Abstract
Picrotoxin, a potent antagonist of the inhibitory central nervous system GABAA and glycine receptors, is believed to interact with residues that line the central ion pore. These pore-lining residues are in the second transmembrane domain (TM2) of each of the five constituent subunits. One of these amino acids, a threonine at the 6′ location, when mutated to phenylalanine, abolishes picrotoxin sensitivity. It has been suggested that this threonine, via hydrogen bonding, directly interacts with the picrotoxin molecule. We previously demonstrated that this mutation, in the α, β or γ subunit, can impart picrotoxin resistance to the GABA receptor. Since the functional pentameric GABA receptor contains two α subunits, two β subunits and one γ subunit, it is not clear how many α and β subunits must carry this mutation to impart the resistant phenotype. In this study, by coexpression of mutant α or β subunits with their wild-type counterparts in various defined ratios, we demonstrate that any single subunit carrying the 6′ mutation imparts picrotoxin resistance. Implications of this finding in terms of the mechanism of antagonism are considered.
That picrotoxin is an antagonist of GABA and glycine receptors has been known for decades (Davidoff & Aprison, 1969; Galindo, 1969; Engberg & Thaller, 1970; Hill et al. 1972). Early evidence suggested that picrotoxin exerts its non-competitive actions by interacting with the chloride-selective pore (Constanti, 1978; Akaike et al. 1985; Inoue & Akaike, 1988; Newland & Cull-Candy, 1992). Sequence comparison between picrotoxin-sensitive and picrotoxin-resistant glycine receptors and homology-driven site-directed mutagenesis provided the first strong evidence of an interaction between the second transmembrane domain (TM2) pore domain and picrotoxin (Pribilla et al. 1992). A subsequent study extended this molecular site of action into the GABA receptor family (Gurley et al. 1995). Since these earlier studies, several amino acids have been identified in the TM2 domain that, when mutated, alter antagonism of the GABA or glycine receptor by picrotoxin (Xu et al. 1995; Zhang et al. 1995; Chang & Weiss, 1998, 2000; Buhr et al. 2001; Shan et al. 2001; Dibas et al. 2002).
In a previous study, we demonstrated that mutation of a highly conserved threonine residue at the 6′ position in the second transmembrane domain of the α1, β2 or γ2 subunits of α1β2γ2 GABA receptors abolished antagonism by picrotoxin (Gurley et al. 1995). At the time of that study, the stoichiometry of the GABA receptor was unknown. Since then, the stoichiometry has been determined to be two α subunits, two β subunits and one γ subunit (Chang et al. 1996; Tretter et al. 1997). Although this implies that the 6′ TM2 mutation in the single γ subunit was sufficient to abolish picrotoxin sensitivity, the same could not be said for the α and β mutations, for which there are two copies in each functional pentameric receptor. In the present study, by coexpressing combinations of wild-type and mutant subunits at defined ratios we directly address this fundamental issue. Our data unambiguously demonstrate that any one of the five subunits carrying this 6′ TM2 mutation can impart picrotoxin resistance. Implications for the mechanism of action and the use of this system for probing native receptor function, stoichiometry and regulation are discussed.
Methods
Clones, site-directed mutagenesis and in vitro transcription
Rat α1, β2 and γ2 cDNAs were subcloned into the pGEMHE vector (Liman et al. 1992). Mutations α1T260F and β2T256F were generated using the overlap extension method (Kammann et al. 1989). All constructs were verified by cDNA sequencing. The DNAs were linearized with Nhe I (New England BioLabs Inc., Ipswich, MA, USA), and run-off capped cRNA was transcribed from linearized cDNAs with standard in vitro transcription procedures using the T7 mMessage mMachine Kit (Ambion, Austin, TX, USA). Integrity and yield of the cRNA were verified on a 0.8% agarose gel.
Xenopus oocyte expression
Xenopus laevis (Xenopus I, Ann Arbor, MI, USA) were anaesthetized by placing them in a 0.2% solution of MS-222. The oocytes were surgically removed and placed in a solution that consisted of 85.5 mm NaCl, 2.5 mm KCl, 5 mm Hepes, 1 mm CaCl2, 1 mm MgCl2, 1 mm Na2HPO4, 50 i.u. ml−1 penicillin and 50 mg ml−1 streptomycin, pH 7.5, adjusted with NaOH. The frog was then allowed to recover fully before being returned to a recovery tank, after which time it was observed daily (for one week) to ensure that no complications had arisen as a result of the surgery. This procedure was in accordance with The Institutional Animal Care and Use Committee of the University of Texas Health Science Center at San Antonio and the NIH office of Laboratory Animal Welfare.
Oocytes were dispersed in this solution without Ca2+, but in the presence of 0.3% collagenase A (Roche Diagnostics, Indianapolis, IN, USA). After isolation, stage VI oocytes were thoroughly rinsed and maintained at 18°C in the above-mentioned solution plus 1 mm Ca2+. Micropipettes for injecting cRNA were pulled on a Sutter P87 horizontal puller. To match the cRNA concentrations, wild-type and mutant cRNAs were diluted 2- to 10-fold with diethyl pyrocarbonate (DEPC)-treated water and electrophoresed in a 0.8% agarose gel. Based on relative intensity of the bands, the cRNA concentrations were readjusted to be equal. The maximum GABA-activated currents recorded from oocytes injected with the same amount of the wild-type and mutant cRNAs were approximately the same. The experiments presented in this study were performed on several different batches of cRNA. For experiments involving coexpression of wild-type and mutant receptors, instead of matching concentrations of cRNA by comparing intensity of cRNA bands on the gel, we compared the maximum GABA-activated currents for control oocytes injected with the same amounts of wild-type or mutant cRNA. The ratios we provide in the figures reflect recalculation based on these maxima.
Voltage clamp of oocytes
Two-electrode voltage-clamp procedures were used for current recording 2 or 3 days after cRNA injection. Each oocytes was placed on a 300 μm nylon mesh suspended in a small-volume chamber (< 100 μl). The oocyte was perfused continuously with a solution containing 92.5 mm NaCl, 2.5 mm KCl, 5 mm Hepes, 1 mm CaCl2 and 1 mm MgCl2, pH 7.5, adjusted with NaOH. The solution was switched to the test solution, which was identical to the perfusion solution with the addition of drug (e.g. GABA). Typical interval between applications (GABA or GABA plus picrotoxin) was 5 min to allow sufficient recovery from desensitization as well as unbinding of picrotoxin. All experiments were performed at room temperature. Recording microelectrodes were pulled on a P87 Sutter horizontal puller and filled with 3 m KCl. The electrode resistances ranged from 1 to 3 MΩ. Standard two-electrode voltage-clamp techniques (GeneClamp 500; Molecular Devices, Sunnyvale, CA, USA) were used to record currents at a holding potential of −70 mV. On-line digitization of the signal at 20 Hz was carried out by using the ITC-16 data acquisition board (Instrutech, Long Island, NY, USA) and Igor software (Wavemetrics, Lake Oswego, OR, USA).
Data analysis
Agonist concentration–response relationships were fitted (least-squares) with the following sigmoidal equation:
| (1) |
where I and Imax represent current at a given agonist concentration (A) and maximal agonist-induced current, respectively. EC50 is the half-maximal effective agonist concentration, and nH is the Hill coefficient.
To assess the sensitivity of block by picrotoxin, the peak amplitudes were measured and normalized to the GABA-activated current in the absence of picrotoxin. The time-dependent decay was a combination of desensitization and block, so the measured block was slightly underestimated. While this approach may alter the accuracy of the determined antagonist sensitivity of the wild-type receptor, the effects would be minimal on the mutant receptors, where little antagonism was observed. The IC50 (concentration of PTX which decreased the agonist-mediated current by 50%) for each receptor was determined by a least-squares fit of the following relationship to the data:
| (2) |
where current (I) is a function of the inhibitor concentration ([PTX]). The IC50 was determined in the presence of 40 μm GABA. All data are presented as means ± s.e.m. with n indicating the number of cells tested.
Statistical comparison of models
All statistical analyses were performed using R-2.3.1 (http://www.R-project.org). The following non-linear models were developed to predict the relative efficiency of inhibition from the proportion of wild-type and mutant isoforms for both α and β subunits:
where Y is the fractional block, Pmm is the estimated proportion of complexes where both subunits are mutant, Pwm is the estimated proportion of complexes where one subunit is mutant and one wild-type, and B1 and B2 are model parameters that are constrained between 0 and 1 in order to prevent the efficiency of inhibition from achieving unrealistic values. The equation used to calculate the fractions was: p2 + 2pq + q2 = 1, where p and q are the fractions of wild type and mutant RNA and 2pq represents the fraction of receptors with both wild type and mutant subunits. In model I and model II, Pmm = q2 and Pwm = 2pq. Model I assumes that both α or both β subunits must be mutant to impart picrotoxin resistance, while model II assumes that only one α or one β subunit must be mutant to impart resistance. Models with a better fit were selected based on values of residual standard errors and results of goodness-of-fit tests (Bates & Watts, 1988; Neter et al. 1996).
Structural modelling
ICM (Molsoft; La Jolla, CA, USA) was used to build a representation of the TM2 region of the GABAA receptor. The homology model was built by working from the 4Å resolution structure of the nicotinic acetylcholine receptor (nAChR) transmembrane domain (Miyazawa et al. 2003). Alignments were constructed by pairing GABA and nAChR subunits on the putative basis of their role in ligand binding. By this model, the two GABAA α1 subunits correspond to nAChR γ and δ, the two GABAA β2 subunits correspond to the two nAChR α subunits and the GABAA γ2 subunit corresponds to the nAChR β subunit. The overall model was then energy minimized to eliminate intra-/intersubunit clashes. Introduction of mutations into the receptor was followed by further energy minimization to ensure that this perturbation did not impart any instability to the TM2 region.
Results
Figure 1A shows GABA-activated currents from occytes expressing wild-type receptors and mutated receptors in which the conserved TM2 threonine at position 260 in the α1 subunit and position 256 in the β subunit were switched to phenylalanine. Both of these residues are in the homologous 6′ position of TM2. In all subsequent discussion, we will refer to the αT260F mutation as αm and the βT256F mutation as βm. Figure 1B plots the GABA dose–response relationship for wild-type receptors (•) as well as the αmβγ ( ) and αβmγ receptors (^). The continuous lines are fits of the Hill equation (eqn (1)) to the dose–response relationships and yielded EC50 values of 40.8 ± 21.0, 65.2 ± 17.0 and 38.6 ± 10.2 for αβγ, αmβγ and αβmγ, respectively (n = 5 for all). This pore mutation did not significantly alter the sensitivity of the receptor to GABA (unpaired t test, n.s.).
) and αβmγ receptors (^). The continuous lines are fits of the Hill equation (eqn (1)) to the dose–response relationships and yielded EC50 values of 40.8 ± 21.0, 65.2 ± 17.0 and 38.6 ± 10.2 for αβγ, αmβγ and αβmγ, respectively (n = 5 for all). This pore mutation did not significantly alter the sensitivity of the receptor to GABA (unpaired t test, n.s.).
Figure 1. TM2 mutations have minimal effect on GABA sensitivity.
A, GABA-activated currents in the presence of increasing concentrations of GABA for α1β2γ (αβγ), α1T260Fβ2γ2 (αmβγ) and α1β2T256Fγ2 (αβmγ). B, current amplitudes are plotted as a function of GABA concentration for the three receptor combinations, and the continuous lines are the best fit of Eqn (1) to the data. The EC50 values were 38.6 ± 10.2, 65.2 ± 17.0 and 40.8 ± 21.0 for αβγ, αmβγ, and αβmγ, respectively (All n= 5).
Figure 2A shows GABA-activated currents (40 μm GABA) in the presence of increasing concentrations of picrotoxin for wild-type, αmβγ and αβmγ receptors. These data are plotted in Fig. 2B, and the continuous line is the best fit of an inhibition function (eqn (2)) that yielded an IC50 for picrotoxin of 1.1 ± 0.3 μm (n = 3) in the wild-type receptor. Note that antagonism by picrotoxin, at concentrations as high as 100 μm, was completely eliminated by the α and β subunit mutations. Similar picrotoxin resistance was seen in the homologous γ subunit mutation (Gurley et al. 1995).
Figure 2. TM2 mutations impart a picrotoxin resistance.
A, GABA-activated currents (40 μm) in the presence of increasing concentrations of picrotoxin for αβγ, αmβγ and αβmγ. Note that the α and β mutations imparted a picrotoxin resistance at concentrations up to 100 μm picrotoxin. B, current amplitudes are plotted as a function of picrotoxin concentration for the three receptor combinations, and the continuous line is the best fit of Eqn (2) to the wild-type data. Since there was no block for the mutants, we could not fit an inhibition function in these two cases. The IC50 for the wild-type receptor was 1.1 ± 0.3 μm (n= 3).
To address the question of how many α and β subunits must carry the mutation to impart a picrotoxin resistance, we coexpressed various ratios of wild-type and mutant subunits. The top of Table 1 shows the possible combinations of receptors when either the wild-type α or β subunits (N6′T) are coexpressed with their corresponding mutants (N6′F). We can consider the possibilities for the α subunit, but the identical argument applies to the β subunit. Functionally, if we coexpress α and αm, there are three combinations to consider: those with two wild-type α subunits, those with two mutant α subunits and those with one wild-type and one mutant α subunit. We can then use the binomial distribution to predict the ratio of these three combinations. In this analysis, we make two assumptions. First, we assume that the subunits assemble with roughly equivalent efficiency and indeed we do observe comparable levels of expression with comparable cRNA injections. In addition, we will describe an analysis later that takes into account any differences in expression between wild-type and mutant subunits. The second assumption is that we can ignore subunit order in terms of the actions of picrotoxin and the binomial calculations. Indeed, these receptors are asymmetric in terms of their subunit order and there is evidence indicating corresponding asymmetric properties (Tretter et al. 1997; Baumann et al. 2002, 2003; Boulineau et al. 2005; Baur et al. 2006). If there are differences in the sensitivity to picrotoxin depending on the position of the single mutant subunit within the pentamer, this should not influence the analysis of our experimental data here, since we are basing our results on the maximal block rather than the picrotoxin sensitivities. It is possible that receptors with, for example, one mutant and one wild-type α subunit show partial block (partial resistance), but as will be demonstrated subsequently, this scenario adds an unnecessary layer of complexity.
Table 1.
Binomial predictions for the relative prevalence of the different combinations of receptors when wild-type and mutant subunits were coexpressed
First, consider the situation in Table 1 when the wild-type to mutant cRNA ratio is 0.11, representing a ninefold excess of αm cRNA compared with wild-type α cRNA. In this case, 0.79 of the receptors will be double mutant, 0.01 would be double wild-type and 0.20 would have one wild-type and one mutant subunit. If two mutant subunits were required to eliminate picrotoxin sensitivity, then 0.79 of the current would be picrotoxin resistant (model II in Methods). Alternatively, if one mutant subunit imparts picrotoxin resistance, then 0.99 of the current would be picrotoxin resistant (model I in Methods). For each ratio of wild-type and mutant cRNAs injected (leftmost column in Table 1), one can predict in this same manner the fraction of current blocked for the model in which only one α subunit needs to be mutant for picrotoxin resistance or the model in which both α subunits must be mutant for picrotoxin resistance.
Figure 3A shows picrotoxin-mediated antagonism for GABA-activated currents (40 μm GABA) with varying ratios of injected wild-type and mutant α cRNA, and Fig. 3B shows picrotoxin-mediated antagonism for GABA-activated currents (40 μm GABA) with varying ratios of injected wild-type and mutant β cRNA. The fraction of wild-type receptors is indicated to the left of each set of traces. Note the decreasing inhibition as the fraction of wild-type subunits decreases (top to bottom). To obtain the fraction of current that is not blocked by picrotoxin, dose–response relationships for the various ratios were fitted with an inhibition function (eqn (2); Fig. 4A and B). Extrapolation of the fitted function provided the fraction of current resistant to picrotoxin. (The resulting IC50 values are provided in Table 1) Again, note that as the relative amount of mutant cRNA was reduced, the fraction of current blocked by picrotoxin decreased.
Figure 3. Picrotoxin-mediated antagonism of receptors in which wild-type and mutant subunits were coexpressed.
A, increasing concentrations of picrotoxin were applied in the presence of 40 μm GABA. The number to the left of each row of current traces is the ratio of wild-type α cRNA to αm cRNA. Note that as the relative fraction of mutant cRNA increases, the fraction of the current blocked by picrotoxin decreases. The top row of traces is all wild-type and the bottom row of traces is all mutant. B, as in A, but for β and βm coexpression.
Figure 4. Dose–response relationships for picrotoxin-mediated antagonism of wild-type and mutant subunit coexpression.
A, the amplitudes of the GABA-activated currents are plotted as a function of picrotoxin concentration. The different ratios of injected cRNAs are represented by different symbols, and the continuous lines represent the best fits of Eqn (2) to the data. The dashed line is the fit to the wild-type data. Values of IC50 obtained from these fits are presented in Table 1. B, as A, but for β and βm coexpression.
We developed and compared non-linear models to predict the relative efficiency of inhibition from different ratios of wild-type and mutant subunits. Model I is the case where both subunits must be mutant for picrotoxin resistance, and model II is the case where only one subunit must be mutant for resistance (see Methods for details). Figure 5A shows the results for the α subunit. The filled circles plot the fractional inhibition as a function of the cRNA ratio (each experiment plotted individually), while the open circles plot the best prediction of model I, and the open triangles plot the best prediction of model II. Figure 5B is a similar graph for the β subunit. Model II clearly fits the data better than model I. This is supported by lower residual standard error values: 0.303 for model I versus 0.077 for model II for the α subunit and 0.429 for model I versus 0.097 for model II for the β subunit. Goodness-of-fit tests indicate that model II is significantly better than model I for both α and β data sets (P values < 2.2 × 10−16). These data, along with our previously published results on the γ subunit, unambiguously support the model in which any one subunit of the α1β2γ2 GABA receptor carrying a mutation in this particular 6′ TM2 threonine residue is resistant to picrotoxin.
Figure 5. Fitting and statistical comparison of models I and II.
A, the filled circles plot the fractional inhibition as a function of the cRNA ratio. The open circles plot the best prediction of model I (both α subunits must be mutant for picrotoxin resistance) and the open triangles plot the best prediction of model II (only one α subunit must be mutant for picrotoxin resistance).
Discussion
Overwhelming evidence suggests that picrotoxin interacts with the TM2 domain of the GABA receptor and therefore binds within the pore region. In fact, studies indicate that picrotoxin can be trapped in the pore when the gate closes (Hawthorne & Lynch, 2005). Interactions in the pore, however, do not necessarily imply that the mechanism of action for picrotoxin is pore occlusion. In fact, although use dependent in its actions, there is strong evidence from kinetic analysis that the antagonism by picrotoxin is allosteric (Smart & Constanti, 1986; Newland & Cull-Candy, 1992; Goutman & Calvo, 2004). In addition, structural rearrangements induced by picrotoxin have been identified using the substituted cysteine accessibility method (Hawthorne & Lynch, 2005), as well as site-directed fluorescence spectroscopy (Chang & Weiss, 2002). In the latter study, picrotoxin-mediated conformational rearrangements were identified in a region surrounding the central vestibule, but well above the plane of the membrane and quite distant from the presumed picrotoxin binding site.
It is worth mentioning that picrotoxin-mediated antagonism of the homologous glycine receptor, in stark contrast to the GABAA receptor, is competitive and not use dependent (Lynch et al. 1995). While this could be interpreted as a fundamental difference in the mechanism and/or site of action of picrotoxin between glycine and GABA receptors, the corresponding 6′ mutation in the glycine receptor does eliminate antagonism by picrotoxin (Hawthorne & Lynch, 2005). This observation suggests a common site of action for GABA and glycine receptors. A possible explanation for the observed differences in the wild-type GABA and glycine receptors (competitive and use dependent) would be that the picrotoxin binding site is more accessible in the closed state for the glycine receptor and more accessible in the open state for the GABA receptor (Hawthorne & Lynch, 2005). In this scenario, the 15′ TM2 mutation (Dibas et al. 2002) and the mutations in the TM2–TM3 extracellular linker (Hawthorne & Lynch, 2005) that convert picrotoxin block of the glycine receptor to use dependent could be explained by a structural rearrangement in TM2 that enhances open state accessibility by the blocker.
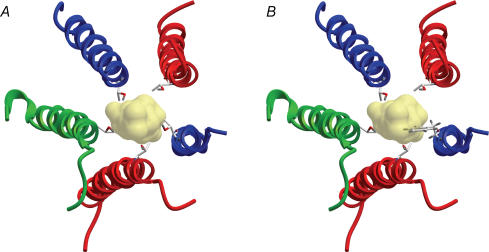
Modelling studies using Monte Carlo minimization give support to this TM2 site of action and postulate that picrotoxin interacts with the cytoplasmic half of the pore and is stabilized by all five TM2 domains (Zhorov & Bregestovski, 2000). In this model, picrotoxin is also stabilized by hydrogen bonds with the 6′ threonine residue. This is the same threonine originally identified to be a major determinant of the action of picrotoxin on GABA and glycine receptors (Pribilla et al. 1992; Gurley et al. 1995) and the same mutation we use in the present study. If a single picrotoxin molecule were stabilized by multiple hydrogen bonds with the 6′ hydroxyl, at least simplistically, one would not predict a complete elimination with the removal of one of the five hydroxyls. Alternatively, the substituted phenylalanine at any one of the five positions could provide a steric hindrance for the pictrotoxin molecule, thereby preventing access to its binding site at a more cytoplasmic position in TM2. In an effort to distinguish these two possibilities, we homology modelled the TM2 domains of the GABAA receptor, based on the homologous nACh receptor (Miyazawa et al. 2003). Figure 6A shows the ribbon structure of the TM2 domains and the 6′ threonine side-chains after optimization (see Methods). The picrotoxin molecule (space filled) is superimposed on this structure to indicate its size relative to this position of the pore. In Fig. 6B, the 6′ threonine of one subunit has been mutated to a phenylalanine. This preliminary structural analysis indicates that the large ringed side-chain of phenylalanine, if positioned as shown in Fig. 6B, could not only prevent access of picrotoxin beyond this position but could also prevent picrotoxin from sitting at this position were it the location of picrotoxin when it blocks the receptor. This could certainly account for the ability of a single phenylalanine mutation in any one of the five subunits to prevent picrotoxin block. A series of mutations of different side-chain sizes at this position are planned in an attempt to test this possibility further.
Figure 6. Structural model of TM2.
A, the TM2 domains of the GABAA receptor were homology modelled based on the homologous acetylcholine receptor (see Methods). The side-chains of the 6′ threonine (shown as ball and stick) were then optimized. The picrotoxin molecule (yellow) was superimposed (not docked) at this position to provide a sense of its size relative to the pore structure in this region. B, same as A, but the threonine in one of the five subunits was mutated to phenylalanine. Note that the phenylalanine side-chain could prevent access of picrotoxin beyond this position, or disrupt its binding were this indeed the location of the picrotoxin binding site.
In summary, we have demonstrated for the first time that a single mutation in any one of the threonines that comprise this ring is sufficient to completely eliminate picrotoxin antagonism. One might propose that this finding was expected, based on the fact that the mutation in the γ subunit, present in one copy per functional pentamer, eliminated picrotoxin sensitivity (Gurley et al. 1995). However, there are certainly well-documented cases where subunit asymmetries exist in the GABA receptor (Baumann et al. 2003; Boulineau et al. 2005) and thus it is necessary to test for symmetry in the actions of picrotoxin. It is convenient that the mutation that eliminates picrotoxin sensitivity has a minimal effect on receptor sensitivity. Expression of a 6′ mutant subunit in neurons or neuronal slices that endogenously express GABA receptors, along with electrophysiological recording in the presence of picrotoxin, can be used to help delineate the role of particular subunit combinations in shaping GABA-mediated inhibition in the brain. Another potential use of these findings is that they may provide a mechanism for delineating relative ratios of native subunits. For example, both GABA and glycine receptors have subunits that impart a native picrotoxin resistance (Pribilla et al. 1992; Zhang et al. 1995). Assessing the fraction of picrotoxin-sensitive current, in conjunction with the binomial analysis we have presented, could enable a determination of the relative expression of the resistance-imparting subunit.
Acknowledgments
This work was supported by NINDS 35291.
References
- Akaike N, Hattori K, Oomura Y, Carpenter DO. Bicuculline and picrotoxin block γ-aminobutyric acid-gated Cl− conductance by different mechanisms. Experientia. 1985;41:70–71. doi: 10.1007/BF02005880. [DOI] [PubMed] [Google Scholar]
- Bates DM, Watts DG. Nonlinear Regression Analysis and its Applications. New York: John Wiley and Sons; 1988. [Google Scholar]
- Baumann SW, Baur R, Sigel E. Forced subunit assembly in α1β2γ2 GABAA receptors: insight into the absolute arrangement. J Biol Chem. 2002;277:46020–46025. doi: 10.1074/jbc.M207663200. [DOI] [PubMed] [Google Scholar]
- Baumann SW, Baur R, Sigel E. Individual properties of the two functional agonist sites in GABAA receptors. J Neurosci. 2003;23:11158–11166. doi: 10.1523/JNEUROSCI.23-35-11158.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baur R, Minier F, Sigel E. A GABAA receptor of defined subunit composition and positioning: concatenation of five subunits. FEBS Lett. 2006;580:1616–1620. doi: 10.1016/j.febslet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Boulineau N, Baur R, Minier F, Sigel E. Consequence of the presence of two different β subunit isoforms in a GABAA receptor. J Neurochem. 2005;95:1724–1731. doi: 10.1111/j.1471-4159.2005.03495.x. [DOI] [PubMed] [Google Scholar]
- Buhr A, Wagner C, Fuchs K, Sieghart W, Sigel E. Two novel residues in M2 of the γ-aminobutyric acid type A receptor affecting gating by GABA and picrotoxin affinity. J Biol Chem. 2001;276:7775–7781. doi: 10.1074/jbc.M008907200. [DOI] [PubMed] [Google Scholar]
- Chang Y, Wang R, Barot S, Weiss DS. Stoichiometry of a recombinant GABAA receptor. J Neurosci. 1996;16:5415–5424. doi: 10.1523/JNEUROSCI.16-17-05415.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y, Weiss DS. Substitutions of the highly conserved M2 leucine create spontaneously opening ρ1 γ-aminobutyric acid receptors. Mol Pharmacol. 1998;53:511–523. doi: 10.1124/mol.53.3.511. [DOI] [PubMed] [Google Scholar]
- Chang Y, Weiss D. Functional domains of GABA receptors. In: Martin DL, Olsen RW, editors. GABA in the Nervous System: the View at Fifty Years. Philadelphia: Lippincott, Williams & Wilkins; 2000. pp. 127–140. [Google Scholar]
- Chang Y, Weiss DS. Site specific fluorescence reveals distinct structural changes with GABA receptor activation and antagonism. Nat Neurosci. 2002;5:1163–1168. doi: 10.1038/nn926. [DOI] [PubMed] [Google Scholar]
- Constanti A. The ‘mixed’ effect of picrotoxin on the GABA dose/conductance relation recorded from lobster muscle. Neuropharmacol. 1978;17:159–167. doi: 10.1016/0028-3908(78)90095-3. [DOI] [PubMed] [Google Scholar]
- Davidoff RA, Aprison MH. Picrotoxin antagonism of the inhibition of interneurons by glycine. Life Sci. 1969;8:107–112. doi: 10.1016/0024-3205(69)90299-9. [DOI] [PubMed] [Google Scholar]
- Dibas MI, Gonzales EB, Das P, Bell-Horner CL, Dillon GH. Identification of a novel residue within the second transmembrane domain that confers use-facilitated block by picrotoxin in glycine α1 receptors. J Biol Chem. 2002;277:9112–9117. doi: 10.1074/jbc.M111356200. [DOI] [PubMed] [Google Scholar]
- Engberg I, Thaller A. On the interaction of picrotoxin with GABA and glycine in the spinal cord. Brain Res. 1970;19:151–154. doi: 10.1016/0006-8993(70)90244-1. [DOI] [PubMed] [Google Scholar]
- Galindo A. GABA–picrotoxin interaction in the mammalian central nervous system. Brain Res. 1969;14:763–767. doi: 10.1016/0006-8993(69)90220-0. [DOI] [PubMed] [Google Scholar]
- Goutman JD, Calvo DJ. Studies on the mechanisms of action of picrotoxin, quercetin and pregnanolone at the GABA ρ1 receptor. Br J Pharmacol. 2004;141:717–727. doi: 10.1038/sj.bjp.0705657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurley D, Amin J, Ross PC, Weiss DS, White G. Point mutations in the M2 region of the α, β, or γ subunit of the GABAA channel that abolish block by picrotoxin. Receptors Channels. 1995;3:13–20. [PubMed] [Google Scholar]
- Hawthorne R, Lynch JW. A picrotoxin-specific conformational change in the glycine receptor M2–M3 loop. J Biol Chem. 2005;280:35836–35843. doi: 10.1074/jbc.M506645200. [DOI] [PubMed] [Google Scholar]
- Hill RG, Simmonds MA, Straughan DW. Antagonism of GABA by picrotoxin in the feline cerebral cortex. Br J Pharmacol. 1972;44:807–809. doi: 10.1111/j.1476-5381.1972.tb07319.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue M, Akaike N. Blockade of γ-aminobutyric acid-gated chloride current in frog sensory neurons by picrotoxin. Neurosci Res. 1988;5:380–394. doi: 10.1016/0168-0102(88)90024-7. [DOI] [PubMed] [Google Scholar]
- Kammann M, Laufs J, Schell J, Gronenborn B. Rapid insertional mutagenesis of DNA by polymerase chain reaction (PCR) Nucl Acid Res. 1989;12:4445–4452. doi: 10.1093/nar/17.13.5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liman ER, Tytgat J, Hess P. Subunit stoichiometry of a mammalian K+ channel determined by construction of multimeric cDNAs. Neuron. 1992;9:861–871. doi: 10.1016/0896-6273(92)90239-a. [DOI] [PubMed] [Google Scholar]
- Lynch JW, Rajendra S, Barry PH, Schofield PR. Mutations affecting the glycine receptor agonist transduction mechanism convert the competitive antagonist, picrotoxin, into an allosteric potentiator. J Biol Chem. 1995;270:13799–13806. doi: 10.1074/jbc.270.23.13799. [DOI] [PubMed] [Google Scholar]
- Miyazawa A, Fujiyoshi Y, Unwin N. Structure and gating mechanism of the acetylcholine receptor pore. Nature. 2003;423:949–955. doi: 10.1038/nature01748. [DOI] [PubMed] [Google Scholar]
- Neter J, Kutner MH, Nachtsheim CJ, Wasserman W. Applied Linear Statistical Models. Chicago: Irwin; 1996. [Google Scholar]
- Newland C, Cull-Candy S. On the mechanism of action of picrotoxin on GABA receptor channels in dissociated sympathetic neurones of the rat. J Physiol. 1992;447:191–213. doi: 10.1113/jphysiol.1992.sp018998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pribilla I, Takagi T, Langosch D, Bormann J, Betz H. The atypical M2 segment of the β subunit confers picrotoxinin resistance to inhibitory glycine receptor channels. EMBO J. 1992;11:4305–4311. doi: 10.1002/j.1460-2075.1992.tb05529.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan Q, Haddrill JL, Lynch JW. A single β subunit M2 domain residue controls the picrotoxin sensitivity of αβ heteromeric glycine receptor chloride channels. J Neurochem. 2001;76:1109–1120. doi: 10.1046/j.1471-4159.2001.00124.x. [DOI] [PubMed] [Google Scholar]
- Smart TG, Constanti A. Studies on the mechanism of action of picrotoxin and other convulsants at the crustacean muscle GABA receptor. Proc Roy Soc Lond Series B Biol Sci. 1986;227:191–216. doi: 10.1098/rspb.1986.0019. [DOI] [PubMed] [Google Scholar]
- Tretter T, Ehya N, Fuchs K, Sieghart W. Stoichiometry and assembly of a recombinant GABAA receptor subtype. J Neurosci. 1997;17:2728–2737. doi: 10.1523/JNEUROSCI.17-08-02728.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Covey DF, Akabas MH. Interaction of picrotoxin with GABAA receptor channel-lining residues probed in cysteine mutants. Biophys J. 1995;69:1858–1867. doi: 10.1016/S0006-3495(95)80056-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Pan Z-H, Brideau AD, Lipton SA. Cloning of a γ-aminobutyric acid type C receptor subunit in rat retina with a methionine residue criticial for picrotoxinin channel block. Proc Natl Acad Sci U S A. 1995;92:11756–11760. doi: 10.1073/pnas.92.25.11756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhorov BS, Bregestovski PD. Chloride channels of glycine and GABA receptors with blockers: Monte Carlo minimization and structure-activity relationships. Biophys J. 2000;78:1786–1803. doi: 10.1016/S0006-3495(00)76729-4. [DOI] [PMC free article] [PubMed] [Google Scholar]