Given the overwhelming complexity and difficulty of the origin of life problem, the most astonishing thing about it is that life actually has evolved on at least one planet in our universe (1). Indeed, it is entirely conceivable that the origin of life involved a series of highly unlikely events, and a substantial part of the explanation for why there is life on earth comes from the anthropic principle (2), i.e., our planet just happens to be one of the extremely rare parts of the universe where such a series of events was realized (3). The anthropic world view, however, by no means frees the students of early evolution from the obligation to explore all possible ways to decrease the improbability of life by demonstrating plausible paths to one or another of the milestones that need to be reached before life actually takes off. The paper by Baaske et al. (4) in this issue of PNAS seems to do just that by describing a simple abiotic system ensuring striking concentration of mono- and polynucleotides in inorganic compartments that might be suitable hatcheries for life.
Spatial compartmentalization is an essential condition for the functioning of biological systems. In modern cells, compartmentalization is provided by an elaborate, highly evolved system of membranes that serve to maintain the integrity of the cell and, at the same time, provide effective means for communication with the extracellular space via diverse transport and signaling systems. For prebiological reactions to produce the minimal complexity required for the origin of first life forms, effective abiogenic compartmentalization is a must, given that high concentrations of organic molecules in the entire primordial ocean are unrealistic, as shown by geochemical extrapolations (5, 6). Russell and coworkers (7–9) have extensively developed the scenario under which networks of inorganic compartments formed, primarily, of iron sulfide and existing in the vicinity of hydrothermal vents constitute a plausible, and perhaps the most likely, cradle of life. Indeed, such formations, which exist at modern vents (10) and have been recovered from fossils of various ages as well (11), provide not only compartments with cell-like dimensions but also the dissipative environment required for the origin of complex structures in the form of thermal and electrochemical gradients and versatile inorganic catalysts (FeS and NiS) for prebiological reactions (9, 12). Moreover, the notion of the early stages of the evolution of life occurring in an abiogenic compartmentalized environment is compatible with the apparent nonhomology of the membranes and the respective biosynthetic machinery in the two prokaryotic domains of life, bacteria and archaea, leading to the proposal that the last universal cellular ancestor (LUCA) was a noncellular, although compartmentalized, entity (13).
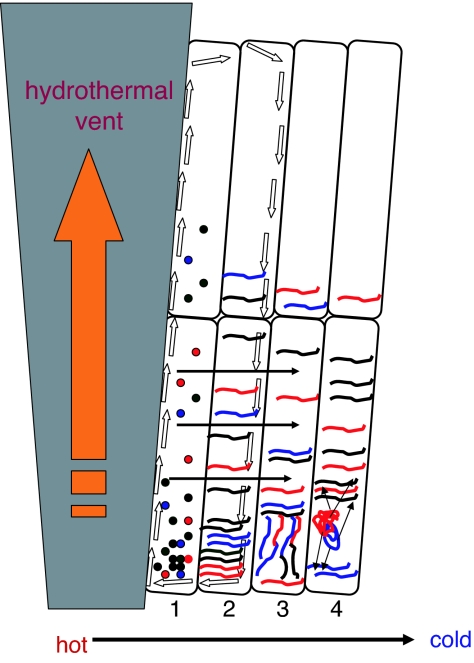
Baaske et al. (4) provide a crucial boost to these scenarios of early evolution by demonstrating in simulation studies that the temperature gradients existing at hydrothermal vents turn the network of pores into a surprisingly effective concentrating device. Two concomitant hydrodynamic processes, namely, thermal convection and thermophoresis along the temperature gradient, are shown to result in a >1,000-fold accumulation of mononucleotides near the bottom of a plugged pore (Fig. 1). Concatenation of pores into networks and/or an increase of the size of the molecules contained in them lead to a dramatic increase in the extent of accumulation such that molar concentrations of ≈100-base RNA or DNA would be readily reached. The concentration effect is shown to be particularly pronounced in long, narrow, vertical clefts (pores with a high aspect ratio), but concatenation of low-aspect-ratio pores into a network is also predicted to yield extremely high concentrations of mono- and polynucleotides.
Fig. 1.
Evolution of an RNA population in a network of inorganic compartments. Open arrows show thermoconvection, and horizontal filled arrows show thermophoresis. Compartment 1, accumulation of mononucleotides; compartment 2, accumulation of abiogenically synthesized RNA molecules; compartment 3, exploration of the RNA sequence space by ligation and recombination of RNA molecules; and compartment 4, emergence of the RNA world. The putative ribozyme replicase is denoted by a “globular” RNA molecule, possibly emerging by the ligation–recombination process. The stack of compartments depicts a contemporaneous, three-dimensional network. However, within the compartments, putative successive stages of evolution are shown, in the direction from the inside (near the vent) to the outside of the network.
This study not only describes a concentration mechanism for abiogenically emerging small molecules and, especially, polymers under plausible conditions of prebiological evolution but also suggests the possibility that networks of inorganic compartments at hydrothermal vents could be veritable “reactors” for RNA synthesis. Indeed, at high concentrations of nucleotides, the equilibrium would be shifted from hydrolysis to polymerization, and the rate of the reaction could be made realistic through the combined effect of the thermal gradient and inorganic catalysis (14, 15). Moreover, the even more efficient accumulation of polynucleotides provides for the particularly intriguing possibility of ligation and recombination of these molecules, reactions that are known to be quite efficiently catalyzed by ribozymes (16) and that would occur spontaneously, given the predicted high RNA concentrations.
Thus, the putative reactors at hydrothermal vents would provide the exact substrate that is needed for the evolution of a primordial RNA world, namely, a dense population of versatile RNA molecules, some of which would possess weak catalytic activities. This system seems to be the best imaginable abiogenic setting for the emergence of ribozyme RNA replicases and other ribozyme activities that are required for the onset of an evolving RNA world (17) (Fig. 1). Furthermore, the compartments would be expected to also accumulate other abiogenic organic molecules, in particular, amino acids, thus resulting in conditions conducive to the evolution of translation and the transition from the RNA world to the RNA–protein world.
Of course, the results of Baaske et al. (4) by no means put away all of the severe difficulties associated with the origin of life. In particular, at the earliest stages of biogenesis, the formation of mononucleotides, in the first place, remains problematic, and when it comes to the more advanced stages, a ribozyme replicase still is a hypothetical entity (18), and the evolutionary path to the translation systems remains essentially uncharted (19). Nevertheless, the intermediate stage, the transition from a solution of small organic molecules to a population of RNAs, now appears much less mysterious than before. Moreover, the hard combinatorial search for the extremely rare RNA sequences capable of catalyzing complex reactions, such as RNA replication, would be substantially facilitated in this setting through ligation of RNA molecules. Best of all, perhaps, the model of Baaske et al. suggests a straightforward experimental design, and such experiments, if successful, could bring us closer to an actual laboratory reproduction of the origin of life than anything done previously.
Footnotes
The author declares no conflict of interest.
See companion article on page 9346.
References
- 1.Davies P. The Fifth Miracle: The Search for the Origin and Meaning of Life. New York: Simon & Schuster; 2000. [Google Scholar]
- 2.Livio M, Rees MJ. Science. 2005;309:1022–1023. doi: 10.1126/science.1111446. [DOI] [PubMed] [Google Scholar]
- 3.Koonin EV. 2007. arXiv: q-bio.PE/0701023.
- 4.Baaske P, Weinert FM, Duhr S, Lemke KH, Russell MJ, Braun D. Proc Natl Acad Sci USA. 2007;104:9346–9351. doi: 10.1073/pnas.0609592104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dose K. BioSystems. 1975;6:224–228. doi: 10.1016/0303-2647(75)90064-7. [DOI] [PubMed] [Google Scholar]
- 6.Mojzsis SJ, Harrison TM, Pidgeon RT. Nature. 2001;409:178–181. doi: 10.1038/35051557. [DOI] [PubMed] [Google Scholar]
- 7.Russell MJ, Daniel RM, Hall AJ, Sherringham J. J Mol Evol. 1994;39:231–243. [Google Scholar]
- 8.Russell MJ, Hall AJ, Cairns-Smith AG, Braterman PS. Nature. 1988;336:117. (lett) [Google Scholar]
- 9.Russell MJ, Hall AJ. J Geol Soc London. 1997;154:377–402. doi: 10.1144/gsjgs.154.3.0377. [DOI] [PubMed] [Google Scholar]
- 10.Kelley DS, Karson JA, Blackman DK, Fruh-Green GL, Butterfield DA, Lilley MD, Olson EJ, Schrenk MO, Roe KK, Lebon GT, Rivizzigno P. Nature. 2001;412:145–149. doi: 10.1038/35084000. [DOI] [PubMed] [Google Scholar]
- 11.Shock EL. Orig Life Evol Biosph. 1992;22:67–107. 191–242. doi: 10.1007/BF01808019. [DOI] [PubMed] [Google Scholar]
- 12.Martin W, Russell MJ. Philos Trans R Soc London B. 2003;358:59–83. doi: 10.1098/rstb.2002.1183. discussion 83–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koonin EV, Martin W. Trends Genet. 2005;21:647–654. doi: 10.1016/j.tig.2005.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ogasawara H, Yoshida A, Imai E, Honda H, Hatori K, Matsuno K. Orig Life Evol Biosph. 2000;30:519–526. doi: 10.1023/a:1026539708173. [DOI] [PubMed] [Google Scholar]
- 15.Ogata Y, Imai E, Honda H, Hatori K, Matsuno K. Orig Life Evol Biosph. 2000;30:527–537. doi: 10.1023/a:1026543825011. [DOI] [PubMed] [Google Scholar]
- 16.Doudna JA, Cech TR. Nature. 2002;418:222–228. doi: 10.1038/418222a. [DOI] [PubMed] [Google Scholar]
- 17.Gesteland RF, Cech TR, Atkins JF. The RNA World. 3rd Ed. Woodbury, NY: Cold Spring Harbor Lab Press; 2006. [Google Scholar]
- 18.Joyce GF. Science. 2007;315:1507–1508. doi: 10.1126/science.1140736. [DOI] [PubMed] [Google Scholar]
- 19.Penny D. Philos Biol. 2005;20:633–671. [Google Scholar]