Abstract
The humanized monoclonal antibody HzKR127 recognizes the preS1 domain of the human hepatitis B virus surface proteins with a broadly neutralizing activity in vivo. We present the crystal structures of HzKR127 Fab and its complex with a major epitope peptide. In the complex structure, the bound peptide forms a type IV β-turn followed by 310 helical turn, the looped-out conformation of which provides a structural basis for broad neutralization. Upon peptide binding, the antibody undergoes a dramatic complementarity determining region H3 lid opening. To understand the structural implication of the virus neutralization, we carried out comprehensive alanine-scanning mutagenesis of all complementarity determining region residues in HzKR127 Fab. The functional mapping of the antigen-combining site demonstrates the specific roles of major binding determinants in antigen binding, contributing to the rational design for maximal humanization and affinity maturation of the antibody.
Keywords: alanine-scanning mutagenesis, crystal structure, humanized antibody, virus neutralization
Human hepatitis B virus (HBV) is a small enveloped DNA virus that causes acute and chronic hepatitis in humans (1–3). There are approximately 400 million carriers of HBV worldwide, posing a serious health threat. The current HBV vaccines have several limitations, including nonresponsiveness, escape mutants, and low immunogenicity (4). Recently, the potential of immunoprophylaxis for virus prevention has been increased by the discovery of monoclonal antibodies that have broad specificity toward various isolates of infectious viruses (5, 6).
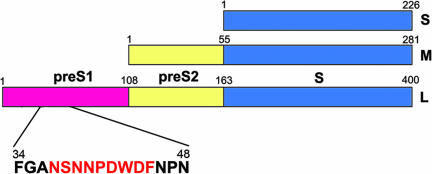
The HBV envelope consists of three surface glycoproteins called the large (L), middle (M), and small (S) proteins. All of these proteins are translated from a single ORF comprising preS1, preS2, and S domains (7). Translation of S, preS2+S, and preS1+preS2+S domains results in S, M, and L proteins, respectively (Fig. 1). The L protein, preferentially localized in infectious HBV particles, plays an essential role in viral infection (7, 8), and its preS1 domain is responsible for cell attachment in viral infection (9–12). In particular, the residue 21–47 region of preS1 has been suggested to contain a specific binding site for hepatocyte receptors and also has been shown to carry virus-neutralizing epitopes (10–13).
Fig. 1.
Domains of HBV surface proteins. The domain structures of S, M, and L proteins are presented with each domain colored differently. The adr subtype preS1 includes an epitope that is recognized by the neutralizing monoclonal antibody HzKR127. The peptide sequence of the epitope region is presented below the preS1 domain, with the epitope colored red.
In the blood of HBV-infected persons, the noninfectious subviral particles with M and S proteins are present in excess amounts over the infectious virus particles containing L protein (7, 8). Thus, neutralizing anti-preS1 antibodies are thought to have advantages over the neutralizing antibodies targeting S protein, which are currently undergoing clinical trials (4). Previously, we generated an anti-preS1 murine antibody KR127 that recognizes adr subtype preS1 (residues 37–45) (15). KR127 and the humanized version of KR127 antibody (HzKR127) bind to the preS1 domain of various clinical HBV isolates, including both adr and ayw subtypes, suggesting their broad neutralizing activity (15, 16). HzKR127 also has been shown to exhibit in vivo neutralizing activity in chimpanzee and to protect the chimpanzee from HBV infection (16), indicating the high potential of the antibody for the immunoprophylaxis of HBV infection and therapy of hepatitis.
An understanding of how the broadly neutralizing antibodies recognize antigens would facilitate design of vaccines by providing information regarding essential antigenic features. In particular, the antigen–antibody complex structure could provide direct information about the native antigen structure. The structural information could lead to the design of structurally constrained peptide antigens for the development of efficient vaccines (17). In HIV-1, the broadly neutralizing antibody b12 binds to the conformationally invariant and functionally conserved CD4-binding site on gp120 (18, 19). The extensive interaction of b12 complementarity determining region (CDR) heavy-chain loops with the CD4-binding loop of gp120 contribute to the high-affinity binding that is required for the effective neutralization of primary HIV-1 isolates (19). The long CDR H3 of another HIV-1-neutralizing antibody, 4E10, has been proposed to contact the viral membrane, which results in effective neutralization (20).
To shed light on the antigen recognition mechanism of the broadly neutralizing antibody of HBV, we determined the crystal structures of antibody HzKR127 in its free and antigen peptide-bound forms. The looped-out conformation of the bound peptide provides a structural basis for broad specificity of the antibody against various clinical HBV isolates. Structural comparison between the free and bound forms reveals a mechanism of lid opening in antibody–antigen recognition. To assess their relative importance for antigen binding, we performed extensive alanine-replacement scan of all CDR residues in HzKR127. The mapping of the functional paratope of HzKR127 would contribute to the generation of improved anti-preS1 humanized antibodies.
Results
Structure Determination and Overall Structure.
The crystal structures of HzKR127 Fab in its free and preS1 peptide-bound forms were determined by the molecular replacement method and refined to 2.5- and 2.6-Å resolutions, respectively (Table 1). In the Fab–peptide complex structure, the clearly defined density was observed for the bound peptide (residues 36–45, 1P–10P) [supporting information (SI) Fig. 5A]. The HzKR127 Fab has the canonical β sandwich Ig fold with four domains: VL, CL, VH, and CH1 (where L indicates light chain; H, heavy chain; V, variable domain; C, constant domain). The elbow angle of the HzKR127 Fab is 153° that falls within the range of 127 to 225° commonly observed for Fab molecules. The CDRs L1, L2, and L3 conform to the canonical conformations belonging to the classifications κL1-3, L2-1, and κL3-1 (21), respectively. The CDRs H1 and H2 belong to canonical classes H1-1 and H2-2 (21), respectively. The short CDR H3 (six residues in length) is folded inside the antigen-binding site that exhibits a large conformational change upon the peptide binding (see below).
Table 1.
Crystallographic and refinement statistics
| Data set | Fab | Fab/epitope complex |
|---|---|---|
| Space group | C2 | C2 |
| Cell constant | ||
| a, Å | 100.12 | 95.34 |
| b, Å | 61.11 | 61.27 |
| c, Å | 77.53 | 83.54 |
| β, ° | 91.49 | 93.25 |
| Resolution, Å | 2.50 (2.64–2.50) | 2.60 (2.74–2.60) |
| Total observation | 44,196 (6,424) | 52,874 (7,721) |
| Unique reflections | 15,758 (2,343) | 14,611 (2,070) |
| Completeness, % | 96.3 (97.9) | 97.5 (96.1) |
| Rmerge,† % | 7.8 (29.4) | 7.5 (28.0) |
| Multiplicity | 2.8 (2.7) | 3.6 (3.7) |
| Average I/σ(I) | 5.9 (2.4) | 6.8 (2.1) |
| Rcryst‡ | 20.7 (31.7) | 19.3 (30.8) |
| Rfree§ | 25.2 (38.0) | 25.8 (40.7) |
| rmsd bond lengths, Å | 0.006 | 0.006 |
| rmsd bond angles, ° | 1.320 | 1.348 |
Values in parentheses are for the highest resolution bin.
†Rmerge = Σ∣Ii − 〈Ii〉∣/Σ∣〈I〉∣, where I is the intensity for the ith measurement of an equivalent reflection with the indices h, k, l.
‡Rcryst = Σ∣Fo − Fc∣/Σ Fo, where Fo and Fc are the observed and calculated structure factor amplitudes, respectively.
§The Rfree value was calculated from 5% of all data that were not used in the refinement.
The structures of constant domains (CL and CH1) are almost the same for both the free and peptide-bound Fabs. The peptide binds to the interface between the light- and heavy-chain variable domains (VL and VH) with the intermolecular interface of 697 Å2 (SI Fig. 5B). The bound peptide is surrounded by light-chain CDRs L1–L3 and heavy-chain CDRs H1–H3 (SI Fig. 5B). The L3 and H3 loops have the closest contacts with the preS1 peptide, providing the most significant contributions to the peptide binding. The L1, H1, and H2 loops form a few interactions with the peptide. The H2 loop is situated distal to the binding pocket behind the H1 loop, maintaining the configuration of the H1 loop adequate for preS1 peptide binding. On the other hand, the L2 loop has no direct interaction with the preS1 peptide.
Peptide Conformation.
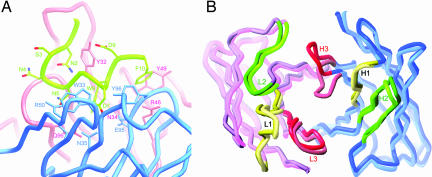
The preS1 peptide comprises a sequence of Ala–Asn–Ser–Asn–Asn–Pro–Asp–Trp–Asp–Phe–Asn (1P–11P, residues 36–46 in the preS1 sequence). In the complex structure, the bound preS1 peptide exhibits two consecutive turns (Fig. 2A). Secondary structure analysis by the program PROMOTIF (22) indicated that residues 2P–5P of the bound peptide form a type IV β turn followed by a 310 helical turn (residues 5P–9P). In the type IV β turn, main-chain φ and ψ angles are −105.3°, −14.3° and −73.1°, −31.4° for the i + 1 (Ser3P) and i + 2 (Asn4P) residues, respectively. The running direction of the backbone is kinked around Asn5P. A total of seven intrapeptide hydrogen bonds stabilize the double-turn conformation. The first turn involves four hydrogen bonds between the side-chain atoms (OD1 and ND2) of Asn2P and the main-chain atoms of Asn4P and Asn5P (SI Table 3). The second turn is maintained by three hydrogen bonds formed between the backbone carbonyl oxygen atoms of Asn5P and Pro6P and the backbone amide nitrogen atoms of Trp8P and Asp9P. The first turn of the peptide is situated proximal to the L3 loop, whereas its second turn has the closest contact with the H3 loop (SI Fig. 5B).
Fig. 2.
Crystal structure of the HzKR127 Fab–preS1 peptide complex. (A) Conformation of the preS1 peptide bound to HzKR127 Fab. Note that the side chains of Ser3P and Asn4P point toward the outside of the binding pocket. The side chains of the Fab residues involved in major interactions with the peptide are drawn and labeled. The light, heavy, and peptide chains are shown in red, blue, and green, respectively. (B) H3 lid opening. The free and bound structures of HzKR127 Fab are superimposed. The backbone structure of the variable domains in the Fab is drawn as thick tubes, and the light and heavy chains are colored pink and blue, respectively. The free and bound forms are shown in lighter and darker colors, respectively. The CDR1, CDR2, and CDR3 loops are colored yellow, green, and red, respectively.
Alanine-scanning mutagenesis of residues 2P–10P of the preS1 epitope showed that mutations in residues 2P and 5P–10P caused a significant decrease in antibody binding, whereas mutations in Ser3P and Asp4P did not have any effect (16). In particular, mutations at Asp7P and Trp8P completely impaired the antibody binding (16). In the complex structure, the residues 5P–10P were placed in the binding pocket of the Fab with Asp7P and Trp8P, located deep inside the pocket (Fig. 2A), explaining their mutational effects. In contrast, the side chains of Ser3P and Asn4P point toward the outside of the binding pocket (Fig. 2A), consistent with no mutational effect of Ser3P and Asp4P on antibody binding.
Conformational Changes upon Peptide Binding.
The superposition of the free and complexed Fab structures reveals conformational changes triggered by the preS1 peptide binding (Fig. 2B). The most noticeable conformational change between the free and complexed structures occurs at the H3 loop (Fig. 2B). The residues Glu95H–Tyr102H of the H3 loop are found to adopt entirely different conformation between the two forms (Fig. 3). The largest atomic displacement in the main chain occurs in Asp97H having a 9.6 Å difference between the two structures. Tyr96H and Glu98H show deviations of 7.7 and 8.0 Å in the main chain, respectively. The side-chain orientation also differs substantially in the free and complexed forms. The side chain of Glu98H rotates from one side of the H3 loop to the other side, showing the largest movement at 15.1 Å. The aromatic side chains of Tyr96H and Tyr102H also flip to the opposite direction, with 7.3 and 6.5 Å shifts, respectively.
Fig. 3.
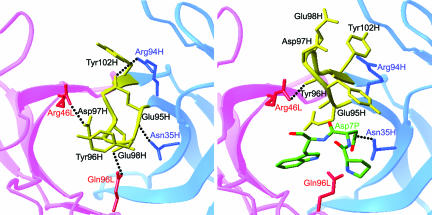
Comparison of antigen-binding site in the free and bound HzKR127 Fabs. Interactions between the H3 lid (yellow) and neighbors in the free (Left) and bound (Right) HzKR127 Fabs. The residues involved in the interactions are drawn, and the major interactions are presented as dotted lines. The light and heavy chains are colored pink and blue (labeled red and blue), respectively. The bound peptide residues Pro6P–Trp8P are presented as sticks with atomic colors.
The CDR H3 lid opening significantly changed the shape of the antigen-binding site. In the free form, no prominent binding groove was present on the surface of the antigen-binding site. The distinct groove connected to a deep binding pocket of HzKR127 became fully exposed only by the CDR H3 lid opening. The prominent groove, with a length of 14 Å, width of 11 Å, and depth of 10 Å, was capable of accommodating the two turns of the bound peptide. The most striking change in the shape of the binding site was the creation of a binding pocket for Trp8P, where the aromatic ring of Trp8P snugly fit into the deep pocket, leading to tight locking-in of the peptide.
The CDR H3 lid opening also resulted in a considerable reconstitution of hydrogen bond network around the antigen-binding site (Fig. 3). In the free structure, the overall H3 loop conformation was maintained by hydrogen bonds between the H3 loop and the rest of the Fab. Upon the preS1 peptide binding, most of these hydrogen bonds were disrupted and a new network of interactions was formed.
Functional Mapping of the Antigen-Combining Site.
To evaluate the relative importance of individual CDR residues in antigen binding, we performed alanine-scanning mutagenesis of all of the residues in the CDRs of HzKR127 Fab (SI Tables 4 and 5) and determined the affinity of each alanine-mutated antibody (SI Fig. 6). The ELISAs on the 50 alanine-replacement mutants, 21 from heavy chain and 29 from light chain, identified 16 hot spots, 7 warm spots, 18 null spots, and 2 affinity-enhanced spots. First, the 16 hot spots (Trp33H, Met34H, Asn35H in CDR H1, Arg50H in CDR H2, Glu95H, Tyr96H in CDR H3, Leu27bL, Ser27eL, Asn28L, Lys30L, Tyr32L, Asn34L in CDR L1, and Gln90L, Gly91L, Phe94L, Pro95L in CDR L3) completely lost the antigen-binding activity (>900-fold decrease in affinity) upon alanine mutation. Second, the 7 warm spots (Tyr52H, Pro52aH in CDR H2, Tyr-27 dl in CDR L1, Leu50L, Asp55L in CDR L2, and Val89L, Gln96L in CDR L3) showed the reduced affinity by >10-fold upon mutation. Third, the 18 null spots (Ser31H, Ser32H in CDR H1, Ile51H, Gly53H, Asp54H, Gly55H, Asp56H, Thr57H, Asn58H in CDR H2, Ser26L, Gln27L, Gly29L, Thr31L in CDR L1, Ser52L, Lys53L, Leu54L, Ser56L in CDR L2, and Thr97L in CDR L3) showed no significant effect on the affinity (<2-fold decrease or slight increase in affinity) upon mutation. Lastly, alanine mutations of Asp97H or both Asp97H and Tyr102H in CDR H3 resulted in significant (>10-fold) increase in affinity.
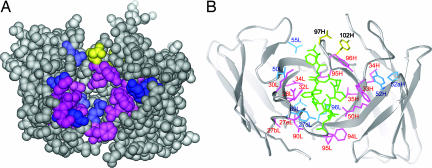
The mapping of the hot spots, warm spots, and affinity-enhanced spots onto the antigen-bound structure of HzKR127 Fab shows the overall distribution of major binding determinants (Fig. 4). The hot spots (colored pink in Fig. 4) mainly surround the binding groove, comprising the antigen-combining site, and thereby most of them involve direct contacts with the preS1 peptide. On the other hand, most warm spots (colored blue in Fig. 4) are situated more remote from the center of the binding site. The null spots are mainly distributed in the CDRs L1, L2, and H2, forming no direct interaction with the peptide. The presence of affinity-enhanced spots (colored yellow in Fig. 4) is restricted to CDR H3, and the spots are not involved in intimate contact with the peptide antigen.
Fig. 4.
Mapping of functionally important residues on the crystal structure of preS1 peptide-bound HzKR127 Fab. (A) Space-filling representation of the interface of the Fab. The variable domain in the Fab is shown in gray. The hot spots, warm spots, and affinity-enhanced spots are colored as follows: pink (>900-fold decreased affinity relative to WT; ΔΔGD, >4 kcal/mol), blue (10- to 900-fold decreased affinity; ΔΔGD, 1.4–4 kcal/mol), and yellow (>10-fold increased affinity; ΔΔGD, less than −1.4 kcal/mol). Tyr102H (labeled in 102H) is colored yellow because the double-alanine mutation of both Asp97H and Tyr102H increased the affinity by 25-fold. (B) Ribbon representation of the HzKR127 Fab bound to preS1 peptide. The color coding is the same as in A. The bound preS1 peptide is drawn as green sticks. Among the hot spots, Gly91L is not displayed because of the absence of its side chain.
The comparison of antigen-binding interaction profile of HzKR127 (Fig. 4A) with other antibodies reveals the distinct characteristics of HzKR127. First, HzKR127 has a very large number of hot spots (16) of ΔGmutant − ΔGWT (ΔΔGD) of >4 kcal/mol, where ΔG indicates free energy change. Thus, formation of HzKR127–preS1 complex is stabilized by the accumulation of many interactions throughout the binding interface, as found in the A6–IFN-γ receptor complex (23) and the VEGF–Fab complex (24). This contrasts with the antibody D1.3–hen egg white lysozyme (HEL) complex (25) and the HyHEL-10–lysozyme complex (26), in which only a small subset of residues dominate the energetics of association (one and three hot spots of ΔΔGD of >4 kcal/mol, respectively). Second, major binding determinants are concentrated in light-chain CDRs (10 of 16 hot spots) (Fig. 4B). The extensive involvement of light-chain CDRs in direct contacts with antigen is contrary to what has been observed for most other peptide–antibody complexes. Generally, the heavy-chain CDRs make more extensive contacts than light-chain CDRs (27). Thus, the HzKR127–preS1 complex is among the few examples that are exceptional to the general trend.
Elucidation of Specific Roles of Major Binding Determinants.
On the basis of the functional mapping on the structure (Fig. 4B), we investigated specific roles of the 23 major binding determinants (16 hot spots and 7 warm spots). Among them, 12 major binding determinants are involved in direct contacts with the antigen (SI Table 3). Consistent with the structural features for the Fab-preS1 complex, alanine substitutions on most directly contacting residues completely abrogated (Trp33H, Asn35H, Arg50H, Glu95H, Tyr96H, Asn28L, Tyr32L, Asn34L, Gly91L, and Phe94L) or significantly impaired (Gln96L, 27-fold reduction; Val89L, 11-fold reduction) the antigen-binding activity (Table 2). Overall, most of the direct Fab–peptide interactions observed in the complex structure are critical for the Fab–peptide binding, exhibiting good correlation between the structure and antigen-binding activity.
Table 2.
Summary of mutagenesis in HzKR127
| Mutant | Kd, nM | KdMUT/KdWT |
|---|---|---|
| CDR H1 | ||
| W33A | >10,000 | >900 |
| M34A | >10,000 | >900 |
| N35A | >10,000 | >900 |
| CDR H2 | ||
| R50A | >10,000 | >900 |
| Y52A | 276.80 ± 23.60 | 25.16 |
| P52aA | 170.30 ± 5.32 | 15.48 |
| CDR H3 | ||
| E95A | >10,000 | >900 |
| Y96A | >10,000 | >900 |
| D97A | 0.57 ± 0.03 | 0.05 |
| D97A/Y102A | 0.44 ± 0.10 | 0.04 |
| CDR L1 | ||
| L27bA | >10,000 | >900 |
| Y27dA | 1032.70 ± 56.10 | 93.90 |
| S27eA | >10,000 | >900 |
| N28A | >10,000 | >900 |
| K30A | >10,000 | >900 |
| Y32A | >10,000 | >900 |
| N34A | >10,000 | >900 |
| CDR L2 | ||
| L50A | >10,000 | >900 |
| G91A | >10,000 | >900 |
| F94A | >10,000 | >900 |
| P95A | >10,000 | >900 |
| Q96A | 293.60 ± 7.13 | 26.70 |
| WT | 11.00 ± 1.66 | |
The dissociation equilibrium constants of HzKR127 mutants (KdMUT) were measured by competitive indirect ELISA and compared with that of HzKR127 WT (KdWT).
The other 11 major binding determinants seem to play a scaffolding role by precisely positioning the contacting residues for optimal interaction with the antigen or by maintaining the CDR loops in a binding-competent configuration. Without direct contact with the antigen, the six scaffolding residues (Met34H, Leu27bL, Ser27eL, Lys30L, Gln90L, and Pro95L) exhibited complete loss of the binding activity, and the five scaffolding residues (Tyr52H, Pro52aH, Tyr-27 dl, Leu50L, and Asp55L) showed 10- to 100-fold reductions in the affinity upon mutation (Table 2). For example, Gln90L stabilizes the canonical conformation of L3 loop by forming a hydrogen bond with the main-chain carbonyl group of Pro95L in CDR L3. In case of Tyr52H, the residue keeps, through hydrophobic interaction, the ring orientation of Trp33H in CDR H1 optimal for direct interaction with preS1 peptide (Fig. 4B). These observations illustrate the functional importance of the scaffolding role in antigen binding.
Discussion
The structures of the HzKR127 Fab and the preS1 peptide complex demonstrate how the antibody is very broadly neutralizing. The sequence of preS1 (residues 37–45) epitope is highly conserved among various different HBV isolates except residues 38 and 39 (15). On the complex structure, the side chains of all of the highly conserved preS1 residues are involved in direct contacts with the antibody. In comparison, the variable residues 38 and 39 point toward the outside of the binding pocket with few contacts with the antibody. This agrees well with the finding that the alanine substitutions on the positions do not affect the reactivity of the preS1 with HzKR127 (16).
So far, structural studies on broad neutralization mechanism of antivirus antibodies have focused mainly on anti-HIV antibodies. The complex structure of anti-HIV antibody 4E10 and its epitope showed why it is broadly neutralizing. The WFXI(T/S) motif of the epitope is highly conserved in all HIV-1 viruses, and variable residues near the epitope are situated on the opposite side of the helical epitope conformation with few contacts with the antibody (20). The ability of anti-HIV antibody 447-52D to bind many different gp120 V3 sequences stems from the mode of peptide–antibody recognition, where the peptide β hairpin forms a three-stranded mixed β sheet with CDR H3, with most of the V3 side chains exposed to solvent (28). Another broadly neutralizing anti-HIV antibody, b12, also interacts with the conserved CD4 epitope through extensive CDR heavy-chain contacts (18, 19). However, there is no information for the broad neutralizing mechanisms for HBV monoclonal antibodies. This study is the first case to uncover a structural basis for a broad neutralization mechanism of an anti-HBV antibody.
The three-dimensional structure of antigen-bound HzKR127 Fab and the comprehensive alanine-scanning mutagenesis of its antigen-combining site identified major binding determinants, including the most critical CDR residues in antigen-binding. Based on the significant structure–function correlation in antigen-combining site, we elucidated their specific roles for antigen binding. This knowledge is crucial for humanization of the antibody by specificity-determining residue grafting. The CDR-grafting, used for constructing HzKR127, is not enough for reducing the immunogenicity of a murine antibody to a minimum because murine CDRs could still elicit antivariable region responses (29, 30). Therefore, humanization of an antibody by grafting only specificity-determining residues onto a human antibody framework may further reduce the immunogenicity. In this process, the framework residues in contact with antigen (Arg46L and Tyr49L in FR2) and the scaffolding residues, which are hot spots but are not in contact with an antigen (Met34H, Leu27bL, Ser27eL, Lys30L, Gln90L, and Pro95L), should be retained.
Although HzKR127 neutralizes HBV in vivo, the humanized antibody of higher affinity would be more effective in clinical use. Structural inspection of affinity-enhanced spots offers a basis of rational design for further affinity maturation. It is noteworthy to find that alanine replacements of Asp97H or both Asp97H and Tyr102H (D97A/Y102A) increased the affinity by 20- and 25-fold, respectively (Table 2). How could the alanine mutations on Asp97H and Tyr102H increase the antigen-binding affinity? Both Asp97H and Tyr102H in CDR H3 are involved not in direct interaction with antigen but in the CDR H3 lid opening (Fig. 3). The Asp97H forms a salt bridge with Arg46L (FR2), which stabilizes the closing of the CDR H3 lid in the free structure (Fig. 3). Obviously, the mutation of Asp97H into alanine would disrupt the salt bridge seen in the structure, making the H3 loop more flexible. This may facilitate the structural transition from closed to open state. On the other hand, Tyr102H is located at the base of the CDR H3 lid, and its aromatic ring flips in the opposite direction upon antigen binding (Fig. 3). The small side chain of alanine is likely to reduce the steric clashes of lid opening more than the bulky rigid aromatic ring of tyrosine. We can therefore conclude that the alanine mutations in both the positions may increase the affinity by facilitating the CDR H3 lid opening, suggesting that the antigen-binding affinity could be further enhanced by modulating the flexibility of the CDR H3 lid.
The three-dimensional structure of HBV surface protein, including preS1, has not yet been determined, and the peptide structure observed in this study may provide an insight into the native structure of preS1 domain. In the complex structure, despite the characteristic secondary structure of the peptide, all Ramachandran angles of the peptide main chain lay on the most favored and additionally allowed regions, and no residue lies in generously allowed or disallowed regions (data not shown). The stable main-chain conformation and the extensive intrapeptide hydrogen bonds (SI Table 3) suggest that the observed peptide conformation may have correlation with its native structure in the surface of HBV.
In summary, we determined the free and preS1 peptide-bound structures of HzKR127 Fab exhibiting in vivo HBV-neutralizing activity. The structures reveal a lid-opening mechanism for antigen-binding, illustrating one of the largest conformational changes in antigen-antibody recognition. In the complex structure, only the conserved peptide residues form tight interaction with the Fab, whereas the varied residues are looped out from the binding pocket, which enables the antibody to recognize various virus isolates. The information regarding the broad neutralizing mechanism and the antigen recognition should contribute to the design of vaccines and the development of immunoprophylaxis strategies against HBV. The extensive alanine-scanning mutagenesis of CDRs identified major binding determinants in antigen binding, offering a basis for maximal humanization of the antibody. Affinity-enhanced mutations in CDR H3 suggest the possibility to further enhance the antigen-binding affinity by modulating the process of CDR H3 lid opening.
Materials and Methods
Preparation of HzKR127 Fab Fragment.
The cDNA encoding the VH and CH1 domains of HzKR127 was synthesized by PCR from humanized heavy-chain plasmid pHKR127HC (16) and cloned into the SalI–NotI sites of pRc/CMV (Invitrogen, Carlsbad, CA) to yield pCMV–HKR127–Fd. This plasmid and humanized light-chain plasmid pKC–dhfr–HKR127 (16) were cotransfected into dihydrofolate reductase-deficient CHO cell line (DG44) and stably transformed cell lines were selected, as previously described (16). The isolated cell line was adapted in nucleosides-minus MEMα medium (Gibco/BRL, Grand Island, NY) supplemented with 10% dialyzed FBS and 20 μM methothrexate (Sigma, St. Louis, MO), then transferred to Cell Factories (Nunc, Roskilde, Denmark) containing CHO-S-SFM II (Gibco/BRL) and 20 μM methothrexate. To purify the Fab, the culture supernatants were harvested, filtered through a 0.22-μm filter, and loaded onto a preS1–Sepharose affinity column, in which GST–preS1 (residues 1–56) (15) was conjugated to CNBr-activated Sepharose FF (Amersham Pharmacia, Piscataway, NJ). The bound HzKR127 Fab fragments were eluted by the addition of 0.2 M glycine–HCl, pH 2.7, followed by immediate neutralization with 1 M Tris–HCl, pH 8.0. The HzKR127 Fab-containing fractions were pooled and dialyzed against 10 mM Hepes–NaOH, pH 7.4. The purity of pooled fraction was analyzed by SDS/PAGE, followed by silver staining, and stored at −70°C until use.
Crystallization and Diffraction Data Collection.
All of the crystals of HzKR127 Fab were obtained at 18°C by the hanging drop vapor diffusion method. An aliquot (1.7 μl) of the protein (20 mg/ml) was mixed with an equal volume of a reservoir solution containing 17% PEG 4000, 10 mM Hepes–NaOH (pH 7.5), and 0.2 M ammonium sulfate. Crystals appeared 4 days after the initial setup, and they belonged to the space group C2 with unit cell dimensions of a = 100.12 Å, b = 61.11 Å, c = 77.53 Å, and β = 91.49°. For the crystallization of the complex, adr subtype HBV preS1 (residues 36–46) peptide (acetyl-ANSNNPDWDFN-amide) was used with a 10-fold molar excess of peptide over Fab. Crystals of the complex grew in the space group C2 with cell dimensions of a = 95.34 Å, b = 61.27 Å, c = 83.54 Å, and β = 93.25°. X-ray diffraction data were collected at room temperature by using a RU-300 rotating anode generator (40 kV and 100 mA; Rigaku, Tokyo, Japan) equipped with an R-AXIS IV++ detector (Rigaku) and an Osmic mirror (Osmic, Troy, MI). Data for phasing and refinement were processed with the programs MOSFLM (31) and CrystalClear (Rigaku).
Structure Determination and Refinement.
The structures of free HzKR127 Fab and the Fab–peptide complex were determined by molecular replacement by using the program AMoRe (32). For the structure determination of free Fab, the anti-hapten (5-para-nitrophenyl phosphonate-pentanoic acid) 48G7 Fab (PDB ID code 1AJ7) that showed the highest sequence homology with HzKR127 Fab was used as the search model. The structure of the Fab–peptide complex was determined by using the refined free form structure as the starting model. The programs O (33) and CNS (34) were used in the model building and refinement, respectively. During the refinement, the randomly selected 5% of data were set aside for the Rfree calculation. The free structure of 2.5 Å resolution was refined to an Rcryst of 20.7% and an Rfree of 25.2%, and the complex structure of 2.6 Å resolution was refined to an Rcryst of 19.3% and an Rfree of 25.8%. Crystallographic and refinement statistics are shown in Table 1. The Ramachandran plots drawn by the program PROCHECK (35) show that 98.6 and 98.9% of all residues in the free and complex structures fall within the most favored and additional allowed regions, respectively. Figures were drawn by using the programs RIBBONS (36) and BOBSCRIPT (37).
Construction of Alanine-Replacement Mutants of HzKR127.
All HzKR127 mutants were constructed by recombinant PCR from the humanized heavy-chain plasmid pHKR127HC (16) or humanized light-chain plasmid pHKR127KC (16) by using mutagenic primers, as described previously (15). The mutated constructs were introduced into Escherichia coli DH5αα and verified by nucleotide sequencing. Finally, the mutated VH or VL was subcloned into the EcoRI–ApaI or HindIII–XbaI sites, respectively, of pdCMV–dhfr–AKA (14) containing human Cγ1 and Cκ to construct heavy or light-chain expression plasmid. WT and mutant HzKR127 were transiently expressed in COS7 cells by cotransfection of the heavy and light-chain expression plasmids by using LipofectAMINE reagent (Life Technologies, Gaithersburg, MD) according to the protocol suggested by the supplier. The culture supernatants were harvested 2 days after transfections and stored at −70°C until use.
ELISA Analysis.
All ELISAs were performed at 37°C, as previously reported (16). To determine the Kd, competition ELISAs were used. The 5- to 10-ng aliquots of antibodies from the culture supernatants were preincubated with various amounts of purified GST–preS1 (residues 1–56) in 400 μl of PBS with Tween 20 that contained 0.2% BSA at 37°C for 3 h, and then mixtures were added to the wells that had been coated with 100 ng per well of GST–preS1 (residues 1–56). Plates were incubated for 30 min, followed by washing and detection.
Supplementary Material
Acknowledgments
This work was supported by the National Creative Research Initiative Program (Korea), the Korea Research Institute of Bioscience and Biotechnology Research Initiative Program, and Ministry of Health Welfare Grant A050260 (Korea).
Abbreviations
- CDR
complementarity determining region
- HBV
hepatitis B virus
- HzKR127
humanized KR127.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 2EH7 (free HzKR127 Fab) and 2EH8 (complexed HzKR127 Fab)].
This article contains supporting information online at www.pnas.org/cgi/content/full/0701279104/DC1.
References
- 1.Dreesman GR, Sanchez Y, Ionescu-Matiu I, Sparrow JT, Six HR, Peterson DL, Hollinger FB, Melnick JL. Nature. 1982;295:158–160. doi: 10.1038/295158a0. [DOI] [PubMed] [Google Scholar]
- 2.Ganem D, Varmus HE. Annu Rev Biochem. 1987;56:651–693. doi: 10.1146/annurev.bi.56.070187.003251. [DOI] [PubMed] [Google Scholar]
- 3.Rehermann B, Nascimbeni M. Nat Rev Immunol. 2005;5:215–229. doi: 10.1038/nri1573. [DOI] [PubMed] [Google Scholar]
- 4.Schodel F. In: Hepatitis B Virus. Koshy R, Caselmann WH, editors. London: Imperial College Press; 1998. pp. 219–250. [Google Scholar]
- 5.Burton DR, Desrosiers RC, Doms RW, Koff WC, Kwong PD, Moore JP, Nabel GJ, Sodroski J, Wilson IA, Wyatt RT. Nat Immunol. 2004;5:233–236. doi: 10.1038/ni0304-233. [DOI] [PubMed] [Google Scholar]
- 6.Ferrantelli F, Ruprecht RM. Curr Opin Immunol. 2002;14:495–502. doi: 10.1016/s0952-7915(02)00362-x. [DOI] [PubMed] [Google Scholar]
- 7.Heermann KH, Goldmann U, Schwartz W, Seyffarth T, Baumgarten H, Gerlich WH. J Virol. 1984;52:396–402. doi: 10.1128/jvi.52.2.396-402.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stibbe W, Gerlich WH. J Virol. 1983;46:626–628. doi: 10.1128/jvi.46.2.626-628.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neurath AR, Kent SB, Strick N, Parker K. Cell. 1986;46:429–436. doi: 10.1016/0092-8674(86)90663-x. [DOI] [PubMed] [Google Scholar]
- 10.Pontisso P, Ruvoletto MG, Gerlich WH, Heermann KH, Bardini R, Alberti A. Virology. 1989;173:522–530. doi: 10.1016/0042-6822(89)90564-3. [DOI] [PubMed] [Google Scholar]
- 11.Ryu CJ, Cho DY, Gripon P, Kim HS, Guguen-Guillouzo C, Hong HJ. J Virol. 2000;74:110–116. doi: 10.1128/jvi.74.1.110-116.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Falco S, Ruvoletto MG, Verdoliva A, Ruvo M, Raucci A, Marino M, Senatore S, Cassani G, Alberti A, Pontisso P, et al. J Biol Chem. 2001;276:36613–36623. doi: 10.1074/jbc.M102377200. [DOI] [PubMed] [Google Scholar]
- 13.Neurath AR, Seto B, Strick N. Vaccine. 1989;7:234–236. doi: 10.1016/0264-410x(89)90235-1. [DOI] [PubMed] [Google Scholar]
- 14.Yoon SO, Lee TS, Kim SJ, Jang MH, Kang YJ, Park JH, Kim KS, Lee HS, Ryu CJ, Gonzales NR, et al. J Biol Chem. 2006;281:6985–6992. doi: 10.1074/jbc.M511165200. [DOI] [PubMed] [Google Scholar]
- 15.Maeng CY, Ryu CJ, Gripon P, Guguen-Guillouzo C, Hong HJ. Virology. 2000;270:9–16. doi: 10.1006/viro.2000.0250. [DOI] [PubMed] [Google Scholar]
- 16.Hong HJ, Ryu CJ, Hur H, Kim S, Oh HK, Oh MS, Park SY. Virology. 2004;318:134–141. doi: 10.1016/j.virol.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 17.Wilson IA, Ghiara JB, Stanfield RL. Res Immunol. 1994;1:73–78. doi: 10.1016/s0923-2494(94)80049-9. [DOI] [PubMed] [Google Scholar]
- 18.Saphire EO, Parren PW, Pantophlet R, Zwick MB, Morris GM, Rudd PM, Dwek RA, Stanfield RL, Burton DR, Wilson IA. Science. 2001;293:1155–1159. doi: 10.1126/science.1061692. [DOI] [PubMed] [Google Scholar]
- 19.Zhou T, Xu L, Dey B, Hessell AJ, Van Ryk D, Xiang SH, Yang X, Zhang MY, Zwick MB, Arthos J, et al. Nature. 2007;445:732–737. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cardoso RM, Zwick MB, Stanfield RL, Kunert R, Binley JM, Katinger H, Burton DR, Wilson IA. Immunity. 2005;22:163–173. doi: 10.1016/j.immuni.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 21.Al-Lazikani B, Lesk AM, Chothia C. J Mol Biol. 1997;273:927–948. doi: 10.1006/jmbi.1997.1354. [DOI] [PubMed] [Google Scholar]
- 22.Hutchinson EG, Thornton JM. Protein Sci. 1996;5:212–220. doi: 10.1002/pro.5560050204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lang S, Xu J, Stuart F, Thomas RM, Vrijbloed JW, Robinson JA. Biochemistry. 2000;39:15674–15685. doi: 10.1021/bi000838z. [DOI] [PubMed] [Google Scholar]
- 24.Muller YA, Chen Y, Christinger HW, Li B, Cunningham BC, Lowman HB, de Vos AM. Structure (London) 1998;6:1153–1167. doi: 10.1016/s0969-2126(98)00116-6. [DOI] [PubMed] [Google Scholar]
- 25.Dall'Acqua W, Goldman ER, Lin W, Teng C, Tsuchiya D, Li H, Ysern X, Braden BC, Li Y, Smith-Gill SJ, et al. Biochemistry. 1998;37:7981–7991. doi: 10.1021/bi980148j. [DOI] [PubMed] [Google Scholar]
- 26.Pons J, Rajpal A, Kirsch JF. Protein Sci. 1999;8:958–968. doi: 10.1110/ps.8.5.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sundberg EJ, Mariuzza RA. Adv Protein Chem. 2003;61:119–160. doi: 10.1016/s0065-3233(02)61004-6. [DOI] [PubMed] [Google Scholar]
- 28.Stanfield RL, Gorny MK, Williams C, Zolla-Pazner S, Wilson IA. Structure (London) 2004;12:193–204. doi: 10.1016/j.str.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 29.Iwahashi M, Milenic DE, Padlan EA, Bei R, Schlom J, Kashmiri SV. Mol Immunol. 1999;36:1079–1091. doi: 10.1016/s0161-5890(99)00094-2. [DOI] [PubMed] [Google Scholar]
- 30.Schneider WP, Glaser SM, Kondas JA, Hakimi J. J Immunol. 1993;150:3086–3090. [PubMed] [Google Scholar]
- 31.Leslie AG. Acta Crystallogr D. 1999;55:1696–1702. doi: 10.1107/s090744499900846x. [DOI] [PubMed] [Google Scholar]
- 32.Navaza J. Acta Crystallogr A. 1994;50:157–163. [Google Scholar]
- 33.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 34.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 35.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 36.Carson M. Methods Enzymol. 1997;277:493–505. [PubMed] [Google Scholar]
- 37.Esnouf RM. J Mol Graphics Model. 1997;15:132–134. doi: 10.1016/S1093-3263(97)00021-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.