Abstract
In mammals, endothelial nitric oxide synthase (eNOS) has the weakest activity, being one-tenth and one-sixth as active as the inducible NOS (iNOS) and the neuronal NOS (nNOS), respectively. The basis for this weak activity is unclear. We hypothesized that a hinge element that connects the FMN module in the reductase domain but is shorter and of unique composition in eNOS may be involved. To test this hypothesis, we generated an eNOS chimera that contained the nNOS hinge and two mutants that either eliminated (P728IeNOS) or incorporated (I958PnNOS) a proline residue unique to the eNOS hinge. Incorporating the nNOS hinge into eNOS increased NO synthesis activity 4-fold, to an activity two-thirds that of nNOS. It also decreased uncoupled NADPH oxidation, increased the apparent KmO2 for NO synthesis, and caused a faster heme reduction. Eliminating the hinge proline had similar, but lesser, effects. Our findings reveal that the hinge is an important regulator and show that differences in its composition restrict the activity of eNOS relative to other NOS enzymes.
Keywords: electron flux, heme reduction, kox
Nitric oxide (NO) is a widespread signal molecule in biology (1, 2). Three nitric oxide synthases (NOSs) generate NO in mammals [inducible NOS (iNOS), neuronal NOS (nNOS), and endothelial NOS (eNOS)]. All three are comprised of an N-terminal heme-containing oxygenase domain, an intervening calmodulin (CaM) binding sequence, and a C-terminal reductase domain that contains FMN and FAD (3). The three NOS enzymes have different gene expression patterns, protein interactions, posttranslational modifications, and catalytic behaviors that enable specific roles in biology (4–8). Of note, the NO synthesis activity of eNOS is one-10th that of iNOS and one-sixth that of nNOS. This activity is associated with a slower heme reduction in eNOS (9, 10). Which protein features enable these differences, and why they evolved, are still unclear.
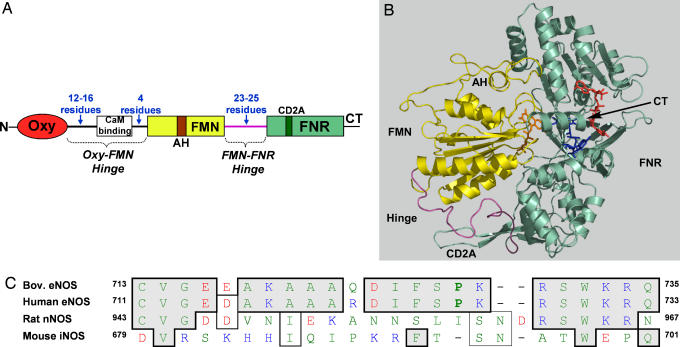
Studies of eNOS and nNOS chimeras established that their NO synthesis activities and heme reduction rates are primarily a function of the reductase domain (9, 11). For example, a chimera comprised of an eNOS oxygenase domain fused to an nNOS reductase domain had an NO synthesis activity and heme reduction rate that were similar to those of wild-type nNOS. This result implies that protein structural elements within the reductase domain are largely responsible for the different catalytic behaviors of eNOS and nNOS. While addressing this issue, we became interested in two hinge elements that connect the FMN subdomain of NOS to the rest of the enzyme (Fig. 1A). During catalysis, these hinge elements position the FMN subdomain to receive electrons from the NADPH–FAD module, and then may guide its interactions with the NOS oxygenase domain for electron transfer to the heme (12–17). In this way, the hinge elements could determine the rate of heme reduction and NO synthesis activity.
Fig. 1.
Hinge element connecting the FMN subdomain in NOS. (A) The two hinge regions (Oxy-FMN and FMN-FNR) that connect the FMN subdomain in NOS enzymes. (B) Structure and subdomain organization of the nNOS reductase domain. The 25-residue FMN–FNR hinge (purple) links the FMN domain (yellow) to the rest of the reductase domain. Cofactors: orange, FMN; blue, FAD; red, NADP. FNR, ferredoxin NADP-reductase; other abbreviations are as described in the text. (C) Alignment of FMN–FNR hinge region of various NOS enzymes.
The hinge that connects the FMN subdomain to the rest of the nNOS reductase domain is visible in the reductase crystal structure (12) (Fig. 1B) and has a varied composition among nNOS, eNOS, and iNOS (Fig. 1C). Specifically, the eNOS hinge is two residues shorter than in nNOS and contains a proline at position 728 in place of other residues found at this position in nNOS or iNOS. To examine whether these differences are responsible for the slow NO synthesis activity of eNOS, we characterized an eNOS mutant that contained the nNOS hinge element in place of its own, and characterized the complementary P728I eNOS and I958P nNOS point mutants (Fig. 1C). Our results establish a primary role for the hinge and reveal that its unique composition in eNOS represses NO synthesis activity.
Results
A flexible hinge that connects the FMN subdomain to the rest of the NOS reductase domain may help control electron transfer to the oxygenase domain heme during catalysis. If the eNOS hinge restricts this process relative to the nNOS hinge, then we may observe increased NO synthesis activity in an eNOS chimera that contains the nNOS hinge element (eNOS-nH), and we may also observe greater activity in P728I eNOS, and less activity in the corresponding I958P nNOS, if the proline residue that is present in the eNOS hinge is a restricting factor.
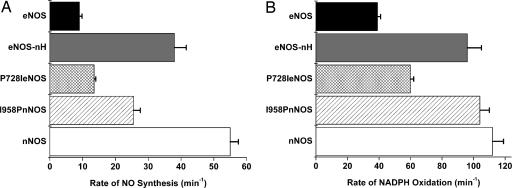
Fig. 2A compares the NO synthesis activities we obtained for the three mutants and for wild-type eNOS and nNOS. Incorporating the nNOS hinge into eNOS significantly increased NO synthesis activity. The eNOS-nH chimera was 4× more active than eNOS and was two-thirds as active as nNOS. This difference suggests that the hinge is important for catalysis and reveals that hinge composition in eNOS limits NO synthesis to a significant extent.
Fig. 2.
Steady-state NO synthesis (A) and concurrent NADPH oxidation (B) activities of wild-type and mutant NOS proteins in the presence of CaM. Measurements were taken at 25°C. Values represent the mean and standard deviations of three independent measurements.
With regard to the proline, P728I eNOS had greater NO synthesis activity relative to eNOS, whereas I958P nNOS had decreased NO synthesis activity relative to nNOS. Thus, a proline present at this hinge position restricted NO synthesis by either enzyme, consistent with proline generally restricting peptide flexibility (18, 19). However, it is clear that the proline accounts for only a fraction of the catalytic repression attributable to the entire eNOS hinge. Thus, its shorter length and/or other unique residues must also contribute to its overall repressive effect.
We also compared how the hinge mutations impact the NADPH oxidation rates of eNOS and nNOS during NO synthesis from l-arginine. In general, the rates of NADPH oxidation followed the NO synthesis activity of each enzyme (Fig. 2B), consistent with the two processes being generally coupled. However, the I958P nNOS did show some uncoupled NADPH oxidation (4.1 NADPH per NO) relative to nNOS (2.0 NADPH per NO). Remarkably, the coupling in eNOS-nH (2.6 NADPH per NO) was more efficient than in eNOS itself (4.3 NADPH per NO). This finding means that that the nNOS hinge works efficiently in eNOS, despite its distinct composition, and enables greater NO synthesis without an increased production of reactive oxygen species.
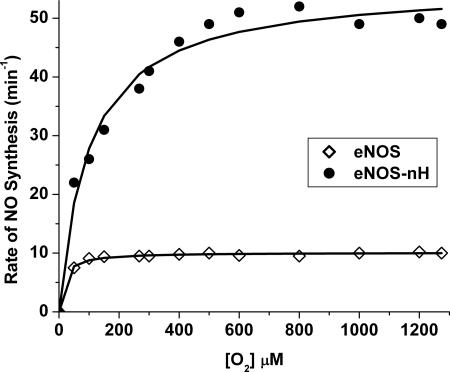
The apparent KmO2 for NO synthesis by nNOS is 350 μM, which is 80× greater than the KmO2 of eNOS (≈4 μM) (7). Given that eNOS-nH is two-thirds as active as nNOS, we were interested in determining its apparent KmO2. Fig. 3 plots the NO synthesis activities of eNOS-nH and wild-type eNOS over a wide range of O2 concentration. The apparent Vmax of eNOS-nH was 56 min−1. This is 5× greater than the apparent Vmax of eNOS, confirming the results we obtained in activity assays run with air-saturated buffer (Fig. 2A). The apparent KmO2 of eNOS-nH was 100 μM, which is an increase of >10× the eNOS value. Thus, the greater NO synthesis activity of eNOS-nH was associated with a significant increase in its apparent KmO2 value.
Fig. 3.
Oxygen activity response curves for eNOS (open diamonds) and the eNOS-nH mutant (filled circles). The initial rates of NO synthesis were determined at each designated O2 concentration. The solid lines are calculated lines of best fit.
We next compared pre-steady-state heme reduction rates in the mutant and wild-type enzymes. Heme reduction rates were determined in stopped-flow reactions by monitoring formation of the enzyme heme–CO complex. The reactions were initiated by mixing either CaM-bound NOS enzymes with excess NADPH or NADPH-reduced NOS enzymes with Ca2+ to trigger CaM binding (9). Given the potential role for the hinge in facilitating heme reduction, and heme reduction limiting NO synthesis by eNOS and nNOS (20, 21), we expected that the heme reduction rates in the mutants would reflect their different NO synthesis activities. The measured rates in Table 1 indicate that this was true within both enzyme groups (eNOS and nNOS). For example, the heme reduction rate was faster in P728I eNOS and in eNOS-nH relative to eNOS, and was slower in I958P nNOS relative to nNOS. However, the rates in the eNOS-based enzymes remained slower than in the nNOS-based enzymes, despite some having similar NO synthesis activities (i.e., I958P nNOS and eNOS-nH).
Table 1.
Heme reduction and flavin reduction of wild-type and mutant proteins
| Protein | Heme reduction, s−1 |
Flavin reduction, s−1 |
||
|---|---|---|---|---|
| NADPH-triggered | Ca2+-triggered | +CaM | −CaM | |
| nNOS | 4.4 ± 0.4 | 4.4 ± 0.3 | k1 = 45 ± 4 | k1 = 5.8 ± 0.3 |
| k2 = 5.2 ± 0.3 | k2 = 1.2 ± 0.08 | |||
| I958P nNOS | 3.6 ± 0.2 | 3.4 ± 0.3 | k1 = 42 ± 4 | k1 = 4.9 ± 0.4 |
| k2 = 4.8 ± 0.4 | k2 = 1.0 ± 0.1 | |||
| eNOS | 0.005 ± 0.003 | 0.0049 ± 0.002 | k1 = 56 ± 4 | k1 = 6.2 ± 0.5 |
| k2 = 4.1 ± 0.3 | k2 = 1.3 ± 0.07 | |||
| P728I eNOS | 0.009 ± 0.004 | 0.010 ± 0.01 | k1 = 51 ± 3 | k1 = 5.4 ± 0.3 |
| k2 = 4.7 ± 0.4 | k2 = 0.8 ± 0.06 | |||
| eNOS-nH | 0.25 ± 0.01 | 0.32 ± 0.02 | k1 = 57 ± 4.5 | k1 = 6.0 ± 0.5 |
| k2 = 3.4 ± 0.4 | k2 = 0.8 ± 0.09 | |||
Measurements were taken under anaerobic conditions at 10°C, as described in Materials and Methods. k1 and k2 refer to the rates for biphasic reactions. The values for flavin and heme reduction in wild-type and mutant NOSs are the mean ± SD of 7–10 individual reactions and are representative of experiments done with two different enzyme preparations.
Because the hinge also helps the FMN subdomain interact with the NADPH–FAD module for FAD-to-FMN electron transfer in NOS (12), it is possible that the hinge mutants might alter the flavin reduction kinetics. However, our measures show that this was not the case (Table 1) and indicate that the hinge mutations do not impact flavin reduction in eNOS or nNOS.
We also compared how the hinge mutations impact electron flux through the NOS heme during steady-state catalysis. This can be done by measuring the rate of NADPH oxidation by each CaM-bound enzyme in the absence of arginine substrate. Under this condition, NADPH-derived electrons pass from the flavins to the NOS heme and enable the enzyme to quickly reduce O2 to superoxide or H2O2 instead of generating NO (22). Thus, the NADPH oxidation rate will primarily reflect the electron flux through the heme and be proportional to the rate of heme reduction, which remains the slow step during the reaction. The NADPH consumption rates we obtained in the absence of arginine or in the presence of the inactive substrate analog agmatine are listed in Table 2 and are compared with the rates of NADPH oxidation we obtained during NO synthesis (i.e., in the presence of arginine). Rates of electron flux measured under the arginine-free condition (or with agmatine) generally followed the NO synthesis activities within each enzyme group. The rate of electron flux through eNOS-nH was well below that of nNOS, except when they were generating NO. These results are consistent with the hinge mutations impacting NO synthesis by changing the heme reduction rates in eNOS and nNOS.
Table 2.
Rate of NADPH oxidation by CaM-bound wild-type and mutant proteins
| Protein | NADPH oxidation rate, min−1 |
||
|---|---|---|---|
| +CaM,+arginine | +CaM,−arginine | +CaM,+agmatine | |
| nNOS | 112 ± 7 | 352 ± 11 | 367 ± 28 |
| I958P nNOS | 104 ± 6 | 225 ± 12 | 246 ± 17 |
| eNOS | 39 ± 2 | 33 ± 2 | 41 ± 3 |
| P728I eNOS | 60 ± 2 | 44 ± 4 | 53 ± 5 |
| eNOS-nH | 96 ± 9 | 82 ± 3 | 94 ± 8 |
The rate of NADPH oxidation was measured at 25°C as described in Materials and Methods. The values represent the mean ± SD for three independent measurements.
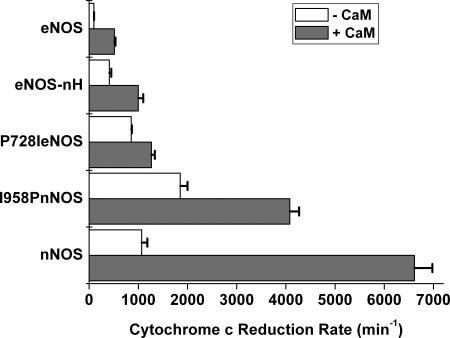
Finally, we tested whether the hinge mutations would alter reductase domain activities that are independent of the NOS heme, such as cytochrome c reductase activity, which involves direct electron transfer from the FMN subdomain to cytochrome c (17, 23). The reductase activities are compared in Fig. 4. All mutants had increased activity relative to wild-type enzymes in the CaM-free state. In the CaM-bound state, P728I eNOS and eNOS-nH had increased activities relative to eNOS, whereas I958P nNOS had a decreased activity relative to nNOS. In general, the mutations altered the reductase activities to a lesser degree. This relationship was particularly true for eNOS-nH, whose reductase activity remained closer to eNOS than to nNOS.
Fig. 4.
Steady-state cytochrome c reductase activities of wild-type and mutant NOS proteins in the absence and presence of CaM. Activities were measured at 25°C in the presence of superoxide dismutase. Values are representative of three measurements taken under identical conditions, using two different enzyme preparations.
Discussion
What protein features cause the NO synthesis activity of eNOS to be so low, and why did this circumstance evolve? Answering these questions might broaden our understanding of NOS's roles in biology. Here, we show that structural differences in a common hinge element are partly responsible for suppressing the NO synthesis activity of eNOS relative to nNOS. It is remarkable that the nNOS hinge, which represents only 3.3% of the entire reductase domain, could have such a large impact when swapped into the eNOS enzyme. Indeed, the NO synthesis activity of our hinge chimera (eNOS-nH) came close to matching that of an eNOS chimera that contains the entire nNOS reductase domain (9, 11). The apparent importance of the hinge in controlling NOS catalysis is generally consistent with hinge elements modulating the activity of a number of multidomain redox enzymes (24–28).
Our data suggest that the hinge modulates NO synthesis activity by influencing the rate of NOS heme reduction and electron flux through the heme. The hinge structure likely helps to determine an interaction between the FMN subdomain and the NOS oxygenase domain that allows electron transfer from the FMN hydroquinone to the heme iron. We already know that this electron transfer step is key for NO synthesis and is under extraordinary control. For example, it takes place only between (not within) the subunits of a NOS dimer and requires that CaM be bound to the NOS subunit that provides the FMN subdomain in the electron transfer reaction (29–31). Model-building with the available NOS crystal structures suggests that the hinge element would allow the FMN subdomain to place its FMN cofactor within 15 Å of the heme in the partner oxygenase domain (12). This distance is at the long end of the range sufficient for electron transfer (32). Domain interactions in electron transfer proteins are also known to be dynamic, with a variety of donor–acceptor positions being sampled in unit time (33, 34). Apparently, the nNOS hinge enables a more probable and/or efficient domain–domain interaction for the heme reduction step than does the eNOS hinge. Our data suggest that both the length and amino acid composition of the hinge may be important; this issue requires further investigation. The fact that the hinge substitution did not completely convert eNOS to nNOS (regarding heme reduction and NO synthesis activity) implies that other structural elements of the eNOS reductase domain must help restrict electron flux to the heme. Among the many potential candidates, one contributor could be the upstream Oxy-FMN hinge element that links the FMN subdomain to the oxygenase domain (Fig. 1A). This “partner hinge” should also define the motions of the FMN subdomain during electron transfer, and its sequence and length differ between eNOS and nNOS.
Hinge composition appears to be less important for repressing the cytochrome c reductase activity of eNOS. This observation is consistent with reductase activity being regulated by distinct structural elements within the reductase domain, including the C-terminal tail, an autoinhibitory helix insert in the FMN subdomain, and an insert in the connecting subdomain (designated CT, AH, and CD2A in Fig. 1B) (10, 35–38). In general, deletion or exchange of these elements in eNOS and nNOS had more prominent effects on their reductase activities and CaM binding affinities than on their NO synthesis activities. Thus, the hinge may be one structural component of the reductase domain that primarily impacts NOS heme reduction and NO synthesis rather than reductase activity.
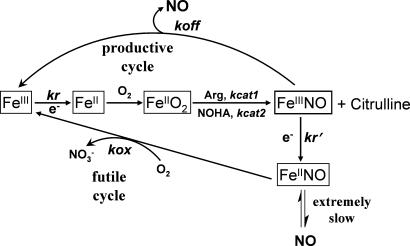
Our measurements of electron flux in the absence of arginine showed that nNOS and I958P nNOS have significantly greater flux than does eNOS-nH. How, then, could eNOS-nH have an NO synthesis activity that is two-thirds that of nNOS and greater than that of I958P nNOS? The key point is that electron flux through all three enzymes becomes similar during NO synthesis (i.e., in reaction mixtures containing arginine). Indeed, NO synthesis by nNOS and I958P nNOS was associated with a 2- to 3-fold decrease in electron flux, whereas electron flux through eNOS-nH remained essentially unaffected by NO synthesis. These different behaviors arise because nNOS and eNOS have different settings for three kinetic parameters (kr, kox, and koff; see Fig. 5) that together determine the NO synthesis behavior of any NOS (Fig. 5). Specifically, the kox value, which is determined by NOS oxygenase domain identity (7), is ≈6× slower in nNOS than in eNOS (7, 21). A slow kox causes a majority of nNOS molecules to build up as an inactive ferrous heme–NO species during NO synthesis (Fig. 5), which diminishes the electron flux and NO release rate in that system (7, 39). In contrast, the faster kox of eNOS means that far fewer enzyme molecules will build up as the ferrous heme–NO species during NO synthesis at any given rate of heme reduction. Thus, a faster kox blunts the impact of NO synthesis on electron flux through eNOS-nH and enables this mutant to achieve a steady-state NO synthesis activity that is close to nNOS while having a slower rate of heme reduction. A similar situation explains the NO synthesis activities of iNOS (21) and the W409F mutant of nNOS (7) because both of these enzymes have slower heme reduction rates than nNOS but have equivalent or greater NO synthesis activities because of their faster kox values.
Fig. 5.
Global kinetic model for NOS catalysis. The biosynthesis of NO and formation of the enzyme ferric heme–NO complex is limited by kr. This product complex partitions between an NO-releasing (productive) cycle and an NO-oxidizing (futile) cycle. Enzyme partitioning and NO release are determined by the relative rates of kr, kox, and koff. NOHA, Nω-hydroxy-l-arginine. Adapted from ref. 7.
Given the above, it is puzzling why eNOS would evolve to have such a slow heme reduction and NO synthesis activity. After all, there is no obvious downside to an eNOS that is more active, and this would be advantageous regarding the efficiency of cellular protein synthesis. The only sacrifice a NOS enzyme makes when it increases its NO synthesis activity by increasing its heme reduction rate is a shift in its oxygen concentration–activity response, which becomes blunted with regard to d(% activity change)/d[O2], as indicated by a shift in apparent KmO2 to higher values. This relationship is explained by the global model (Fig. 5), which stipulates that faster heme reduction enables greater NO production but also causes more enzyme molecules to partition into the ferrous heme–NO complex during NO synthesis. This, in turn, increases the influence of kox on the overall oxygen response of the system and shifts the apparent KmO2 to a higher value (7, 21, 40). Thus, faster heme reduction in eNOS-nH causes its apparent KmO2 to increase and makes its oxygen concentration–activity response become more like nNOS than eNOS. In this regard, eNOS has the steepest oxygen concentration–activity response, given that its near-total percentage change in activity takes place across a smaller range of oxygen concentrations than does either iNOS or nNOS. It is tempting to speculate that the unique oxygen response profiles of eNOS, iNOS, and nNOS are matched in some way to their specific biological functions. For example, eNOS and nNOS both function in tissues where oxygen tension can change dramatically during ischemia or high mitochondrial activity. Their unique oxygen response profiles would dictate widely different NO release patterns under these circumstances. How this may relate to their biological functions is worthy of further investigation.
The activity of eNOS is increased upon serine phosphorylation of its reductase domain (41) or when eNOS interacts with dynamin-2 or HSP-90 (42, 43). Although the mechanisms of activation are unknown, the global model (7) and previous results (40, 44) predict that they likely involve an increase in eNOS heme reduction rate. Boosting eNOS activity through these means is expected to add to the increase we observed through hinge substitution. These strategies can now be used to create a “super-eNOS” with enhanced NO synthesis activity for use in gene replacement therapies involving eNOS (45–49).
Materials and Methods
Reagents.
All reagents and materials were obtained from Sigma (St. Louis, MO) or from sources previously reported (50, 51).
Molecular Biology.
Restriction digestions, cloning, and bacterial growth were performed using standard procedures. Transformations were performed using a TransformAid bacterial transformation kit (Fermentas, Hanover, MD). Oligonucleotides used to construct site-directed mutants in nNOS were obtained from Integrated DNA Technologies (Coralville, IA). Site-directed mutagenesis was performed using a QuikChange XL site-directed mutagenesis kit (Stratagene, La Jolla, CA). Mutations (bold) and their corresponding oligonucleotides were as follows: I958PnNOSFl (sense), ACTCCCTCCCTAGCAATGATCGATCCTGGAAGAGGAAC; I958PnNOSFl (antisense), GTTCCTCTTCCAGCTTCGGTCATTGCTAGGGAGGGAGTT; P728I eNOSFl (sense), GATATCTTCAGCATCAAACGGAGC; and P728IeNOSFl (antisense), GCTCCGTTTGATGCTGAAGATATC. To replace the eNOS hinge with the corresponding residues of nNOS, we used PCR oligonucleotides containing a Bsm1 (5′ oligonucleotide) and a KpnI (3′ oligonucleotide) site to amplify nNOS between bases corresponding to amino acids 943 and 967 (5′ oligonucleotide-TTTTAAGGCGGCATTCCAGGCCTCCTGCGAGACGTTCTGCGTGGGGGATGAC; 3′ oligonucleotide TTCCGGTACCTGTTCCTCTTCCAGCTTCGGTC; restriction sites are shown in bold and sequence corresponding to nNOS are underlined). Both the PCR product and the pCW-eNOS vector were digested with Bsm1 and KpnI and the PCR fragment then inserted into the vector replacing the eNOS hinge region. The hybrid constructs were confirmed by DNA sequencing at the Cleveland Clinic Genomics Core.
Expression and Purification of Wild-Type and Mutant Proteins.
Wild-type and mutant proteins had a His6 tag attached to their N termini to aid purification. They were overexpressed in Escherichia coli strain BL21(DE3) and purified by sequential chromatography on Ni2+-NTA and CaM-Sepharose resin (16). The ferrous heme–CO adduct absorbing at 444 nm was used to measure heme protein content with an extinction coefficient of ε444 = 74 mM−1·cm−1 (A444–A500).
NO Synthesis, NADPH Oxidation, and Cytochrome c Reduction.
Steady-state activities of wild-type and mutant proteins were determined separately at 25°C by using spectrophotometric assays that were described in detail in refs. 50 and 51. For the electron flux measurement through the NOS heme during steady-state catalysis, we measured the rate of NADPH oxidation by each CaM-bound enzyme in the absence of arginine substrate or in the presence of 2 mM agmatine (22).
Measurement of Apparent KmO2.
Apparent Km and Vmax values were calculated by fitting the initial rates of NO synthesis measured at various oxygen concentrations (52) to a hyperbolic equation (53) by using Origin software (OriginLab, Northampton, MA). Reactions were run in septum-sealed cuvettes that contained 0.2 μM nNOS, 40 mM 4-(2-hydroxyethyl)-1-piperazinepropanesulfonic acid (EPPS) (pH 7.6), 150 mM NaCl, 0.3 mM DTT, 4 μM FAD, 4 μM FMN, 10 μM (6R)-tetrahydrobiopterin, 10 mM l-arginine, 1 mg/ml BSA, 0.8 mM CaCl2, 0.6 mM EDTA, 0.9 μM CaM, 100 units/ml catalase, 25 units/ml superoxide dismutase, and 5 μM oxyhemoglobin diluted in various ratios of N2-, air-, or O2-saturated buffer solutions. The reaction was initiated with 300 μM NADPH (final volume, 1 ml). The initial O2 concentration in each reaction was calculated based on the solution mixing ratio and the O2 concentration of air- or O2-saturated buffer at 25°C (≈0.26 and 1.26 mM, respectively).
Heme and Flavin Reduction.
The kinetics of heme reduction were analyzed at 10°C as described previously (9, 40, 50, 51), by using a stopped-flow apparatus and diode array detector (Model SF-61 Hi-Tech Scientific, Bradford, U.K.) equipped for anaerobic analysis. Ferric heme reduction was followed by formation of the ferrous heme–CO complex at 444 nm. Reactions were initiated by rapidly mixing an anaerobic, buffered, CO-saturated solution containing either 50 μM NADPH or 5 mM CaCl2 with an anaerobic, buffered, CO-saturated solution containing wild-type or mutant NOS (2 μM), 100 mM 4-(2-hydroxyethyl)-1-piperazinepropanesulfonic acid (pH 7.6), 100 mM NaCl, 10 μM (6R)-tetrahydrobiopterin, 2 mM l-arginine, 0.3 mM DTT, 6 μM CaM, and either 1 mM CaCl2 when triggered with NADPH or 50 μM NADPH when triggered with Ca2+. Signal-to-noise ratios were improved by averaging 8–10 individual mixing experiments. Flavin reduction was monitored under the same conditions at 485 nm. The time course of the absorbance change was fit to single or multiple exponential equations by using a nonlinear least-squares method provided by Hi-Tech Scientific.
Acknowledgments
This work was supported by National Institutes of Health Grants GM51491 and CA53914 (to D.J.S.) and HL076491 (to S. Hazen and D.J.S.).
Abbreviations
- NOS
NO synthase
- eNOS
endothelial NOS
- iNOS
inducible NOS
- nNOS
neuronal NOS
- CaM
calmodulin
- eNOS-nH
an eNOS chimera that contains the FMN–ferredoxin NADP-reductase hinge element from nNOS.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Moncada S, Bolanos JP. J Neurochem. 2006;97:1676–1689. doi: 10.1111/j.1471-4159.2006.03988.x. [DOI] [PubMed] [Google Scholar]
- 2.Schmidt HH, Walter U. Cell. 1994;78:919–925. doi: 10.1016/0092-8674(94)90267-4. [DOI] [PubMed] [Google Scholar]
- 3.Knowles RG. Biochem Soc Trans. 1997;25:895–901. doi: 10.1042/bst0250895. [DOI] [PubMed] [Google Scholar]
- 4.Fulton D, Gratton JP, Sessa WC. J Pharmacol Exp Ther. 2001;299:818–824. [PubMed] [Google Scholar]
- 5.Forstermann U, Boissel JP, Kleinert H. FASEB J. 1998;12:773–790. [PubMed] [Google Scholar]
- 6.Michel T, Feron O. J Clin Invest. 1997;100:2146–2152. doi: 10.1172/JCI119750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stuehr DJ, Santolini J, Wang ZQ, Wei CC, Adak S. J Biol Chem. 2004;279:36167–36170. doi: 10.1074/jbc.R400017200. [DOI] [PubMed] [Google Scholar]
- 8.Kleinert H, Pautz A, Linker K, Schwarz PM. Eur J Pharmacol. 2004;500:255–266. doi: 10.1016/j.ejphar.2004.07.030. [DOI] [PubMed] [Google Scholar]
- 9.Adak S, Aulak KS, Stuehr DJ. J Biol Chem. 2001;276:23246–23252. doi: 10.1074/jbc.M102509200. [DOI] [PubMed] [Google Scholar]
- 10.Roman LJ, Martasek P, Miller RT, Harris DE, de La Garza MA, Shea TM, Kim JJ, Masters BS. J Biol Chem. 2000;275:29225–29232. doi: 10.1074/jbc.M004766200. [DOI] [PubMed] [Google Scholar]
- 11.Nishida CR, Ortiz de Montellano PR. J Biol Chem. 1998;273:5566–5571. doi: 10.1074/jbc.273.10.5566. [DOI] [PubMed] [Google Scholar]
- 12.Garcin ED, Bruns CM, Lloyd SJ, Hosfield DJ, Tiso M, Gachhui R, Stuehr DJ, Tainer JA, Getzoff ED. J Biol Chem. 2004;279:37918–37927. doi: 10.1074/jbc.M406204200. [DOI] [PubMed] [Google Scholar]
- 13.Crane BR, Arvai AS, Ghosh DK, Wu C, Getzoff ED, Stuehr DJ, Tainer JA. Science. 1998;279:2121–2126. doi: 10.1126/science.279.5359.2121. [DOI] [PubMed] [Google Scholar]
- 14.Ghosh DK, Holliday MA, Thomas C, Weinberg JB, Smith SM, Salerno JC. J Biol Chem. 2006;281:14173–14183. doi: 10.1074/jbc.M509937200. [DOI] [PubMed] [Google Scholar]
- 15.Roman LJ, Masters BS. J Biol Chem. 2006;281:23111–23118. doi: 10.1074/jbc.M603671200. [DOI] [PubMed] [Google Scholar]
- 16.Konas DW, Zhu K, Sharma M, Aulak KS, Brudvig GW, Stuehr DJ. J Biol Chem. 2004;279:35412–35425. doi: 10.1074/jbc.M400872200. [DOI] [PubMed] [Google Scholar]
- 17.Craig DH, Chapman SK, Daff S. J Biol Chem. 2002;277:33987–33994. doi: 10.1074/jbc.M203118200. [DOI] [PubMed] [Google Scholar]
- 18.Schimmel PR, Flory PJ. J Mol Biol. 1968;34:105–120. doi: 10.1016/0022-2836(68)90237-4. [DOI] [PubMed] [Google Scholar]
- 19.MacArthur MW, Thornton JM. J Mol Biol. 1991;218:397–412. doi: 10.1016/0022-2836(91)90721-h. [DOI] [PubMed] [Google Scholar]
- 20.Miller RT, Martasek P, Omura T, Siler Masters BS. Biochem Biophys Res Commun. 1999;265:184–188. doi: 10.1006/bbrc.1999.1643. [DOI] [PubMed] [Google Scholar]
- 21.Santolini J, Meade AL, Stuehr DJ. J Biol Chem. 2001;276:48887–48898. doi: 10.1074/jbc.M108666200. [DOI] [PubMed] [Google Scholar]
- 22.Adak S, Crooks C, Wang Q, Crane BR, Tainer JA, Getzoff ED, Stuehr DJ. J Biol Chem. 1999;274:26907–26911. doi: 10.1074/jbc.274.38.26907. [DOI] [PubMed] [Google Scholar]
- 23.Klatt P, Heinzel B, John M, Kastner M, Bohme E, Mayer B. J Biol Chem. 1992;267:11374–11378. [PubMed] [Google Scholar]
- 24.White P, Manson FD, Brunt CE, Chapman SK, Reid GA. Biochem J. 1993;291:89–94. doi: 10.1042/bj2910089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Govindaraj S, Poulos TL. Biochemistry. 1995;34:11221–11226. doi: 10.1021/bi00035a031. [DOI] [PubMed] [Google Scholar]
- 26.Davis CA, Crowley LJ, Barber MJ. Arch Biochem Biophys. 2004;431:233–244. doi: 10.1016/j.abb.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 27.Blazyk JL, Lippard SJ. J Biol Chem. 2004;279:5630–5640. doi: 10.1074/jbc.M308940200. [DOI] [PubMed] [Google Scholar]
- 28.Nett JH, Hunte C, Trumpower BL. Eur J Biochem. 2000;267:5777–5782. doi: 10.1046/j.1432-1327.2000.01650.x. [DOI] [PubMed] [Google Scholar]
- 29.Sagami I, Daff S, Shimizu T. J Biol Chem. 2001;276:30036–30042. doi: 10.1074/jbc.M104123200. [DOI] [PubMed] [Google Scholar]
- 30.Panda K, Ghosh S, Stuehr DJ. J Biol Chem. 2001;276:23349–23356. doi: 10.1074/jbc.M100687200. [DOI] [PubMed] [Google Scholar]
- 31.Siddhanta U, Presta A, Fan B, Wolan D, Rousseau DL, Stuehr DJ. J Biol Chem. 1998;273:18950–18958. doi: 10.1074/jbc.273.30.18950. [DOI] [PubMed] [Google Scholar]
- 32.Moser CC, Page CC, Dutton PL. Philos Trans R Soc London B. 2006;361:1295–1305. doi: 10.1098/rstb.2006.1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leys D, Basran J, Talfournier F, Sutcliffe MJ, Scrutton NS. Nat Struct Biol. 2003;10:219–225. doi: 10.1038/nsb894. [DOI] [PubMed] [Google Scholar]
- 34.Kang SA, Crane BR. Proc Natl Acad Sci USA. 2005;102:15465–15470. doi: 10.1073/pnas.0505176102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lane P, Gross SS. J Biol Chem. 2002;277:19087–19094. doi: 10.1074/jbc.M200258200. [DOI] [PubMed] [Google Scholar]
- 36.Daff S, Sagami I, Shimizu T. J Biol Chem. 1999;274:30589–30595. doi: 10.1074/jbc.274.43.30589. [DOI] [PubMed] [Google Scholar]
- 37.Nishida CR, de Montellano PR. J Biol Chem. 2001;276:20116–20124. doi: 10.1074/jbc.M101548200. [DOI] [PubMed] [Google Scholar]
- 38.Knudsen GM, Nishida CR, Mooney SD, Ortiz de Montellano PR. J Biol Chem. 2003;278:31814–31824. doi: 10.1074/jbc.M303267200. [DOI] [PubMed] [Google Scholar]
- 39.Santolini J, Adak S, Curran CM, Stuehr DJ. J Biol Chem. 2001;276:1233–1243. doi: 10.1074/jbc.M006858200. [DOI] [PubMed] [Google Scholar]
- 40.Adak S, Santolini J, Tikunova S, Wang Q, Johnson JD, Stuehr DJ. J Biol Chem. 2001;276:1244–1252. doi: 10.1074/jbc.M006857200. [DOI] [PubMed] [Google Scholar]
- 41.Fleming I, Busse R. Am J Physiol. 2003;284:R1–R12. doi: 10.1152/ajpregu.00323.2002. [DOI] [PubMed] [Google Scholar]
- 42.Cao S, Yao J, McCabe TJ, Yao Q, Katusic ZS, Sessa WC, Shah V. J Biol Chem. 2001;276:14249–14256. doi: 10.1074/jbc.M006258200. [DOI] [PubMed] [Google Scholar]
- 43.Garcia-Cardena G, Fan R, Shah V, Sorrentino R, Cirino G, Papapetropoulos A, Sessa WC. Nature. 1998;392:821–824. doi: 10.1038/33934. [DOI] [PubMed] [Google Scholar]
- 44.McCabe TJ, Fulton D, Roman LJ, Sessa WC. J Biol Chem. 2000;275:6123–6128. doi: 10.1074/jbc.275.9.6123. [DOI] [PubMed] [Google Scholar]
- 45.Yu J, deMuinck ED, Zhuang Z, Drinane M, Kauser K, Rubanyi GM, Qian HS, Murata T, Escalante B, Sessa WC. Proc Natl Acad Sci USA. 2005;102:10999–11004. doi: 10.1073/pnas.0501444102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qian HS, Liu P, Huw LY, Orme A, Halks-Miller M, Hill SM, Jin F, Kretschmer P, Blasko E, Cashion L., et al. Gene Ther. 2006;13:1342–1350. doi: 10.1038/sj.gt.3302781. [DOI] [PubMed] [Google Scholar]
- 47.Elrod JW, Duranski MR, Langston W, Greer JJ, Tao L, Dugas TR, Kevil CG, Champion HC, Lefer DJ. Circ Res. 2006;99:78–85. doi: 10.1161/01.RES.0000231306.03510.77. [DOI] [PubMed] [Google Scholar]
- 48.Elrod JW, Greer JJ, Bryan NS, Langston W, Szot JF, Gebregzlabher H, Janssens S, Feelisch M, Lefer DJ. Arterioscler Thromb Vasc Biol. 2006;26:1517–1523. doi: 10.1161/01.ATV.0000224324.52466.e6. [DOI] [PubMed] [Google Scholar]
- 49.Bivalacqua TJ, Musicki B, Usta MF, Champion HC, Kadowitz PJ, Burnett AL, Hellstrom WJ. Curr Pharm Des. 2005;11:4059–4067. doi: 10.2174/138161205774913345. [DOI] [PubMed] [Google Scholar]
- 50.Adak S, Ghosh S, Abu-Soud HM, Stuehr DJ. J Biol Chem. 1999;274:22313–22320. doi: 10.1074/jbc.274.32.22313. [DOI] [PubMed] [Google Scholar]
- 51.Panda K, Haque MM, Garcin-Hosfield ED, Durra D, Getzoff ED, Stuehr DJ. J Biol Chem. 2006;281:36819–36827. doi: 10.1074/jbc.M606129200. [DOI] [PubMed] [Google Scholar]
- 52.Abu-Soud HM, Rousseau DL, Stuehr DJ. J Biol Chem. 1996;271:32515–32518. doi: 10.1074/jbc.271.51.32515. [DOI] [PubMed] [Google Scholar]
- 53.Dowd JE, Riggs DS. J Biol Chem. 1965;240:863–869. [PubMed] [Google Scholar]