Abstract
Both exercise training and cold acclimatization induce muscle remodelling in vertebrates, producing a more aerobic phenotype. In ectothermic species exercise training and cold-acclimatization represent distinct stimuli. It is currently unclear if these stimuli act through a common mechanism or if different mechanisms lead to a common phenotype. The goal of this study was to survey responses that represent potential mechanisms responsible for contraction- and temperature-induced muscle remodelling, using an ectothermic vertebrate. Separate groups of adult zebrafish (Danio rerio) were either swim trained or cold acclimatized for 4 weeks. We found that the mitochondrial marker enzyme citrate synthase (CS) was increased by 1.5× in cold and by 1.3× with exercise (P < 0.05). Cytochrome c oxidase (COx) was increased by 1.2× following exercise training (P < 0.05) and 1.2× (P = 0.07) with cold acclimatization. However, only cold acclimatization increased β-hydroxyacyl-CoA dehydrogenase (HOAD) compared to exercise-trained (by 1.3×) and pyruvate kinase (PK) relative to control zebrafish. We assessed the whole-animal performance outcomes of these treatments. Maximum absolute sustained swimming speed (Ucrit) was increased in the exercise trained group but not in the cold acclimatized group. Real-time PCR analysis indicated that increases in CS are primarily transcriptionally regulated with exercise but not with cold treatments. Both treatments showed increases in nuclear respiratory factor (NRF)-1 mRNA which was increased by 2.3× in cold-acclimatized and 4× in exercise-trained zebrafish above controls. In contrast, peroxisome proliferator-activated receptor (PPAR)-α mRNA levels were decreased in both experimental groups while PPAR-β1 declined in exercise training only. Moreover, PPAR-γ coactivator (PGC)-1α mRNA was not changed by either treatment. In zebrafish, both temperature and exercise produce a more aerobic phenotype, but there are stimulus-dependent responses (i.e. HOAD and PK activities). While similar changes in NRF-1 mRNA suggest that common responses might underlie aerobic muscle remodelling there are distinct changes (i.e. CS and PPAR-β1 mRNA) that contribute to specific temperature- and exercise-induced phenotypes.
Skeletal muscle plays important roles not only in locomotion but also in whole-body metabolic homeostasis. Within a species, skeletal muscle shows a high degree of phenotypic plasticity, both developmentally and in response to energetic and environmental stress. This plasticity may involve remodelling the muscle into a more aerobic phenotype through changes in mitochondrial mass and the upregulation of metabolic pathways involved in aerobic metabolism. Whether there is remodelling of either the anaerobic or aerobic machinery of muscle depends on the type and duration of the stress. Moreover, the molecular mechanisms involved in remodelling may differ, depending on many factors including the energetic status of the animal. It has been known for decades that repeated contractions associated with endurance exercise training can lead to a more aerobic phenotype of skeletal muscle in mammals (e.g. Gollnick & King, 1969; Gollnick et al. 1972). Indeed, in mammals, contraction-induced muscle metabolic remodelling and mitochondrial biogenesis are thought to occur through a series of cellular events that begin with the activation of various signalling pathways (reviewed in Moyes & Hood, 2003). Calcium signalling appears to play a major role in contraction-induced gene expression acting through the Ca2+/calmodulin-dependent kinase (CaMK) pathway (Chin, 2005), but multiple signals including changes in ATP turnover and reactive oxygen species (ROS) are likely to also be involved (Adhihetty et al. 2003; Moyes & Hood, 2003). The corresponding increased expression of transcription factors eventually elevates mRNA for genes encoding enzymes of various metabolic pathways and of mitochondrial proteins (Hood, 2001). Transcription factors shown to be important in mammals for differentiation of muscle fibres to a more aerobic phenotype include the nuclear respiratory factors (NRF)-1 and -2, peroxisome proliferators-activated receptors (PPAR)-α, -β/δ, -γ and PPAR-γ cofactor (PGC)-1α (Lin et al. 2002). Along with chronic exercise there are other stimuli known to induce muscle remodelling in mammals, including shivering thermogenesis, chronic electrical stimulation, and hyperthyroidism (reviewed in Leary & Moyes, 2000). Although there are some differences in how remodelling is induced (Weitzel et al. 2003), a common stimulus (hypermetabolism) leads to similar muscle phenotypes. In non-mammalian ectothermic vertebrates, quite different stimuli can produce similar changes in muscle phenotype. For example, in muscle a more aerobic phenotype can occur in response to chronic cold temperature exposure, yet this can coincide with a decrease in metabolic rate. Many fish species have been found to increase muscle mitochondrial content to compensate for the deceleration effect of low temperature on metabolism to maintain suitable levels of muscular activity for feeding and reproducing. In fact, in teleost fishes, cold-induced skeletal muscle remodelling (Battersby & Moyes, 1998) occurs to the same extent as exercise training (Johnston & Moon, 1980; Farrell et al. 1991). Relatively few studies have investigated the potential molecular mechanisms responsible for temperature-induced muscle remodelling in ectotherms (e.g. Battersby & Moyes, 1998; Lucassen et al. 2003; Malek et al. 2004). No studies have investigated these mechanisms in response to exercise. It is possible that there are conserved mechanisms that lead to a more aerobic muscle phenotype regardless of the initial stimuli. Alternatively, different stimuli may rely on unique molecular mechanisms but lead to the same phenotype. In addition, it is unclear how these different stimuli affect qualitative aspects of mitochondrial metabolism. Specifically, which stimuli trigger increases in mitochondrial volume density versus the ability of mitochondria to preferentially oxidize fatty acids over pyruvate, amino acids or lactate? Current evidence suggests that the temperature-induced muscle remodelling in fish may occur through a separate set of molecular mechanisms than contraction-induced remodelling and different from those seen in mammals (Battersby & Moyes, 1998; Malek et al. 2004). Moreover, adult zebrafish (Danio rerio) represent a potentially powerful model organism for making these comparisons. The zebrafish holds many advantages for the study of muscle physiology, having a comprehensive genomic database, homologous muscle fibre masses, ability to exercise in large groups and amenability to large-scale screening for examination of heritable aspects of muscle performance.
The goals for this study were to exploit the many advantages that already exist for zebrafish as a model organism and to extend this to the study of adult vertebrate muscle phenotypic plasticity, to determine: (i) the changes in key metabolic and mitochondrial enzymes used as indices of skeletal muscle remodelling and whether they are similar with cold acclimatization and exercise training; (ii) the changes in transcription for genes that encode these enzymes and in the transcription factors implicated in muscle remodelling in mammals; and (iii) the whole-organism performance using a standardized critical swimming speed test after exercise training and cold acclimatization. We hypothesize that contraction and cold-induced muscle remodelling will produce a similar phenotype, but the patterns of gene expression will differ due to the divergent nature of the initial stimulus.
Methods
Animals
The present study was approved by the McMaster University Animal Research Ethics Board (AREB). Adult zebrafish (Danio rerio) were obtained from a local supplier (PetsMart, Ancaster, ON, Canada) and control fish were housed in 3 l tanks (Aquatic Habitats, Apopka, FL, USA) in recirculating dechlorinated Hamilton tap water. Fish were fed commercial tropical fish food (Top Fin, Phoenix, AZ, USA) 1–2 times per day. Unless otherwise noted, water temperature was maintained at 28°C and the room was kept on a 12 h: 12 h light: dark cycle. Water quality was monitored weekly. These experiments were carried out over a 14-month period using two separate groups of zebrafish. Group 1 was used for the enzyme activities and gene expression measurements and group 2 for the swimming performance (Ucrit) and metabolite measurements. Since body mass can affect metabolism (Hochachka, 1987) and body length can affect swimming power output, we assessed both in all groups after treatments. Physical characteristics of these two groups at the time of tissue sampling (see below) appear in Table 1. Condition factor (k) is a measure of the condition of fish based on the cubes of weight and length, because growth in weight are proportional to growth in volume (Fulton, 1902). If k < 1.0 a fish is in poor condition and if k > 1.4, a fish is in good to excellent condition. This was calculated using the following equation:
Table 1.
Physical characteristics of all adult zebrafish from Group 1 used for enzyme activity and gene expression measurements and Group 2 used for swimming performance and muscle metabolite measurements
| All fish | Group | Weight (g) | Length (cm) | Condition factor, k |
|---|---|---|---|---|
| Control | 1 | 0.55 ± 0.03 (36) | 3.31 ± 0.06 (31) | 1.52 ± 0.05 (31) |
| 2 | 0.41 ± 0.01 (29)‡ | 2.84 ± 0.04 (29)‡ | 1.74 ± 0.09 (29) | |
| Cold acclimatized | 1 | 0.58 ± 0.02 (33) | 3.29 ± 0.04 (33) | 1.61 ± 0.04 (33) |
| 2 | 0.52 ± 0.02 (30)* | 3.07 ± 0.06 (30)*‡ | 1.79 ± 0.06 (30) | |
| Exercise trained | 1 | 0.48 ± 0.02 (32)† | 3.00 ± 0.04 (32)*† | 1.77 ± 0.05 (32) |
| 2 | 0.54 ± 0.03 (26)* | 3.13 ± 0.05 (26)*‡ | 1.81 ± 0.09 (26) |
The total number of individuals in each category appears in parentheses. Values are means ±s.e.m.
Significantly different from controls from the same group
significantly different from cold acclimatized from the same group
significantly different from the same treatment in group 1 (P < 0.05).
| (1) |
where mass was measured in g and length in cm.
Exercise training
Approximately 60 fish (group 1) were transferred to a Brett-style recirculating swim flume (Brett, 1964) kept at 28°C, and were allowed to acclimatize to the tunnel for 1 week. Several litres of water were drained from the flume daily and were replaced with fresh 28°C dechlorinated water. Fish were exercised twice daily for 3 h, 6 days a week with a 2 h rest period when the flow was turned off and the fish were fed. Starting at a speed of 6.0 cm s−1 (equivalent to 2 BL s−1, where BL is body length), speed was increased by 1 BL s−1 each week to a final speed of 15.0 cm s−1 (5 BL s−1) and training lasted for a total of 4 weeks. The water current was kept at a speed of 3 cm s−1 (∼1 BL s−1) at all other times (i.e. non-training and overnight). Based on previous measures of maximal sustained swimming speed (Plaut, 2001), these training speeds represent low to moderate exercise intensity and aerobic training for zebrafish (∼25–30% maximal sustainable swimming speed). We experienced ∼30% mortality in the trained fish (compared to ∼15% for controls) as a result of a single occurrence of a dislodged air stone in the swim tunnel at 3 weeks. In the first set of experiments, the fish were removed from the swim flume and killed by flick of the finger to the head; length and weight were recorded, and fish were then decapitated. The skin, tail, and gut were removed, leaving the muscle mass which was quickly immersed in liquid nitrogen and then stored at −80°C for future analysis.
Temperature acclimatization
Approximately 50 fish were transferred to two 40 l aquaria kept at 28°C in a 10°C cold room. The temperature was lowered 2°C day−1 over 1 week to 18°C, and fish were kept at this temperature for a further 3 weeks. At the end of the 4 week period, muscle samples were taken as described above. Control fish were maintained at 28°C.
Enzyme activities
Muscles from 10 fish in each group were powdered using a liquid nitrogen-cooled mortar and pestle. Maximal enzyme activities were measured on individual fish by taking approximately 50 mg from each individual and homogenizing in 20 volumes of extraction buffer consisting of 20 mm Hepes (pH 7.0), 1 mm EDTA, and 0.1% Triton X-100, using a glass homogenizer. The activities (in U g−1) of cytochrome c oxidase (COx), citrate synthase (CS), β-hydroxyacyl-CoA dehydrogenase (HOAD), pyruvate kinase (PK) and lactate dehydrogenase (LDH) were assayed in these crude homogenates at 28°C as previously described (McClelland et al. 2005). Analysis was carried out in 96-well format on either a Spectramax 340pc or a Spectramax Plus 384 microplate reader (Molecular Devices, Sunnyvale, CA, USA). Optical path lengths were determined for each machine according to Brooks (1994).
Total genomic DNA per gram of muscle tissue was determined on enzyme homogenates as previously described (McClelland et al. 2005). Briefly, aliquots of homogenates were digested overnight at 55°C using digestion buffer consisting of (mm): 100 NaCl, 10 Tris-HCl (pH 8.0), 25 EDTA (pH 8.0) 0.5% SDS and proteinase K (0.2 mg ml−1). DNA was quantified using PicoGreen dsDNA quantification reagent (Molecular Probes, Eugene, OR, USA), using a standard curve constructed from a known amount of zebrafish DNA isolated from additional fish as described above. Samples and standards were loaded on a 96-well microplate, and fluorescence was measured on a fluorescence spectrophotometer (excitation at 485 nm and emission at 538 nm on a Spectramax Gemini Spectrofluorometer, Molecular Devices, Sunnyvale, CA, USA). Total protein was measured by the Bradford method (Bio-Rad, Mississauga, ON, Canada) on the same homogenates used for enzyme activity and DNA measurement.
mRNA analysis by real-time PCR
Total RNA was extracted from ∼50 mg of skeletal muscle from additional fish in group 1 using Trizol reagent (Invitrogen Life Technologies, Calsbad, CA, USA). Muscle was first powdered using a liquid nitrogen-cooled mortar and pestle, and placed in Trizol and homogenized using a motorized homogenizer (Fisher Scientific). RNA was then further purified and DNase treated (Qiagen DNase I) using RNeasy columns (Qiagen Sciences, Mississauga, ON, Canada). Total RNA was quantified at 260 nm, and purity was verified by absorbance (A)260/280 ratios greater than 1.6 (normally 1.8–2.0). Random RNA samples were also denatured and separated on 1% agarose-formaldehyde gels, and ribosomal RNA stained with ethidium bromide to determine if any degradation occurred during tissue storage and processing. First-strand cDNA was synthesized from 2 μg of total RNA using Stratascript reverse transcriptase (RT, Stratagene, La Jolla, CA, USA), RNase inhibitor (Amersham Biosciences, Piscataway, NJ, USA) and random primers (Promega Corp., Madison, WI, USA) for 5–6 individuals from each group. mRNA expression for genes of interest was measured in duplicate with a Stratagene MX3000P real-time PCR machine using SYBR green with ROX as a reference dye (either Bio-Rad iTaq SYBR green supermix (Bio-Rad Laboratories Inc., Mississauga, ON, Canada), or Stratagene SYBR green supermix (Stratagene, La Jolla, CA, USA)). Primers (Table 2) were designed using Primer 3 software (http://biotools.umassmed.edu/bioapps/primer3_www.cgi) or using previously published primer sequences (Ton et al. 2003; Taylor et al. 2004). Specificity of all primer pairs was tested by RT-PCR and gel electrophoresis using DNA size standards (Fermentas GeneRuler). In addition a SYBR green dissociation curve was run to ensure a single fluorescence peak at the appropriate temperature. Expression levels are relative to a randomly selected control sample (calibrator), and are calculated by the delta-delta threshold cycle (Ct) method (Livak & Schmittgen, 2001) using the Stratagene software:
Table 2.
Primer sequences used for RT-PCR and real-time PCR analysis of mRNA expression in zebrafish
| Gene | Forward primer | Reverse primer | Amplicon size (bp) |
|---|---|---|---|
| (Ascension no.) | (5′–3′) | (5′–3′) | |
| CS | agcgctgctatgaatggtct | ctgaggaagacagaccctcg | 199 |
| (BC045362) | |||
| CYC-1* | acttagccaaccaggagcac | gggtggaagaagtcagaagc | 163 |
| (AY996924) | |||
| β-actin* | ttctggtcggtactactggtattgtg | atcttcatcaggtagtctgtcaggt | 170 |
| (AF057040) | |||
| HOAD | ccacaggacattcagtggtg | gtcagtgccatgaacgacag | 200 |
| (ZGI TC279717) | |||
| LDH-A* | cacagttgaagatggcctca | tgtgcgtcttgagaaacagg | 201 |
| (NM131246) | |||
| Dlat† (AY188775) | tggaactacctgccttgtcc | ctccaatgggcacatctctt | 201 |
| PGC-1α | tgaggaaaatgaggccaact | agcttcttcagcagggaagg | 200 |
| (XM678107) | |||
| PPAR-α (U93473) | gttcttcaggcggacgatt | agggggaaacacttctgaaa | 121 |
| PPAR-β1 (AF342937) | gcgtaagctagtcgcaggtc | tgcaccagagagtccatgtc | 204 |
| NRF-1 (NM131680) | aggccctgaggactatcgtt | gctccagtgccaacctgtat | 202 |
| sMyHC (AY333451) | agcccagaagcaactcaaga | gttcctgctcagccagtttc | 203 |
| fMyHC (AF165817) | aaagcaaaggccaatcttga | tgttttcctcgtgtgagctg | 200 |
CS, citrate synthase; CYC-1, cytochrome c oxidase subunit 1; HOAD, β-hydroxyacyl-CoA dehydrogenase; LDH-A, lactate dehydrogenase A subunit; dLat, pyruvate dehydrogenase E2 subunit; PPAR, peroxisome proliferator-activated receptor; PGC, PPAR-γ cofactor; sMyHC, myosin heavy chain slow isoform; fMyHC, myosin heavy chain fast isoform; NRF, nuclear respiratory factor.
| (2) |
where ΔΔCt is the ΔCt for the unknown −ΔCt for the calibrator sample. ΔCt is the difference between the Ct for the target gene and the housekeeping gene β-actin. A value of 2 is used for 100% efficiency, but was modified to reflect true efficiencies determined from standard curves. Negative controls included RNA processed for cDNA in the absence of RT (no RT) and reactions run with RNase-, DNase-free H2O in place of cDNA template (no template).
Total RNA per gram of muscle was quantified on a separate set of five fish from control and treatment groups in group 1 by extracting RNA from 40–100 mg of tissue in 1 ml of Trizol, and sampling the aqueous phase without further purification. Quantification of 2 μl of each sample was via Ribogreen (Molecular Probes, Eugene, OR, USA) compared to an RNA standard curve (excitation at 485 nm and emission at 520 nm on a Spectramax Gemini Spectrofluorometer, Molecular Devices, Sunnyvale, CA, USA).
Critical swimming tests (Ucrit)
The critical swimming speed test in fish involves a stepwise increase in swimming speed until fatigue. This test is used to assess prolonged swimming capacity, and to estimate maximum sustained swimming speed. Fish in group 2 underwent the same training and cold acclimatization protocols as described above. Critical swimming tests were carried out in a 30 cm × 8 cm section of a Brett-style swim tunnel at either 18°C or 28°C as previously described for zebrafish (Plaut, 2000). Fish were placed in the swim tunnel in groups of 10–15 and allowed to acclimatize to the tunnel for 48 h. The exercise training groups were tested 48 h after their last training session. Swimming speed was progressively increased from 1 BL s−1 (3 cm s−1) every 5 min (as used by other studies on small fish; Plaut, 2000; Hammill et al. 2004) until individuals could not hold their position in the current and rested on a grid at the back of the swimming chamber. The final swimming speed achieved prior to fatigue was defined as the maximal sustainable critical swimming speed (Ucrit) and calculated by the following formula:
| (3) |
where Vls is the final speed (cm s−1) completed, ts is the time spent at the final speed, ti is the time increment (min), and Vi is the speed increment (Plaut, 2001; Richards et al. 2002). At fatigue, each individual fish was siphoned out of the swim flume, and their skeletal muscle quickly dissected and frozen using liquid nitrogen-cooled tongs. These samples were used to assess post-exercise (Ucrit) metabolite levels at fatigue. Separate control individuals were sampled for resting baseline metabolite levels after anaesthetic (MS222) overdose, then muscle quickly frozen using liquid nitrogen-cooled tongs. All samples were placed at −80°C for future metabolite analysis (see below).
Metabolites
Muscle samples were powdered using a liquid nitrogen-cooled mortar and pestle. Metabolites were extracted and quantified according to standard methods (Bergmeyer, 1974) adapted to 96-well format for the Spectramax Plus 384 microplate reader (Molecular Devices, Sunnyvale, CA, USA). Briefly, samples were homogenized in 600 μl of cold 8% perchloric acid, and homogenized using a power homogenizer. The homogenate was centrifuged at 10 000 g for 10min at 4°C, and an aliquot of the supernatant was removed and stored at −80°C for lactate determination. Final assay conditions for lactate were 2 mm NAD+ in glycine/hydrazine buffer pH 9.2 (Sigma-Aldrich, Oakville, ON, USA) and the assay was started with 5 U LDH (Sigma from bovine muscle). A separate aliquot was neutralized with 3 m K2CO3 and centrifuged at 10 000 g for 10 min at 4°C. The supernatant was assayed for creatine phosphate (CrP) and ATP on the same day as homogenization. Final assay conditions for CrP and ATP were (mm) 5 MgCl2, 20 Tris (pH 8.0), 5 glucose, 2 NAD+, 1 U ml−1 glucose-6-phophate dehydrogenase (G6PDH, Roche, from L. mesenteriodes), and for CrP only 1 mm ADP and 1.5 U ml−1 hexokinase (HK, Roche, from yeast). For ATP the reaction was started with 1 U of HK, and for CrP the reaction was started with 2.5 U of creatine phosphokinase (CPK, Sigma, from rabbit muscle).
Statistical analysis
Data presented are means ± s.e.m. Data were analysed using a combination of ANOVA, post hoc tests (Tukey's) and t tests with Bonferoni correction. Data with unequal variance were analysed using a Mann–Whitney rank sum test.
Results
Muscle total RNA, protein and DNA
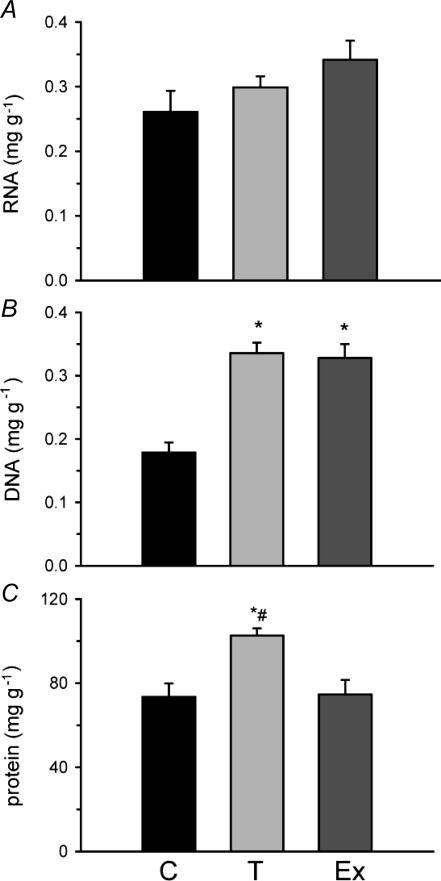
Changes in total RNA, DNA and protein per gram of muscle were determined in response to 4 weeks of exercise training or 4 weeks of cold-acclimatization in group 1 fish. There was no significant difference in total RNA between the groups (Fig. 1A). However, both cold acclimatization (0.34 ± 0.02 mg g−1) and exercise training (0.33 ± 0.02 mg g−1) showed significant increases in total DNA content relative to controls (0.18 ± 0.02 mg g−1; Fig. 1B). Total protein content was greater in cold-acclimatized (103 ± 4 mg g−1) than either exercise-trained (75 ± 7 mg g−1) or control (73 ± 6 mg g−1) fish muscle (Fig. 1C). Exercise-trained fish from group 1 were significantly lighter and shorter than cold-exposed fish. Exercised fish were also shorter but did not differ in weight from controls. Exercise-trained and cold-acclimatized fish from group 2 were heavier and longer than controls. All fish in both groups 1 and 2 were in good physical condition with k values all over 1.5. (Table 1).
Figure 1. Total skeletal muscle RNA (A) DNA (B) and protein (C) from zebrafish cold acclimatized (T) to 18°C for 4 weeks, exercise-trained (Ex) for 4 weeks at 28°C compared to 28°C controls (C) from group 1 fish.
N= 9—10 for DNA and protein measured on the same samples; n = 5 for RNA from separate fish. *Significantly different from controls, #significantly different from exercise-trained fish. Error bars represents s.e.m.
Enzyme activities
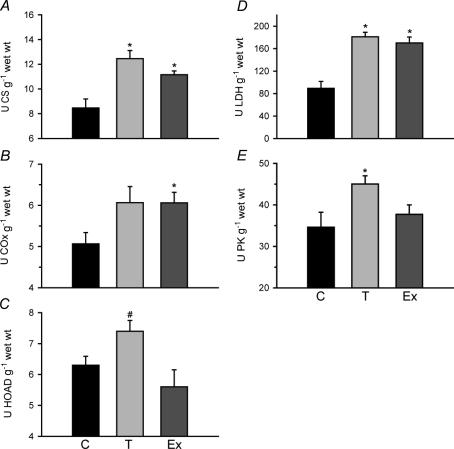
Activities for several enzymes in muscle metabolism were measured at 28°C (Fig. 2). Zebrafish muscle responded to prolonged cold exposure and exercise training with increases in CS by 1.5× and 1.3×, respectively. Exercise increased COx activity 1.2× (P < 0.05), while cold acclimatization showed a non-significant rise (P = 0.07) of 1.2× above controls. Only cold exposure increased the activity of the β-oxidation enzyme HOAD used as an index of fatty acid oxidation potential, by 1.3× above that measured in trained fish muscle (P < 0.05; Fig. 2C). Both cold acclimatization and exercise increased the activity of LDH (Fig. 2D). Cold also increased the activity of PK, but exercise training did not affect this enzyme (Fig. 2E).
Figure 2. Skeletal muscle enzyme activities (assayed at 28°C) in adult zebrafish (Danio rerio) after exposure to cold (18°C, T) for 2 and 4 weeks and after 4 weeks of exercise training (Ex) at 28°C, compared to 28°C controls (C).
Values are presented for A, citrate synthase (CS) B, cytochrome oxidase (COx), C, β-hydroxyacyl-CoA dehydrogenase (HOAD), D, lactate dehydrogenase (LDH) and E, pyruvate kinase (PK) All measurements are from group 1 n= 10, except HOAD from group 2, n= 5. *Significantly different from controls (P < 0.05). U, μmol min−1. Error bars represent s.e.m.
Gene expression
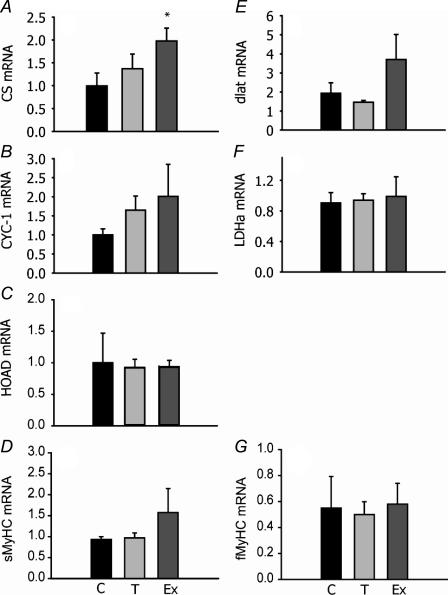
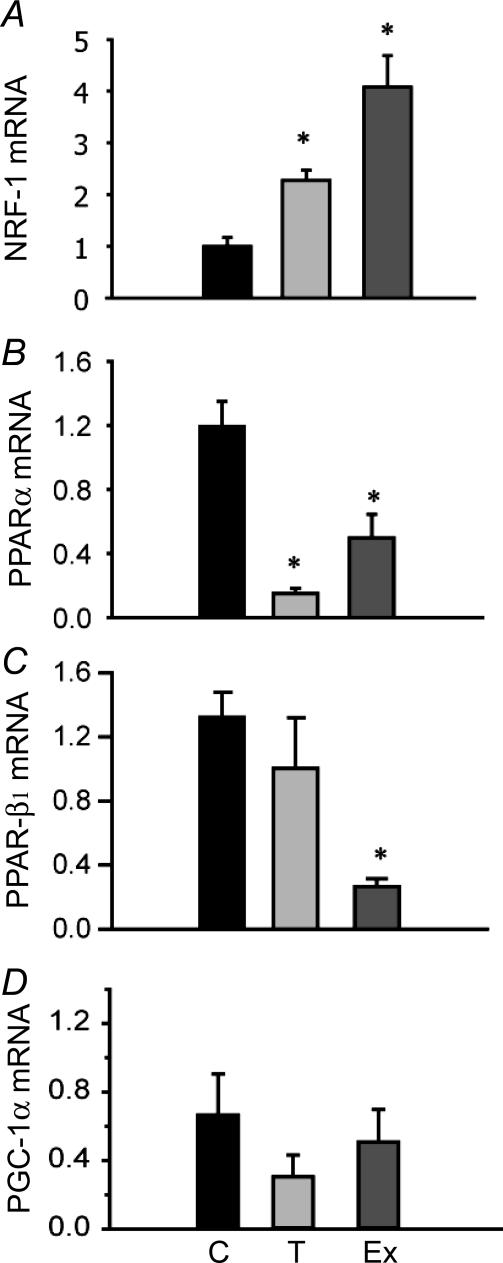
mRNA expression was assessed using real-time PCR for genes encoding mitochondrial enzymes involved in electron transport (CYC-1), the Kreb's cycle (CS), carbohydrate oxidation (pyruvate dehydrogenase E2 subunit, dLat), β-oxidation of fatty acids (HOAD) and glycolysis (LDH) plus contractile elements (MyHC). Only mRNA levels for CS in exercise-trained individuals increased significantly (Fig. 3A). All other mRNA levels examined did not change significantly with exercise training or cold exposure relative to controls. mRNA expression was also measured for transcription factors implicated in muscle aerobic remodelling in mammals. There was a significant increase in NRF-1 expression relative to controls by 4× with exercise and 2.3× with cold acclimatization (P < 0.05). There were also significant decreases in PPAR-α in both experimental groups and PPAR-β1 in exercise-trained zebrafish (Fig 4B and C). Levels of mRNA for PGC-1α did not change significantly after 4 weeks' training or cold exposure (Fig. 4D).
Figure 3. Relative gene expression levels (mRNA) for citrate synthase (CS: A), cytochrome c oxidase subunit I (CYC-1; B), β-hydroxyacyl-CoA dehydrogenase (HOAD; C), myosin heavy chain slow isoform (sMyHC; D), pyruvate dehydrogenase E2 subunit (dLat; E), lactate dehydrogenase (LDH; F) and fast (fMyHC; G) isoform in adult zebrafish exposed to cold temperatures (18°C, T) for 4 weeks and exercise trained for 4 weeks (Ex), compared to 28°C controls (C).
Expression levels were measured by real-time PCR using SYBR green. Results are relative to a randomly selected control sample and normalized to β-actin. n= 5 except for MyHC (n= 3). *Significantly different from controls (P < 0.05). Error bars represent s.e.m.
Figure 4. Gene expression levels (mRNA) for transcription factors: nuclear respiratory factor (NRF)-1 (A), peroxisome proliferator activator receptor (PPAR)-α (B), PPAR-β1 (C) and PPAR-γ cofactor (PGC)-1α (D) in adult zebrafish exposed to cold temperatures (18°C, T) for 4 weeks and exercise trained for 4 weeks (Ex), compared to 28°C controls.
Expression levels were measured by real-time PCR using SYBR green. Results are relative to a randomly selected control sample and normalized to β-actin. n= 5 for all except PCG-1α (n= 6). *Significantly different from control (P < 0.05). Error bars represent s.e.m.
Critical swimming speed (Ucrit)
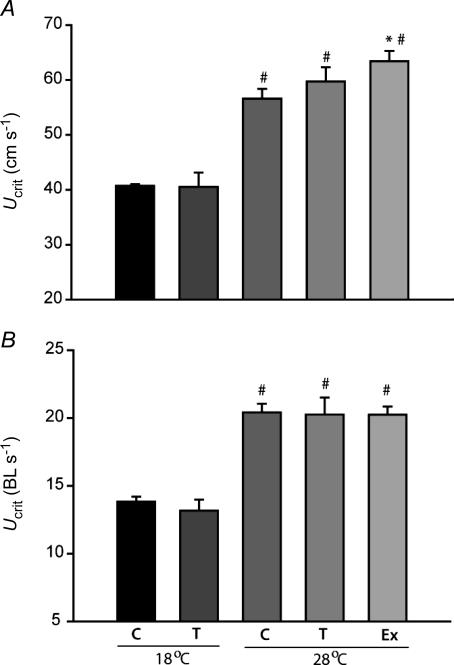
Critical swimming tests revealed that only exercise training resulted in a significant increase in absolute maximum sustainable swimming speed (Ucrit; Fig. 5A) at 28°C. Exercise-trained zebrafish reached an average sustainable swimming speed at 28°C of 63 ± 2 cm s−1 compared to 57 ± 2 cm s−1 for controls at the same temperature. Both controls and cold-acclimatized zebrafish had significantly decreased swimming performance at 18°C. Moreover, prolonged exposure to 18°C in the cold acclimatized group did not improve swimming performance at 28°C relative to controls or exercise-trained fish (Fig. 5). Group 2 cold acclimatized (T) and exercise-trained (Ex) zebrafish were heavier and longer than control fish (Table 2). This resulted in similar relative Ucrit values to controls when standardized to individual body length (as BL s−1, Fig. 5B).
Figure 5. Critical swimming speeds (Ucrit) in absolute speeds (cm s−1; A) or relative Ucrit in body lengths per second (BL s−1; B) at 18°C and 28°C in adult zebrafish exposed to cold temperatures (18°C, T) for 4 weeks and exercise trained for 4 weeks (Ex), compared to controls (C).
*Significantly different from 28°C controls, #significantly different from 18°C. n= 15 (C, T and Ex at 28°C) and n= 10 (C, T at 18°C). Error bars represent s.e.m.
Metabolites
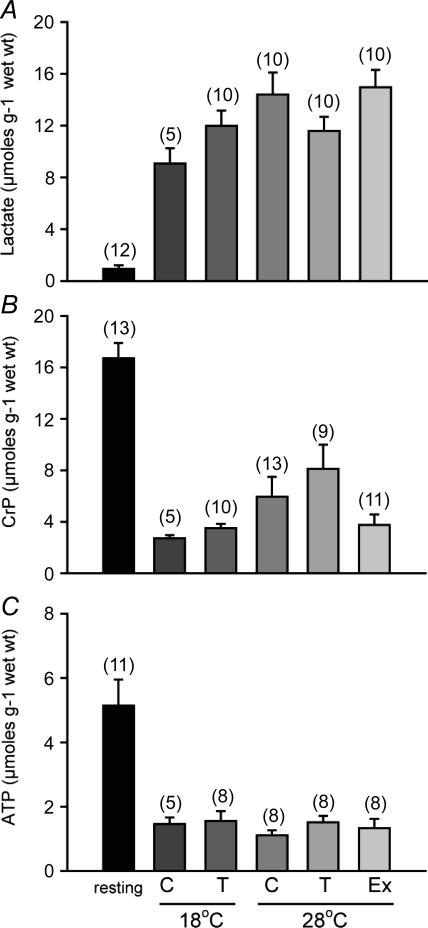
Depletions of metabolites when fatigue was reached were compared to resting control fish at 28°C. All swimming tests in all groups resulted in large accumulations of lactate in the muscle. The highest levels at 15 ± 1 mm were seen in the exercise-trained fish, with resting levels in controls of 0.9 ± 0.3 mm (Fig. 6A). Significant depletions in CrP and ATP were seen in all trials. There was no significant difference in the ATP levels between the groups after exercise, but CrP reached a lower level in exercise-trained fish relative to cold acclimatized fish at 28°C (P < 0.05, t test) (Fig. 6B and C).
Figure 6. Mixed muscle concentrations of A, lactate (A), phosphocreatine (B) and ATP in μmol (g wet weight)−1 (C).
Resting values from 28°C controls are compared to fatigue values at the end of the critical swimming (Ucrit) tests at 18°C and 28°C in adult zebrafish exposed to cold temperatures (18°C, T) for 4 weeks and exercise trained for 4 weeks (Ex), compared to controls (C). Samples sizes appear above each bar. All post-exercise data are significantly different from resting controls. #Significantly different from T exercised at 28°C. Error bars represent s.e.m.
Discussion
Adult zebrafish muscle remodelling
Adult zebrafish muscle demonstrates considerable phenotypic plasticity to stresses that produce a more aerobic phenotype. Both cold-induced and contraction-induced muscle remodelling show similar increases in biochemical markers (Fig. 2) of mitochondrial density (CS) and pyruvate to lactate flux capacity (LDH). However, only in the exercise-trained fish did mRNA levels for CS increase significantly (Fig. 3). The transcription factor NRF-1 was also elevated in cold-acclimatized (2.3 ×) but to a greater extent in trained (4 ×) fish compared to controls (Fig. 4). Important differences exist with muscle response to temperature (Fig. 2), stimulating the β-oxidation machinery (HOAD) as well as potential for glycolytic flux (PK) not observed in the exercise-trained fish. Muscle mRNA levels for PPAR-α, a transcription factor important for the regulation of the fatty acid pathway in mammals (Gulick et al. 1994; Vredendaal et al. 1998), were downregulated with both experimental treatments compared to controls. Another isoform of this factor, PPAR-β1 had mRNA levels that were downregulated in the exercise-trained but not in the cold-acclimatized group (Fig. 4). Surprisingly, the cofactor PGC-1α, important for enhancing DNA binding of PPARs (Lemberger et al. 1996), for mitochondrial biogenesis and for producing an aerobic muscle phenotype in mammals (Koves et al. 2005), did not change in expression after either treatment (Fig. 4D). Chronic cold temperature and exercise both produce a more aerobic phenotype in adult zebrafish muscle. Interestingly, similar changes in NRF-1 mRNA suggest this as a common response that might underlie aerobic muscle remodelling because similar changes are seen in endurance-trained mammalian muscle. However, there are stimulus-dependent changes in enzyme profiles, transcription factors and other gene-expression profiles, as well as absolute sustained swimming speeds, which suggest distinct responses contribute to the development of temperature- and exercise-induced muscle phenotypes.
Enzyme activities in cold versus exercise
Enzyme activities can be used as biochemical indices of mitochondrial density (CS; Reichmann et al. 1985), the electron transport chain (COx), fatty acid oxidation capacity (HOAD) and capacity of glycolytic flux (PK, LDH). Previous research on other fish models has studied muscle biochemical and structural changes in separate studies either with cold-acclimatization or with prolonged exercise. These studies have demonstrated increases in mitochondrial and cytosolic enzymes in muscle, following cold acclimatization (mainly trout: Cordiner & Egginton, 1997; Battersby & Moyes, 1998; Bouchard & Guderley, 2003; McClelland et al. 2005) and exercise training (coalfish Pallachius virens: Johnston & Moon, 1980; and rainbow trout: Farrell et al. 1991). Proliferation of the mitochondria results in increased surface area over which diffusion between the mitochondria and cytosolic compartments would occur (Guderley & St-Pierre, 2002), and a reduction in intermitochondrial diffusion distances (Portner, 2002). This is beneficial for both life in the cold to compensate for the deceleration effect on metabolism, and for the increased muscle aerobic capability important for exercise. Here we directly compare these stressors at the level of both muscle biochemistry and gene expression changes. The significant increase in maximal CS activities seen in cold-acclimatized and exercise-trained zebrafish (Fig. 2A) confirms that both stimuli increase muscle mitochondrial mass in fish muscle. Seasonal acclimatization to cold in trout also increases LDH in white muscle (Cordiner & Egginton, 1997) as observed here for mixed muscle in zebrafish (Fig. 2). Previous studies have found that exercise training also increases HOAD in both red and white muscle (Johnston & Moon, 1980; Farrell et al. 1991). Perhaps this is explained by the greater training intensity (∼60% Ucrit) and duration used in these studies compared to the 25–30% Ucrit used in the current study. Moreover, muscle remodelling in zebrafish with cold and exercise either produce qualitatively different mitochondria (specialized for fat oxidation in cold-acclimatization) or there is a fibre-specific response to each treatment.
Gene expression
As a window into some of the potential molecular mechanisms that govern cold-induced and contraction-induced muscle remodelling, expression changes of genes encoding metabolic enzymes and key transcription factors were quantified. We used 4 week endpoints for both our cold-acclimatization and exercise-training treatments to maximize phenotypic changes. This sampling protocol may have missed earlier changes in gene expression. For instance, cold-acclimatization did not change levels of mRNA of genes for key enzymes in metabolism (Fig. 3). However, mRNA levels for genes of interest may have their own distinct patterns of expression over the course of our treatments. For example, hepatic mRNA levels for CS peak at approximately 5 days of cold acclimatization while CS activity peaks at 12 days and remains elevated in a eurythermic eelpout (Zoarces viviparus) (Lucassen et al. 2003). Although mRNA levels with exercise training have not been measured before in fish, elevated CS mRNA seen after 4 weeks of training in this study suggests that either expression of this gene peaks late in training or, more likely, it remains elevated throughout the training period. Future studies may choose to examine the temporal patterns in mRNA levels with these treatments to elucidate times of peak expression. We assessed changes in total muscle RNA with our treatments to determine estimates of protein synthesis capacity (>98% of total RNA being ribosomal). Storch et al. (2005) found cold-acclimatizaton results in increased RNA: protein ratios indicative of increased protein synthesis capacities. In contrast, our cold-acclimatization data suggest a decrease in RNA: protein ratios and therefore a decrease in protein synthesis capacity. It is possible that translational efficiency is modified in cold-acclimatized zebrafish, but this was not directly measured. Interestingly, the half-life of some genes with exercise training in mammals is thought to increase (reviewed in Fluck & Hoppeler, 2003). Whether this occurs in zebrafish is unknown, but our data do suggest that RNA: protein ratios and protein synthesis capacity increase after training. After the removal of a single outlier (0.91 mg g−1) from the control fish data, cold acclimatization and exercise -training showed significant increases in DNA content per gram of tissue. Although this suggests gene dosage as a potential form of regulation, cold acclimatization and exercise training increased DNA content equally, while the mRNA expression for genes of interest showed distinct patterns between these groups. Higher myonuclear content in muscle fibres has also been seen after training in the common carp, with nuclei increasing in relation to fibre diameter (Johnston, 2006). However, our own data (K. Dhekney and G. B. McClelland, unpublished observations) are beginning to show that total red and white muscle area is unchanged with either exercise or cold in zebrafish.
There is very little information available for the regulation of transcription factors in fish muscle but increases in NRF-1 seen in this study suggest a conserved response between cold and exercise in fish and exercise in mammals (Adhihetty et al. 2003). However, mammalian studies also find increases in gene expression for all three PPAR (α, γ and β/δ) isoforms and PGC-1α after endurance exercise (60–85% V˙O2max, Mahoney et al. 2005) not seen here for cold or exercise in zebrafish (Fig. 4). Again, it is possible that differences in training intensity and duration might explain some of the differences in exercise-trained zebrafish compared to the training response in mammals. It is important to note that many transcription factors can be regulated by kinase pathways such as the p38-MAPK and ERK-MAPK pathways, at least in mammals (e.g. Barger et al. 2000, 2001). Future studies using fish should determine changes in DNA binding of these and other transcription factors with different stressors. It is also important to note that tetraploid zebrafish have more copies of genes than mammals (e.g. Amores et al. 1998). For example zebrafish have at least two isoforms of PPAR-β (β1 and β2; Robinson-Rechavi et al. 2001) but it is unclear if they respond in the same way to exercise or cold.
Swimming performance
To assess any potential whole-animal performance consequences of these treatments, we employed a standard swimming trial used extensively in fish research. Critical swimming speed (Ucrit) is used as a way to assess swimming capacity of fish by increasing water speeds until the fish can no longer keep its position in the current and fatigues (Brett, 1964). Oxygen consumption increases up to fatigue (Brett, 1964), and it is assumed that maximum oxygen consumption occurs at the critical swimming speed (Hammer, 1995). Moreover, absolute critical swimming speed increases with size while relative (to body length, BL) critical swimming speed decreases with size. Previously, it has been found that zebrafish are capable of unusually high swimming speeds compared to most other fish species for which this measure is documented (Plaut & Gordon, 1994; Plaut, 2000). Zebrafish in the current study had maximum sustained swimming speeds at 28°C of 56–63 cm s−1 or 20–22 BL s−1 in agreement with previous measurements in this species (Plaut & Gordon, 1994; Plaut, 2000). Only exercise-trained zebrafish showed significant increases in absolute maximum sustained swimming speeds at 28°C used as an indicator of whole-body swimming capacity (Fig. 5A). When standardized relative to fish length, relative critical swimming speeds were the same between all fish since training stimulated growth in group 2 fish (Table 2). Zebrafish also show a typical decrease in maximum sustained swimming speeds with temperature (Fig. 5A and B) due to declines in muscle power output associated with a drop in temperature (Day & Butler, 2005). Previously, muscle HOAD activities and maximum sustained swimming speeds were found to be highly correlated in trout (Farrell et al. 1991), and increased with training (Farrell et al. 1990). In contrast, trained zebrafish had a greater maximum absolute sustained swimming speed (Fig. 5) despite lower HOAD activities (Fig. 2) compared to cold-acclimatized and control fish. In fish, anaerobic metabolism is initiated at approximately 70–80% Ucrit and at 100% Ucrit, pronounced changes in muscle metabolites can occur as fish fatigue (Burgetz et al. 1998). We assessed post-exercise metabolite levels and found significant increases in lactate and significant decreases in CrP and ATP in all groups with exercise compared to non-exercised controls (Fig. 6), in agreement with previous studies (Burgetz et al. 1998). Interestingly, training in coalfish increased the activity of the enzyme creatine phosphokinase (CPK) (Johnston & Moon, 1980), and this may explain why trained zebrafish are able to deplete muscle CrP levels below the levels in cold-acclimatized fish muscle (Fig. 6).
Muscle morphology
A major advantage of using fish for muscle research is that they have anatomically separated slow (red) and fast (white) muscle masses, which allows the measurement of fibre-type-specific responses to stress. However, due to their small size, mixed muscle samples were used for all of the biochemical and gene expression measurements in this initial study. Past studies on fish have found that cold acclimatization (Johnston & Lucking, 1978) and exercise training (Johnston & Moon, 1980; Hammill et al. 2004) can alter the distribution of muscle fibre types, with both stimuli increasing the proportion of red muscle mass. Moreover, exercise training in trout has been found to stimulate somatic growth and muscle fibre hypertrophy of fast fibres (Martin & Johnston, 2005). No significant differences in the mRNA levels for fast or slow myosin heavy chains were detected with either cold or exercise (Fig. 3C and F), but it is unclear if this translates to different protein levels. Our own measurement using myosin-ATPase histochemistry are beginning to show that red muscle mass does not increase either in absolute area or in the proportion of total cross-sectional area with either cold exposure or exercise training (K. Dhekney and G. B. McClelland, unpublished observations). This is supported by protein: DNA ratios (Fig. 1B and C) that are not increased with either treatment. It is possible that muscle fibre types respond in a similar way or perhaps to different degrees to stress. For example, in a continuous 3 week training experiment, coalfish showed similar changes in metabolic enzyme activities in both red and white muscle (Johnston & Moon, 1980). In contrast, rainbow trout exercised 18 h a day at 60% of their maximal sustainable swimming speed showed a greater plasticity in enzyme activities in white versus red muscles (Farrell et al. 1991). Cold acclimatization in rainbow trout has been found to induce similar metabolic changes in red and white muscle (McClelland et al. 2005). Experiments under way will determine fibre-specific changes in gene expression to cold and to exercise in zebrafish.
Conclusions and perspectives
Cold acclimatization and exercise training represent different stimuli that induce aerobic phenotypes in zebrafish muscle. The present data suggest that there may be different mechanisms responsible for muscle remodelling between fish and mammals and between different modes of aerobic phenotype induction in zebrafish. However, certain responses (NRF-1 mRNA) could be classified as essential for the generation of an aerobic phenotype, a common response to different stimuli, and evolutionarily conserved. Moreover cold acclimatization may represent a reliable and straightforward surrogate to exercise-training protocols for the study of conserved muscle remodelling events. Other responses (PPAR-β1) may be more specialized to different stimuli and result in the development of distinct phenotypes. Direct causation between these responses and changes in muscle phenotype remain to be established. It is still unclear what mechanisms ultimately trigger this remodelling in response to cold or exercise. There is currently some controversy regarding the importance of calcium (Adhihetty et al. 2003; Martin & Johnston, 2005) and reactive oxygen species (ROS) signalling mechanisms (Abele et al. 2002) in other systems, and so a comprehensive picture of the mechanisms involved is currently lacking.
The zebrafish has been used extensively as a model of vertebrate development, including the formation of different muscle fibre types (e.g. Bryson-Richardson et al. 2005). We show here that zebrafish can also be used as a model of adult vertebrate muscle plasticity. They offer the added advantages of the ability to perform genetic screens or the use of molecular techniques to establish direct connections between mechanisms of remodelling and muscle phenotypes (e.g. morpholinos and iRNA). Clearly many fundamental aspects regarding the mechanisms of muscle plasticity can be readily assessed using the adult zebrafish system.
Acknowledgments
The authors would like to thank Dr Fern Galvez for invaluable technical advice regarding PCR analysis. We also acknowledge Dr Chris Wood for use of his facilities, particularly during our lab renovations and setup. Special thanks go to Dr Y. Wang for technical advice on metabolite measurements. Additional technical assistance was provided by Déanne Malenfant, and Ayaz Hyder. This work was made possible by funding to GBM though a Natural Science and Engineering Research Council of Canada (NSERC) Discovery Grant, McMaster University and a Canadian Foundation for Innovation (CFI) New Opportunities Award. PMC is the recipient of an NSERC postgraduate award. The authors thank the two anonymous reviewers for their very helpful constructive criticisms and suggestions.
References
- Abele D, Heise K, Portner HO, Puntarulo S. Temperature-dependence of mitochondrial function and production of reactive oxygen species in the intertidal mud clam Mya arenaria. J Exp Biol. 2002;205:1831–1841. doi: 10.1242/jeb.205.13.1831. [DOI] [PubMed] [Google Scholar]
- Adhihetty PJ, Irrcher I, Joseph A-M, Ljubicic V, Hood DA. Plasticity of skeletal muscle mitochondria in response to contractile activity. Exp Physiol. 2003;88:99–107. doi: 10.1113/eph8802505. [DOI] [PubMed] [Google Scholar]
- Amores A, Force A, Yan Y-L, Joly L, Amemiya C, Fritz A, Ho RK, Langeland J, Prince V, Wang Y-L, Westerfield M, Ekker M, Postlethwait JH. Zebrafish hox clusters and vertebrate genome evolution. Science. 1998;282:1711–1714. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- Barger PM, Brandt JM, Leone TC, Weinheimer CJ, Kelly DP. Deactivation of peroxisome proliferator-activated receptor-α during cardiac hypertrophic growth. J Clin Invest. 2000;105:1723–1730. doi: 10.1172/JCI9056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barger PM, Browning AC, Garner AN, Kelly DP. p38 Mitogen-activated protein kinase activates peroxisome proliferator-activated receptor alpha. J Biol Chem. 2001;276:44495–44501. doi: 10.1074/jbc.M105945200. [DOI] [PubMed] [Google Scholar]
- Battersby BJ, Moyes CD. Influence of acclimation temperature on mitochondrial DNA, RNA, and enzymes in skeletal muscle. Am J Physiol Regul Integr Comp Physiol. 1998;275:R905–R912. doi: 10.1152/ajpregu.1998.275.3.R905. [DOI] [PubMed] [Google Scholar]
- Bergmeyer HU. Methods of Enzymatic Analysis. Vol. 3. New York: Academic Press; 1974. [Google Scholar]
- Bouchard P, Guderley H. Time course of the response of mitochondria from oxidative muscle during thermal acclimation of rainbow trout. Oncorhynchus Mykiss J Exp Biol. 2003;206:3455–3465. doi: 10.1242/jeb.00578. [DOI] [PubMed] [Google Scholar]
- Brett JR. The respiratory metabolism and swimming performance of young sockeye salmon. J Fish Res Board Can. 1964;21:1183–1226. [Google Scholar]
- Brooks SPJ. A program for analyzing enzyme rate data obtained from a microplate reader. Biotechniques. 1994;17:1154–1161. [PubMed] [Google Scholar]
- Bryson-Richardson RJ, Daggett DF, Cortes F, Neyt C, Keenan DG, Currie PD. Myosin heavy chain expression in zebrafish and slow muscle composition. Dev Dyn. 2005;233:1018–1022. doi: 10.1002/dvdy.20380. [DOI] [PubMed] [Google Scholar]
- Burgetz I, Rojas-Vargas A, Hinch S, Randall D. Initial recruitment of anaerobic metabolism during sub-maximal swimming in rainbow trout (Oncorhynchus mykiss) J Exp Biol. 1998;201:2711–2721. doi: 10.1242/jeb.201.19.2711. [DOI] [PubMed] [Google Scholar]
- Chin ER. Role of Ca2+/calmodulin-dependent kinases in skeletal muscle plasticity. J Appl Physiol. 2005;99:414–423. doi: 10.1152/japplphysiol.00015.2005. [DOI] [PubMed] [Google Scholar]
- Cordiner S, Egginton S. Effects of seasonal temperature acclimatization on muscle metabolism in rainbow trout Onchorhynchus mykiss. Fish Physiol Biochem. 1997;16:333–343. [Google Scholar]
- Day N, Butler PJ. The effects of acclimation to reversed seasonal temperatures on the swimming performance of adult brown trout Salmo trutta. J Exp Biol. 2005;208:2683–2692. doi: 10.1242/jeb.01669. [DOI] [PubMed] [Google Scholar]
- Farrell A, Johansen JA, Moyes CD, West TG, Suarez RK. Effects of exercise training and coronary ablation on swimming performance, heart size, and cardiac enzymes in rainbow trout, Onchorhynchus mykiss. Can J Zool. 1990;68:1174–1179. [Google Scholar]
- Farrell AP, Johansen JA, Suarez RK. Effects of exercise-training on cardiac performance and muscle enzymes in rainbow trout, Oncorhynchus mykiss. Fish Physiol Biochem. 1991;9:303–312. doi: 10.1007/BF02265151. [DOI] [PubMed] [Google Scholar]
- Fluck M, Hoppeler H. Molecular basis of skeletal muscle plasticity – from gene to form and function. Rev Physiol Biochem Pharmacol. 2003;146:159–216. doi: 10.1007/s10254-002-0004-7. [DOI] [PubMed] [Google Scholar]
- Fulton T. Rate of growth of seas fishes. Sci Invest Fish Division Scot Rept. 1902;20:1035–1039. [Google Scholar]
- Gollnick PD, Armstong RB, Saubert CWIV, Piehl K, Saltin B. Enzyme activity and fiber composition in skeletal muscle of untrained and trained men. J Appl Physiol. 1972;33:312–319. doi: 10.1152/jappl.1972.33.3.312. [DOI] [PubMed] [Google Scholar]
- Gollnick PD, King DW. Effect of exercise and training on mitochondria of rat skeletal muscle. Am J Physiol. 1969;216:1502–1509. doi: 10.1152/ajplegacy.1969.216.6.1502. [DOI] [PubMed] [Google Scholar]
- Guderley H, St-Pierre J. Going with the flow or life in the fast lane: contrasting mitochondrial responses to thermal change. J Exp Biol. 2002;205:2237–2249. doi: 10.1242/jeb.205.15.2237. [DOI] [PubMed] [Google Scholar]
- Gulick T, Cresci S, Caira T, Moore D, Kelly D. The peroxisome proliferator-activated receptor regulates mitochondrial fatty acid oxidative enzyme gene expression. Proc Natl Acad Sci U S A. 1994;91:11012–11016. doi: 10.1073/pnas.91.23.11012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer C. Fatigue and exercise tests with fish. Comp Biochem Physiol A. 1995;112:1–20. [Google Scholar]
- Hammill E, Wilson RS, Johnston IA. Sustained swimming performance and muscle structure are altered by thermal acclimation in male mosquitofish. J Therm Biol. 2004;29:251–257. [Google Scholar]
- Hochachka PW. Limits: how fast and how slow muscle metabolism can go. In: Benzi G, editor. Advances in Myochemistry. Vol. 1. John Libbey Eurotext Ltd; 1987. pp. 3–12. [Google Scholar]
- Hood DA. Plasticity in skeletal, cardiac, and smooth muscle invited review: Contractile activity-induced mitochondrial biogenesis in skeletal muscle. J Appl Physiol. 2001;90:1137–1157. doi: 10.1152/jappl.2001.90.3.1137. [DOI] [PubMed] [Google Scholar]
- Johnston IA. Environment and plasticity of myogenesis in teleost fish. J Exp Biol. 2006;209:2249–2264. doi: 10.1242/jeb.02153. [DOI] [PubMed] [Google Scholar]
- Johnston I, Lucking M. Temperature induced variation in distribution of different types of muscle-fiber in goldfish (Carassius auratus) J Comp Physiol. 1978;124:111–116. [Google Scholar]
- Johnston IA, Moon TW. Endurance exercise training in the fast and slow muscles of a teleost fish (Pollachius virens) J Comp Physiol. 1980;135:147–156. [Google Scholar]
- Koves TR, Li P, An J, Akimoto T, Slentz D, Ilkayeva O, Dohm GL, Yan Z, Newgard CB, Muoio DM. Peroxisome proliferator-activated receptor-γ co-activator 1α-mediated metabolic remodeling of skeletal myocytes mimics exercise training and reverses lipid-induced mitochondrial inefficiency. J Biol Chem. 2005;280:33588–33598. doi: 10.1074/jbc.M507621200. [DOI] [PubMed] [Google Scholar]
- Leary SC, Moyes CD. The effects of bioenergetic stress and redox balance on the expression of genes critical to mitochondrial function. In: Storey KB, Storey J, editors. Environmental Stressors and Gene Responses. New York: Elsevier Science; 2000. pp. 209–229. [Google Scholar]
- Lemberger T, Desvergne B, Wahli W. Peroxisome proliferator-activated receptors: a nuclear receptor signaling pathway in lipid physiology. Annu Rev Cell Dev Biol. 1996;12:335–363. doi: 10.1146/annurev.cellbio.12.1.335. [DOI] [PubMed] [Google Scholar]
- Lin J, Wu H, Tarr PT, Zhang C-Y, Wu Z, Boss O, Michael LF, Pulgserver P, Isotani E, Olson EN, Lowell BB, Bassell-Duby R, Spiegelman BM. Transcriptional co-activator PGC-1α drives the formation of slow-twitch muscle fibres. Nature. 2002;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lucassen M, Schmidt A, Eckerle LG, Portner H-O. Mitochondrial proliferation in the permanent vs. temporary cold: enzyme activities and mRNA levels in Antarctic and temperate zoarcid fish. Am J Physiol Regul Integr Comp Physiol. 2003;285:R1410–R1420. doi: 10.1152/ajpregu.00111.2003. [DOI] [PubMed] [Google Scholar]
- Mahoney DJ, Parise G, Melov S, Safdar A, Tarnopolsky MA. Analysis of global mRNA expression in human skeletal muscle during recovery from endurance exercise. FASEB J. 2005;19:1498–1500. doi: 10.1096/fj.04-3149fje. [DOI] [PubMed] [Google Scholar]
- Malek RL, Sajadi H, Abraham J, Grundy MA, Gerhard GS. The effects of temperature reduction on gene expression and oxidative stress in skeletal muscle from adult zebrafish. Comp Biochem Physiol C. 2004;138:363–373. doi: 10.1016/j.cca.2004.08.014. [DOI] [PubMed] [Google Scholar]
- Martin CI, Johnston IA. The role of myostatin and the calcineurin-signalling pathway in regulating muscle mass in response to exercise training in the rainbow trout Oncorhynchus mykiss Walbaum. J Exp Biol. 2005;208:2083–2090. doi: 10.1242/jeb.01605. [DOI] [PubMed] [Google Scholar]
- McClelland GB, Dalziel AC, Fragoso NM, Moyes CD. Muscle remodeling in relation to blood supply: implications for seasonal changes in mitochondrial enzymes. J Exp Biol. 2005;208:515–522. doi: 10.1242/jeb.01423. [DOI] [PubMed] [Google Scholar]
- Moyes CD, Hood DA. Origins and consequences of mitochondrial variation in vertebrate muscle. Annu Rev Physiol. 2003;65:177–201. doi: 10.1146/annurev.physiol.65.092101.142705. [DOI] [PubMed] [Google Scholar]
- Plaut I. Effects of fin size on swimming performance, swimming behaviour and routine activity of zebrafish Danio rerio. J Exp Biol. 2000;203:813–820. doi: 10.1242/jeb.203.4.813. [DOI] [PubMed] [Google Scholar]
- Plaut I. Critical swimming speed: its ecological relevance. Comp Biochem Physiol A. 2001;131:41–50. doi: 10.1016/s1095-6433(01)00462-7. [DOI] [PubMed] [Google Scholar]
- Plaut I, Gordon M. Swimming metabolism of wild-type and cloned zebrafish Brachydanio rerio. J Exp Biol. 1994;194:209–223. doi: 10.1242/jeb.194.1.209. [DOI] [PubMed] [Google Scholar]
- Portner HO. Physiological basis of temperature-dependent biogeography: trade-offs in muscle design and performance in polar ectotherms. J Exp Biol. 2002;205:2217–2230. doi: 10.1242/jeb.205.15.2217. [DOI] [PubMed] [Google Scholar]
- Reichmann H, Hoppeler H, Mathieu-Costello O, Bergen F, Pette D. Biochemical and ultrastructural changes of skeletal muscle mitochondria after chronic electrical stimulation in rabbits. Pflugers Arch. 1985;404:1–9. doi: 10.1007/BF00581484. [DOI] [PubMed] [Google Scholar]
- Richards JG, Mercado AJ, Clayton CA, Heigenhauser GJF, Wood CM. Substrate utilization during graded aerobic exercise in rainbow trout. J Exp Biol. 2002;205:2067–2077. doi: 10.1242/jeb.205.14.2067. [DOI] [PubMed] [Google Scholar]
- Robinson-Rechavi M, Marchand O, Escriva H, Bardet P-L, Zelus D, Hughes S, Laudet V. Euteleost fish genomes are characterized by expansion of gene families. Genome Res. 2001;11:781–788. doi: 10.1101/gr.165601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storch D, Lannig G, Portner HO. Temperature-dependent protein synthesis capacities in Antarctic and temperate (North Sea) fish (Zoarcidae) J Exp Biol. 2005;208:2409–2420. doi: 10.1242/jeb.01632. [DOI] [PubMed] [Google Scholar]
- Taylor MR, Hurley JB, Van Epps HA, Brockerhoff SE. A zebrafish model for pyruvate dehydrogenase deficiency: rescue of neurological dysfunction and embryonic lethality using a ketogenic diet. Proc Natl Acad Sci U S A. 2004;101:4584–4589. doi: 10.1073/pnas.0307074101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton C, Stamatiou D, Liew C-C. Gene expression profile of zebrafish exposed to hypoxia during development. Physiol Genomics. 2003;13:97–106. doi: 10.1152/physiolgenomics.00128.2002. [DOI] [PubMed] [Google Scholar]
- Vredendaal PJ, van den Berg IET, van der Stroobants AKADL, Malingre HE, Berger R. Structural organization of the human short-chain 3-hydroxyacyl-CoA dehydrogenase gene. Mamm Genome. 1998;9:763–768. doi: 10.1007/s003359900860. [DOI] [PubMed] [Google Scholar]
- Weitzel JM, Iwen KAH, Seitz HJ. Regulation of mitochondrial biogenesis by thyroid hormone. Exp Physiol. 2003;88:121–128. doi: 10.1113/eph8802506. [DOI] [PubMed] [Google Scholar]