 6.Geymonat M, Spanos A, Walker PA, Johnston LH, Sedgwick SG. In vitro regulation of budding yeast Bfa1/Bub2 GAP activity by Cdc5. J. Biol. Chem. 2003;278:14591–14594. doi: 10.1074/jbc.C300059200. [DOI] [PubMed] [Google Scholar]
- 7.Sakchaisri K, et al. Coupling morphogenesis to mitotic entry. Proc. Natl. Acad. Sci. USA. 2004;101:4124–4129. doi: 10.1073/pnas.0400641101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Futcher B. Transcriptional regulatory networks and the yeast cell cycle. Curr Opin Cell Biol. 2002;14:676–683. doi: 10.1016/s0955-0674(02)00391-5. [DOI] [PubMed] [Google Scholar]
- 9.Koranda M, Schleiffer A, Endler L, Ammerer G. Forkhead-like transcription factors recruit Ndd1 to the chromatin of G2/M-specific promoters. Nature. 2000;406:94–98. doi: 10.1038/35017589. [DOI] [PubMed] [Google Scholar]
- 10.Kumar R, et al. Forkhead transcription factors, Fkh1p and Fkh2p, collaborate with Mcm1p to control transcription required for M-phase. Curr. Biol. 2000;10:896–906. doi: 10.1016/s0960-9822(00)00618-7. [DOI] [PubMed] [Google Scholar]
- 11.Pic A, et al. The forkhead protein Fkh2 is a component of the yeast cell cycle transcription factor SFF. EMBO J. 2000;19:3750–3761. doi: 10.1093/emboj/19.14.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu G, et al. Two yeast forkhead genes regulate the cell cycle and pseudohyphal growth. Nature. 2000;406:90–94. doi: 10.1038/35017581. [DOI] [PubMed] [Google Scholar]
- 13.Breeden LL. Cyclin transcription: Timing is everything. Curr. Biol. 2000;10:R586–R588. doi: 10.1016/s0960-9822(00)00634-5. [DOI] [PubMed] [Google Scholar]
- 14.Pic-Taylor A, Darieva Z, Morgan BA, Sharrocks AD. Regulation of cell cycle-specific gene expression through cyclin-dependent kinase-mediated phosphorylation of the forkhead transcription factor Fkh2p. Mol. Cell. Biol. 2004;24:10036–10046. doi: 10.1128/MCB.24.22.10036-10046.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Darieva Z, et al. Cell cycle regulated regulated transcription through the FHA domain of Fkh2p and the coactivator Ndd1p. Curr. Biol. 2003;13:1740–1745. doi: 10.1016/j.cub.2003.08.053. [DOI] [PubMed] [Google Scholar]
- 16.Reynolds D, et al. Recruitment of Thr 319-phosphorylated Ndd1p to the FHA domain of Fkh2p requires Clb kinase activity: a mechanism for CLB cluster gene activation. Genes Dev. 2003;17:1789–1802. doi: 10.1101/gad.1074103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng L, Hunke L, Hardy CF. Cell cycle regulation of the Saccharomyces cerevisiae polo-like kinase cdc5p. Mol. Cell. Biol. 1998;18:7360–7370. doi: 10.1128/mcb.18.12.7360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakajima H, Toyoshima-Morimoto F, Taniguchi E, Nishida E. Identification of a consensus motif for Plk (Polo-like kinase) phosphorylation reveals Myt1 as a Plk1 substrate. J. Biol. Chem. 2003;278:25277–25280. doi: 10.1074/jbc.C300126200. [DOI] [PubMed] [Google Scholar]
- 19.Song S, Lee KS. A novel function of Saccharomyces cerevisiae CDC5 in cytokinesis. J. Cell. Biol. 2001;152:451–469. doi: 10.1083/jcb.152.3.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Althoefer H, Schleiffer A, Wassmann K, Nordheim A, Ammerer G. Mcm1 is required to coordinate G2-specific transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 1995;15:5917–5928. doi: 10.1128/mcb.15.11.5917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spellman PT, et al. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simon I. Serial regulation of transcriptional regulators in the yeast cell cycle. Cell. 2001;106:697–708. doi: 10.1016/s0092-8674(01)00494-9. [DOI] [PubMed] [Google Scholar]
- 23.Asano S, et al. Concerted mechanism of Swe1/Wee1 regulation by multiple kinases in budding yeast. EMBO J. 2005;24:2194–2204. doi: 10.1038/sj.emboj.7600683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zilahi E, Salimova E, Simanis V, Sipiczki M. The S. pombe sep1 gene encodes a nuclear protein that is required for periodic expression of the cdc15 gene. FEBS Lett. 2000;481:105–108. doi: 10.1016/s0014-5793(00)01990-6. [DOI] [PubMed] [Google Scholar]
- 25.Buck V, et al. Fkh2p and Sep1p regulate mitotic gene transcription in fission yeast. J. Cell Sci. 2004;117:5623–5632. doi: 10.1242/jcs.01473. [DOI] [PubMed] [Google Scholar]
- 26.Bulmer R, et al. The forkhead transcription factor Fkh2 regulates the cell division cycle of Schizosaccharomyces pombe. Euk. Cell. 2004;3:944–954. doi: 10.1128/EC.3.4.944-954.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rustici G, et al. Periodic gene expression program of the fission yeast cell cycle. Nat. Genet. 2004;36:809–817. doi: 10.1038/ng1377. [DOI] [PubMed] [Google Scholar]
- 28.Alvarez B, Martinez AC, Burgering BM, Carrera AC. Forkhead transcription factors contribute to execution of the mitotic programme in mammals. Nature. 2001;413:744–747. doi: 10.1038/35099574. [DOI] [PubMed] [Google Scholar]
- 29.Laoukili J, et al. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat. Cell. Biol. 2005;7:126–136. doi: 10.1038/ncb1217. [DOI] [PubMed] [Google Scholar]
- 30.Anderson M, et al. Plo1(+) regulates gene transcription at the M-G(1) interval during the fission yeast mitotic cell cycle. EMBO J. 2002;21:5745–5755. doi: 10.1093/emboj/cdf564. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.