Abstract
Activation of the checkpoint kinase Rad53 is a critical response to DNA damage that results in stabilization of stalled replication forks, inhibition of late-origin initiation, up-regulation of dNTP levels, and delayed entry to mitosis. Activation of Rad53 is well understood and involves phosphorylation by the protein kinases Mec1 and Tel1 as well as in trans autophosphorylation by Rad53 itself. However, deactivation of Rad53, which must occur to allow the cell to recover from checkpoint arrest, is not well understood. Here, we present genetic and biochemical evidence that the type 2A-like protein phosphatase Pph3 forms a complex with Psy2 (Pph3–Psy2) that binds and dephosphorylates activated Rad53 during treatment with, and recovery from, methylmethane sulfonate-mediated DNA damage. In the absence of Pph3–Psy2, Rad53 dephosphorylation and the resumption of DNA synthesis are delayed during recovery from DNA damage. This delay in DNA synthesis reflects a failure to restart stalled replication forks, whereas, remarkably, genome replication is eventually completed by initiating late origins of replication despite the presence of hyperphosphorylated Rad53. These findings suggest that Rad53 regulates replication fork restart and initiation of late firing origins independently and that regulation of these processes is mediated by specific Rad53 phosphatases.
Keywords: checkpoint, phosphorylation, YBL046W, cell cycle
Faithful maintenance of genome integrity is essential for cellular viability. Failure to efficiently recognize and repair DNA damage can lead to genomic instability and, in higher eukaryotes, cancer (1). To coordinate the response to genotoxic stress, eukaryotic cells use intricate signaling networks, called checkpoints, that control cell cycle progression, transcription of DNA damage response genes, activation of DNA repair pathways, and recruitment of proteins to sites of damage (2). An efficient checkpoint response is especially important during S phase, as replication forks are particularly vulnerable to DNA damage (3, 4).
The response to DNA damage in S phase is orchestrated by the intra-S cell cycle checkpoint response and has been particularly well characterized in Saccharomyces cerevisiae, where it is mediated by the overlapping replication checkpoint and DNA damage checkpoint pathways (1). A central component of both intra-S pathways is the protein kinase Rad53. Activation of Rad53 depends on the ATM- and ATR-like kinases Mec1 and Tel1 as well as a host of adapters and damage sensors, such as the replication stress-specific proteins, Mrc1 and Tof1 and the DNA damage-specific proteins Rad9, Mec3/Ddc1/Rad17 (PCNA-like complex), and Rad24/Rfc2–5 (RFC-like complex) (2). After activation by Mec1 and/or Tel1, Rad53 amplifies its own activity by autophosphorylation in trans (5).
Once activated by phosphorylation in S-phase, Rad53 protects damage-stalled replication forks from collapse and inhibits activation of late-firing origins of replication (3, 4). Fork stabilization is thought to occur by reinforcing the association of the replisome components with the fork and inhibiting the activity of recombination enzymes at these sites. These events are likely regulated through Rad53-mediated phosphorylation of different targets such as RPA, Mrc1, Srs2, Mus81, and the DNA polymerase α–primase complex (4). The mechanism of origin inhibition may include Rad53-dependent phosphorylation of Dbf4, which inhibits the kinase activity as well as chromatin association of the Dbf4-dependent kinase, Dbf4-Cdc7, which is required for replication initiation (6).
In contrast to its activation and its roles in the checkpoint response, relatively little is known about deactivation of Rad53. New protein synthesis is not required for the appearance of unphosphorylated Rad53 during recovery from checkpoint arrest (7), indicating that the activated kinase is dephosphorylated, not degraded. This model is supported by the observation that Ptc2 and Ptc3, two PP2C-like phosphatases, are required for Rad53 dephosphorylation after prolonged exposure to a persistent double-strand break (8). Recently, it was suggested that Rad53 deactivation during prolonged recovery from a double-strand break depends on γH2AX (H2A phosphorylated at Ser-129) dephosphorylation by the type 2A-like phosphatase, Pph3, in conjunction with both Psy2 and Ybl046w (9).
We now report that Pph3 and Psy2 form a phosphatase complex (Pph3–Psy2) that negatively regulates Rad53 activity independent of Ybl046w. We present in vitro and in vivo evidence suggesting that Pph3–Psy2 performs this function by directly binding and dephosphorylating Rad53. We also find that psy2Δ and pph3Δ cells do not efficiently resume DNA synthesis during recovery from DNA damage and attribute this to the defective restart of stalled forks. However, DNA replication in psy2Δ and pph3Δ cells is completed by the initiation of late firing replication origins during recovery from DNA damage despite the presence of hyperphosphorylated Rad53.
Results
Identification of the Pph3–Psy2 Phosphatase Complex.
We originally identified PSY2 as a gene involved in tolerating and/or regulating the cellular response to DNA damage (10). To better understand the function of Psy2, we used multidimensional protein identification technology (MudPIT) (11) to identify proteins that associate with a Psy2-TAP fusion protein expressed at endogenous levels from the PSY2 promoter. Consistent with previous reports, three potential binding partners of Psy2 were identified: Pph3, Ybl046w, and Spt5 [see supporting information (SI) Table 1] (12–14). Coimmunoprecipitation experiments confirmed that Psy2 interacts with Pph3 and Ybl046w (Fig. 1A). Spt5 did not immunoprecipitate with Psy2 and was not further characterized.
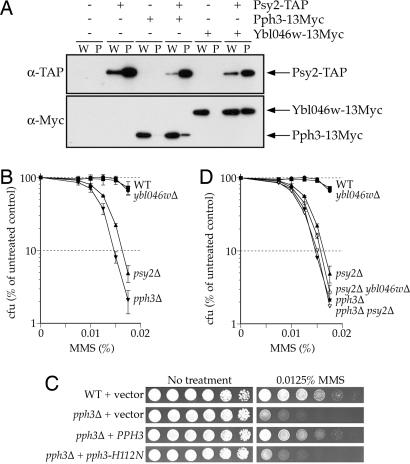
Fig. 1.
Pph3–Psy2 contributes to MMS resistance. (A) Whole-cell extracts were prepared from yeast strains expressing Psy2-TAP, Pph3–13Myc, and/or Ybl046w-13Myc and incubated with IgG-Sepharose. Whole-cell extracts (W, 10 μg of protein) and precipitated proteins (P, 5% of precipitated fraction) were analyzed by Western blotting with the indicated antibodies. (B–D) Survival assays of yeast containing wild-type or null alleles of PSY2, PPH3, and/or YBL046W. (B and D) Colony forming units (cfu) were counted 3 days after deposition onto YPD-agar media containing the indicated concentration of MMS. Strains are as listed in SI Table 2 and are isogenic with BY4741. (C) Fivefold serial dilutions of wild-type (BY4741) or pph3Δ (FR1046) cells harboring YEplac195 or YEplac195 carrying a wild-type or mutant allele of PPH3 were deposited on SC-URA plates containing MMS.
We disrupted PPH3 and YBL046W to determine whether these genes contribute to MMS resistance, as observed for PSY2 (10), and found that pph3Δ cells are hypersensitive to MMS (Fig. 1B), but ybl046wΔ cells are not. The MMS sensitivity of pph3Δ is complemented by wild-type PPH3 but not by pph3-H112N, which encodes a catalytically inactive mutant of Pph3 (Fig. 1C), demonstrating that the phosphatase activity of Pph3 is required for tolerance of MMS. We also examined the genetic interactions among psy2Δ, pph3Δ, and ybl046wΔ to determine whether these genes function in the same pathways. psy2Δ pph3Δ cells are no more MMS sensitive than pph3Δ cells (Fig. 1D), indicating that PSY2 and PPH3 function in the same pathway(s). psy2Δ ybl046wΔ cells are marginally more sensitive to MMS than psy2Δ cells, which suggests that YBL046W does not function with PSY2 and PPH3 in the response to MMS (Fig. 1D). Together, these data suggest that the Pph3–Psy2 phosphatase complex promotes viability after MMS-induced damage and that this function is independent of Ybl046w.
Pph3–Psy2 Antagonizes the DNA Damage Checkpoint Response in a Rad53-Dependent Manner.
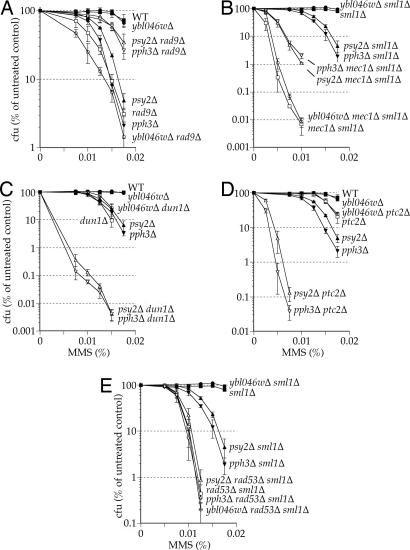
To study the contribution of Pph3–Psy2 to replication and/or cell cycle control, we examined the genetic interactions of PSY2 and PPH3 with genes involved in the DNA damage and replication checkpoints. Loss of PSY2 or PPH3 results in a dramatic decrease in the MMS sensitivity of strains deficient for genes that function upstream of Rad53 activation (RAD9, RAD17, RAD24, or MEC1), a synergistic increase in the sensitivity of strains lacking genes that function downstream of Rad53 (DUN1 or PTC2), and no effect in cells lacking RAD53 itself (Fig. 2 and SI Fig. 5). However, it should also be noted that Dun1 has Rad53-independent functions (15). The epistatic relationship between psy2Δ or pph3Δ and rad53Δ is particularly interesting because it suggests that Pph3–Psy2 does not function outside of the Rad53-mediated checkpoint pathway to promote viability in response to MMS damage. These data indicate that Pph3–Psy2 acts downstream of Rad53 to antagonize the activation of the DNA damage checkpoint.
Fig. 2.
PSY2 and PPH3 genetically interact with cell cycle checkpoint components. Survival assays of yeast containing wild-type or null alleles of PSY2, PPH3, and/or YBL046W in combination with wild-type or null alleles of cell cycle checkpoint components. Colony forming units (cfu) were counted 3 days after deposition onto YPD-agar media containing the indicated concentration of MMS. Strains are as listed in SI Table 2 and are isogenic with BY4741 (A and D) or RDKY3615 (B, C, and E).
Pph3–Psy2 Interacts with and Directly Dephosphorylates Rad53.
Although not detected by MudPIT, published yeast two-hybrid studies indicate that Psy2 interacts with Rad53 (14, 16). To confirm this interaction and determine whether it is domain-specific, we cloned PSY2 into the bait vector pGBT9 and expressed Rad53 from the prey vector pACTIIst as full-length protein, kinase domain, or individual Forkhead-associated (FHA) phosphothreonine-binding domains, FHA1 and FHA2 (8). An interaction was detected between Psy2 and the Rad53 kinase domain as well as between Psy2 and the full-length protein but not between Psy2 and either FHA domain (Fig. 3A). The interaction could not be detected by coimmunoprecipitation experiments from undamaged or MMS-treated cells (data not shown). The ability to detect a Pph3–Psy2-Rad53 complex only under the high protein concentrations of the yeast two-hybrid system is consistent with an enzyme–substrate interaction, for which only modest affinities are expected.
Fig. 3.
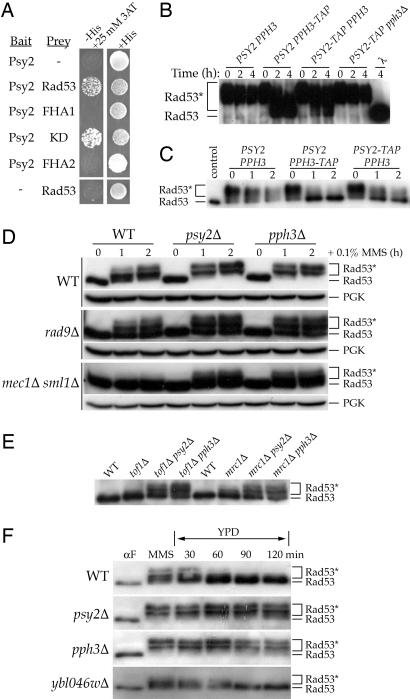
Pph3–Psy2 regulates the phosphorylation state of Rad53. (A) Yeast two-hybrid analysis of Psy2 interaction with Rad53. pGBT9 expressing the Gal4BD-Psy2 fusion protein was introduced into the Y190 tester strain along with pACTIIst expressing the Gal4AD fused to full-length Rad53 or fragments encompassing the Rad53 FHA1, kinase (KD) or FHA2 domains. (B–F) Western blot analysis of Rad53 phosphorylation state. (B and C) In vitro assay of Rad53 dephosphorylation by Pph3–Psy2. Incubation of Rad53 isolated from E. coli (B) or MMS-treat yeast (C) cells with immunoprecipitates from whole-cell lysates of the indicated yeast strains (isogenic with W1588–4C). (D–F) In vivo regulation of Rad53 by Pph3–Psy2. (D) Log phase cultures of the indicated strains (isogenic with BY4741 or RDKY3615) were treated with 0.1% MMS for 0, 1, and 2 h. Phosphoglycerate kinase (PGK) is a loading control. (E) Analysis of undamaged log phase cultures of the indicated strains (isogenic with BY4741). (F) The indicated strains (isogenic with BY4741) were synchronized in G1 at 30°C, released into 0.033% MMS for 1 h, and then shifted to YPD. Rad53* denotes phosphorylated Rad53.
We next determined whether the Pph3–Psy2 complex can directly dephosphorylate Rad53 in vitro. Using recombinant expression in Escherichia coli, we obtained Rad53 that was autophosphorylated on many of the sites induced by MMS in vivo (17, 18) and incubated it with immunoprecipitates of whole-cell extracts from yeast containing untagged or C-terminally TAP-tagged Psy2 or Pph3. Immunoprecipitate from untagged cells results in no observable Rad53 dephosphorylation; however, immunoprecipitate from Pph3-TAP cells results in the efficient dephosphorylation of Rad53 (Fig. 3B). Immunoprecipitate from Psy2-TAP cells also dephosphorylates Rad53 but not when purified from pph3Δ cells. Finally, immunoprecipitate from Psy2-TAP pph3Δ cells harboring a plasmid expressing wild-type Pph3, but not the catalytically inactive H112N mutant, dephosphorylates Rad53 (SI Fig. 6). Immunoprecipitates from Pph3-TAP and Psy2-TAP cells also dephosphorylate Rad53 isolated from MMS-treated yeast cells (Fig. 3C). Taken together, these results suggest that Pph3–Psy2 is a Rad53 phosphatase.
Pph3–Psy2 Promotes Dephosphorylation of Activated Rad53 During Recovery from MMS Treatment.
We next examined the role of Pph3–Psy2 in regulating Rad53 phosphorylation in vivo. Asynchronous wild-type cells accumulate partially phosphorylated Rad53 after 1 h of treatment with MMS and fully phosphorylated Rad53 after 2 h (Fig. 3D). Loss of PSY2 or PPH3 results in the more rapid accumulation of fully phosphorylated Rad53, relative to wild-type cells. Consistent with the observed suppression of MMS sensitivity, loss of PSY2 or PPH3 partially restores the MMS-induced phosphorylation of Rad53 in rad9Δ and mec1Δ mutants (Fig. 3D). Additionally, disruption of PSY2 or PPH3 increases the amount of phosphorylated Rad53 in undamaged tof1Δ and mrc1Δ cells (Fig. 3E), which are known to have elevated levels of spontaneous DNA damage because of defective replication forks (19–23). These effects are consistent with previously observed genetic relationships between tof1Δ or mrc1Δ and psy2Δ or pph3Δ mutants (9, 10). To determine whether Pph3–Psy2 regulates Rad53 activation specifically in the intra-S phase checkpoint, we synchronized cells in G1 and released them into S phase in the presence of MMS. pph3Δ and psy2Δ mutants accumulate more hyperphosphorylated Rad53 than wild-type cells (Fig. 3F), consistent with a hyperactive intra-S checkpoint response.
We next examined the phosphorylation state of Rad53 in cells recovering from MMS treatment in S phase. After removal of MMS from the media, virtually full dephosphorylation of Rad53 was observed in wild-type cells by 60 min (Fig. 3F). In contrast, Rad53 remained hyperphosphorylated in pph3Δ and psy2Δ mutants throughout the entirety of the 2-h experiment. These data demonstrate that the Pph3–Psy2 phosphatase is required for the dephosphorylation of Rad53 during recovery from the intra-S DNA damage checkpoint.
Pph3–Psy2 Is Required for Efficient DNA Synthesis During Recovery from MMS Treatment.
To determine the impact of misregulated Rad53 on the kinetics of DNA replication, we monitored DNA content by flow cytometry during and after exposure to MMS. Fig. 4A shows that wild-type, psy2Δ, and pph3Δ cells synchronized in G1 and released into yeast extract–peptone–dextrose (YPD) in the absence of MMS progress similarly through S-phase. Cells were also synchronized in G1, released into YPD containing 0.033% MMS for 1 h, and then transferred to YPD. All three strains enter S-phase in the presence of MMS with similar kinetics based on budding morphology (SI Fig. 7). As expected, DNA synthesis is inhibited soon after entry into S-phase in wild-type, psy2Δ, and pph3Δ cells (Fig. 4B). However, psy2Δ and pph3Δ cells synthesize significantly less DNA than wild-type cells during incubation in MMS, consistent with hyperactivation of the checkpoint (Fig. 4B, time = 60/0). In addition, psy2Δ and pph3Δ cells require significantly more time than wild-type cells to complete replication after removal of MMS, with wild-type cells largely completing replication by 60 min, and cells lacking Pph3–Psy2 requiring at least 180 min (Fig. 4B and data not shown). psy2Δ and pph3Δ cells also require significantly more time than wild-type cells to reenter the cell cycle (SI Fig. 8). These data demonstrate that cells deficient in Pph3–Psy2 elicit a hyperactive intra-S DNA damage response and fail to resume DNA synthesis and reenter the cell cycle normally after the removal of MMS.
Fig. 4.
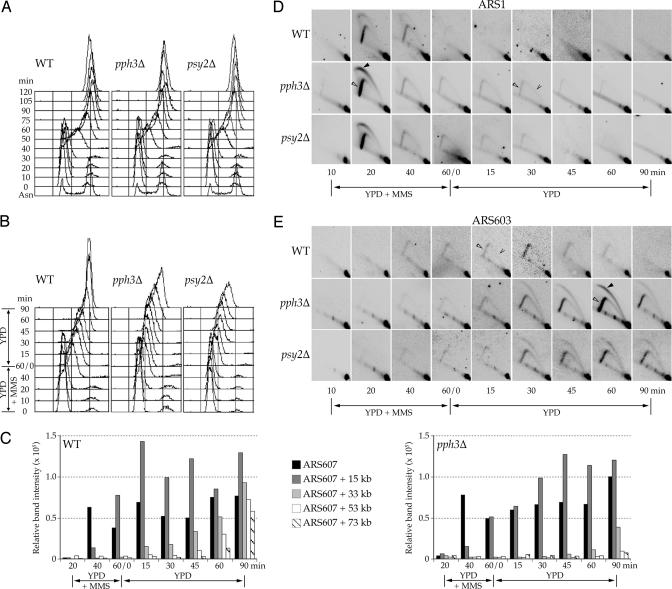
Pph3–Psy2 promotes efficient replication restart during recovery from MMS exposure in S phase. (A and B) DNA content analysis by flow cytometry of wild-type (SSy187), pph3Δ (SSy188), and psy2Δ (SSy189) cells that were blocked in G1 with α-factor and released into S-phase in YPD (A) or YPD with 0.033% MMS for 1 h, and then shifted to YPD (B). All steps were conducted at 30°C. (C) BrdU incorporation analysis of wild-type (SSy205) and pph3Δ (SSy210) cells treated as in B but grown at 23°C. Immunoprecipitated DNA sequences at ARS607 and at distances of 15, 33, 53, and 73 kb were detected by PCR amplification. (D and E) Two-dimensional gel analysis of samples collected in B that were digested with BamHI and NcoI. Blots were probed for ARS1 (D) or ARS603 (E). Filled arrowheads indicate large replication bubbles, open arrowheads indicate large replication forks, and caret indicates small and medium replication forks.
Pph3–Psy2 Promotes Restart of Stalled Replication Forks.
Rad53 activation during S-phase inhibits the initiation of unfired replication origins and prevents the collapse of replication forks encountering a damaged template, presumably by maintaining the association of replisome components with the fork structure (24–28). These functions of Rad53 enable the rapid resumption of DNA synthesis after removal of DNA damage, which is normally correlated with dephosphorylation of Rad53 (Fig. 3F and ref. 7). Thus, we hypothesized that dephosphorylation of Rad53 by Pph3–Psy2 facilitates the resumption of normal DNA synthesis after removal of MMS by regulating replication fork restart and/or the firing of replication origins. To directly examine replication fork progression in the absence of Pph3–Psy2 function, we used BrdU incorporation to monitor a replication fork that initiates at ARS607 and progresses across a 70-kb, origin-free region of chromosome VI. Upon entry into S-phase, BrdU is efficiently incorporated into ARS607 in wild-type and pph3Δ cells, reflecting normal entry into S-phase and initiation at this early origin (Fig. 4C). During incubation with MMS, both wild-type and pph3Δ cells incorporate BrdU at a DNA sequence 15 kb from ARS607 but do not incorporate BrdU at more distal sites, consistent with a reduced rate of elongation in the presence of DNA damage. Upon release from MMS, BrdU incorporation in wild-type cells at DNA sequences 33, 53, and 73 kb from the origin is detected at ≈15, 45, and 60 min, respectively. In pph3Δ cells, BrdU incorporation at these sequences is delayed at least 30 min, with replication at the most distal sequences beginning only after 90 min. These results demonstrate that Pph3–Psy2 promotes the resumption of DNA synthesis after MMS-induced fork arrest and suggest that this function is mediated by dephosphorylation of Rad53.
We next analyzed initiation of both early and late origins during and after exposure to MMS. As above, cells were released synchronously into S-phase in the presence of 0.033% MMS. Initiation of the early origin ARS1 occurs with similar timing and efficiency in wild-type, psy2Δ, and pph3Δ cells (Fig. 4D). In addition, initiation of the late origin ARS603 is blocked in wild-type, psy2Δ, and pph3Δ cells (Fig. 4E), demonstrating that Pph3–Psy2 is not required to inhibit late origin firing. Upon release of wild-type cells from MMS, ARS603 replication occurs “passively” by replication fork restart (Fig. 4E). In contrast, upon release of psy2Δ and pph3Δ cells from MMS, ARS603 replication is delayed. Replication in these mutants occurs not by restart of stalled replication forks but, rather, by initiation of ARS603 establishing new forks (Fig. 4E). Some replication forks persist at ARS1 in psy2Δ and pph3Δ cells (Fig. 4D), consistent with delayed replication restart of forks that initially stall near ARS1 during MMS treatment. Nevertheless, these forks appear stable, because we observed no evidence of fork collapse, such as broken bubbles or forks, which would appear as aberrantly migrating forms on the 2D gels. Together, these data strongly suggest that replication forks established at early origins in psy2Δ and pph3Δ cells are stable but unable to efficiently resume DNA synthesis after removal of MMS, and, instead, new forks are initiated from late origins to complete genome replication, despite the persistence of hyperphosphorylated Rad53.
γH2AX-Independent Regulation of Rad53 by Pph3–Psy2.
During the course of this study, it was reported that Psy2 forms a complex with Pph3 and Ybl046w that dephosphorylates γH2AX and that this function is required to inactivate Rad53 during prolonged recovery from a double-strand break (9). To further examine the suggested connection between γH2AX and Rad53 dephosphorylation, we examined the MMS sensitivity of an H2AX mutant that cannot be converted to γH2AX by phosphorylation (hta1-S129A). An additive increase in MMS sensitivity is observed when PPH3 is disrupted in the hta1-S129A mutant, demonstrating that Pph3 has γH2AX-independent functions (SI Fig. 9A). In addition, deletion of YBL046W, which is required for γH2AX dephosphorylation, does not impart the phenotypes observed with deletion of PPH3 or PSY2, including: sensitivity to MMS (Fig. 1B), genetic interactions with cell cycle checkpoint mutants (rad9Δ, mec1Δ, dun1Δ, or ptc2Δ; Fig. 2 A–D), and delayed reentry into the cell cycle during recovery from MMS treatment (SI Fig. 8). Most importantly, ybl046wΔ does not show a Rad53 dephosphorylation defect (Fig. 3F), despite its γH2AX dephosphorylation defect (ref. 9 and SI Fig. 9B). Thus, whereas Pph3–Psy2 requires Ybl046w to dephosphorylate γH2AX, it does not require Ybl046w to dephosphorylate Rad53, mechanistically separating dephosphorylation of Rad53 from that of γH2AX.
Discussion
Activation of Rad53 by phosphorylation and the initiation of cell cycle checkpoints are essential for the maintenance of genome integrity. Although the phosphorylation of Rad53 has been intensively studied, its deactivation, which is thought to be necessary for cells to reenter the cell cycle, is less understood. To date, the only phosphatases directly implicated in Rad53 deactivation are Ptc2 and Ptc3 (8). In their absence, cells suffering a persistent double-strand break are unable to adapt to, or recover from Rad53-mediated G2/M arrest. In addition, Rad53 dephosphorylation after induction of a repairable but long-lived double-strand break has been suggested to depend on dephosphorylation of γH2AX by a complex of Pph3, Psy2, and Ybl046w (9).
We demonstrate that Pph3 and Psy2 form a complex that regulates Rad53 activity in MMS-treated cells, in a manner independent of its role with Ybl046w in γH2AX dephosphorylation. Our results suggest that Ybl046w is a specificity factor for the Pph3–Psy2 phosphatase complex. That Rad53 dephosphorylation after repair of a double-strand break by means of single-stand annealing is independent of Pph3 in h2a-S129A cells (9) may reflect switching to an alternative, Pph3–Psy2-independent repair pathway (29, 30), perhaps an adaptation pathway involving Ptc2 and/or Ptc3 (8). This is consistent with the synergistic increase in MMS sensitivity of ptc2Δ cells caused by disruption of PSY2 or PPH3. That Pph3–Psy2 regulates Rad53 is supported by a body of evidence that includes: genetic analyses placing Pph3–Psy2 downstream of Rad53 in a role that antagonizes the DNA damage checkpoint response; yeast two-hybrid data indicating that Psy2 interacts with Rad53; and biochemical results demonstrating that Pph3–Psy2 can directly dephosphorylate Rad53 in vitro and that Pph3–Psy2 is required for dephosphorylation in vivo of activated Rad53 during recovery from MMS treatment in S-phase.
DNA damage checkpoint-mediated activation of Rad53 regulates DNA synthesis by stabilizing replication forks that are already initiated and by delaying the initiation of late firing origins. Both functions appear normal in cells bereft of Pph3–Psy2 (Fig. 4 D and E). However, replication forks fail to restart normally in psy2Δ or pph3Δ cells after removal of the genotoxic stress. The defect in replication fork restart may result from Rad53 remaining in a hyperphosphorylated state; however, a previous study indicated that fork progression rates are unaffected by the presence or absence of Rad53 (28), suggesting that dephosphorylation of other factors may also be involved. DNA replication eventually resumes through firing of late origins. This late-origin firing is consistent with, and likely indirectly enabled by, the delayed restart of early established forks that would otherwise passively replicate the late origin, as is observed with wild-type cells and is particularly remarkable, given the persistence of activated Rad53. These results suggest a functional uncoupling of the Rad53-dependent processes of fork stabilization and late-origin control. Precedence for such an uncoupling is provided by studies of the mec1–100 hypomorphic allele, which is proficient in stabilizing replication forks, but defective in preventing late-origin firing (7).
How might activated Rad53 independently regulate replication fork restart and late-origin firing? One possibility is that fork stabilization and the inhibition of late-origin firing are mediated by distinct Rad53 phosphorylation patterns and that these different phosphorylation patterns are recognized and regulated by different phosphatases. According to this model, Pph3–Psy2 recognizes a subset of Rad53 phosphorylation sites that are required to stabilize stalled replication forks, and thus, it is required to restart forks. However, the efficient activation of late origins during recovery from DNA damage appears to result from regulation of different Rad53 phosphorylation sites by another phosphatase(s). This model is supported by work that has shown that different facets of the checkpoint response controlled by Rad53 are mediated by different domains of the protein (31–33). Additional support for this model is provided by the observation that different Rad53 residues are phosphorylated in response to different types of DNA damage (18, 34). Thus, specific Rad53 phosphorylation sites may be induced by different types of damage, be regulated by different mechanisms, and mediate different aspects of the DNA damage response.
Another plausible mechanism separating Rad53-dependent fork stabilization from origin control is that the hyperphosphorylated Rad53 apparent in the absence of Pph3–Psy2 is sequestered at stalled forks, and thus unavailable to act elsewhere. Therefore, newly synthesized Dbf4 (and perhaps other factors) might escape inhibition by activated Rad53 and thus promote late-origin firing. This model is analogous to that proposed for hypomorphic Mec1–100, which is proficient for the stabilization of stalled forks but defective in late-origin control (7).
Checkpoint pathways are well conserved among eukaryotes. Rad53 is the S. cerevisiae homolog of the mammalian tumor suppressor Chk2 (35). In the presence of DNA damage, Chk2 is phosphorylated and activated by ATM (35) and then phosphorylates downstream effectors, including the tumor suppressors p53, BRCA1, and Cdc25 (35). Homologs of Pph3 and Psy2 have also been identified in higher eukaryotes; in humans the homologs are encoded by PP4 and PP4R3, respectively (14, 36). Interestingly, PP4R3 complements the cisplatin hypersensitivity of yeast lacking PSY2, indicating functional conservation through evolution (14). Furthermore, a conserved role of PP4 and PP4R3 as a Chk2 phosphatase complex is suggested by a yeast two-hybrid interaction between PP4R3 and Rad53 (14).
Further understanding the regulation of Rad53 in S. cerevisiae will provide additional insights into the checkpoint response. In particular, how different phosphorylation patterns of Rad53 are induced by different forms of genotoxic stress, how these sites are regulated (i.e., sequence specifically or by localization to specific replication structures), and their functional significance are of great interest. Further characterizing the role of Pph3–Psy2 in these processes, and determining whether Pph3–Psy2 dephosphorylates additional replication factors is also crucial to gaining a clearer understanding of the mechanisms of fork stabilization and origin control.
Materials and Methods
General Yeast Methods.
Yeast strains used in this study were constructed by standard methods (10) and are listed in SI Table 2. Experiments were carried out in rich media (YPD) supplemented with 30 mg of adenine at 30°C unless otherwise indicated. For G1 synchrony experiments, cells were arrested in YPD media containing α-factor (American Peptide, Sunnyvale, CA) for 2 or 2.5 h (pH was adjusted to 4.5 for BAR1 strains). Colony survival assays were performed by counting cfu 3 days after deposition onto YPD-agar media containing the indicated concentration of MMS (Figs. 1 and 2). Each point represents the mean of at least three independent experiments. Error bars represent standard deviations of the independent experiments. The Matchmaker yeast two-hybrid system (Clontech, Mountain View, CA) was used to analyze Psy2–Rad53-binding interactions. Details about the construction of plasmids used in this study are available upon request.
Protein Methods.
Tandem affinity purification, MudPIT, coimmunoprecipitation, and in vitro phosphatase assays were performed as described in SI Methods. Whole-cell protein extracts were prepared according to standard protocols (10) and normalized for total protein concentration by using Bio-Rad Protein Assay reagent (Bio-Rad, Hercules, CA). Protein samples were separated by SDS/PAGE, transferred to PVDF, and analyzed by Western blotting (described in SI Methods). Rad53 phosphorylation state was measured as changes in the electrophoretic mobility of Rad53.
Analysis of DNA Replication.
Sample preparation, flow cytometry, 2D gel analysis and BrdU incorporation experiments were performed as reported (22). Replication structure comparisons were performed on samples prepared concurrently to minimize any errors that might result from differential transfer or hybridization efficiencies. Sequences of primers used for PCR analyses of BrdU incorporation are available upon request.
Supplementary Material
Acknowledgments
We thank J. Chin and J. Heierhorst for critically reading the manuscript; M. C. Marsolier-Kergoat (Service de Biochimie et de Génétique Moléculaire, Gif-sur-Yvette, France) for Rad53 yeast two-hybrid vectors; R. Kolodner (University of California at San Diego, La Jolla, CA) and D. Durocher (University of Toronto, Toronto, ON, Canada) for yeast strains; J. Diffley (Cancer Research U.K., London, U.K.) for Rad53 expression vector; C. Viggiani (University of Southern California) for the BrdU incorporation vector; and M. Keogh (Albert Einstein College of Medicine, Bronx, NY) for α-γH2AX antibody. This work was supported by Tobacco-Related Disease Research Program Grant 14DT-0137 and the Achievement Rewards for College Scientists Foundation (B.M.O.) and National Institutes of Health Grants GM068569 (to F.E.R.) and GM065494 (to O.M.A.).
Abbreviation
- MMS
methylmethane sulfonate.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0703252104/DC1.
References
- 1.Kolodner RD, Putnam CD, Myung K. Science. 2002;297:552–557. doi: 10.1126/science.1075277. [DOI] [PubMed] [Google Scholar]
- 2.Nyberg KA, Michelson RJ, Putnam CW, Weinert TA. Annu Rev Genet. 2002;36:617–656. doi: 10.1146/annurev.genet.36.060402.113540. [DOI] [PubMed] [Google Scholar]
- 3.Lambert S, Carr AM. Biochimie. 2005;87:591–602. doi: 10.1016/j.biochi.2004.10.020. [DOI] [PubMed] [Google Scholar]
- 4.Branzei D, Foiani M. Curr Opin Cell Biol. 2005;17:568–575. doi: 10.1016/j.ceb.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 5.Ma J-L, Lee S-J, Duong JK, Stern DF. J Biol Chem. 2006;281:3954–3963. doi: 10.1074/jbc.M507508200. [DOI] [PubMed] [Google Scholar]
- 6.Duncker BP, Brown GW. Mut Res. 2003;532:21–27. doi: 10.1016/j.mrfmmm.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 7.Tercero JA, Longhese MP, Diffley JF. Mol Cell. 2003;11:1323–1336. doi: 10.1016/s1097-2765(03)00169-2. [DOI] [PubMed] [Google Scholar]
- 8.Leroy C, Lee SE, Vaze MB, Ochsenbien F, Guerois R, Haber JE, Marsolier-Kergoat MC. Mol Cell. 2003;11:827–835. doi: 10.1016/s1097-2765(03)00058-3. [DOI] [PubMed] [Google Scholar]
- 9.Keogh MC, Kim JA, Downey M, Fillingham J, Chowdhury D, Harrison JC, Onishi M, Datta N, Galicia S, Emili A, et al. Nature. 2006;439:497–501. doi: 10.1038/nature04384. [DOI] [PubMed] [Google Scholar]
- 10.O'Neill BM, Hanway D, Winzeler EA, Romesberg FE. Nucleic Acids Res. 2004;32:6519–6530. doi: 10.1093/nar/gkh994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Washburn MP, Wolters D, Yates JR., III Nat Biotech. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 12.Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, et al. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 13.Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, et al. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 14.Gingras A-C, Caballero M, Zarske M, Sanchez A, Hazbun TR, Fields S, Sonenberg N, Hafen E, Raught B, Aebersold R. Mol Cell Proteomics. 2005;4:1725–1740. doi: 10.1074/mcp.M500231-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Bashkirov VI, Bashkirova EV, Haghnazari E, Heyer W-D. Mol Cell Biol. 2003;23:1441–1452. doi: 10.1128/MCB.23.4.1441-1452.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uetz P, Giot L, Cagney G, Mansfield T, Judson R, Knight J, Lockshon D, Narayan V, Srinivasan M, Pochart P, et al. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 17.Gilbert CS, Green CM, Lowndes NF. Mol Cell. 2001;8:129–136. doi: 10.1016/s1097-2765(01)00267-2. [DOI] [PubMed] [Google Scholar]
- 18.Smolka MB, Albuquerque CP, Chen S-h, Schmidt KH, Wei XX, Kolodner RD, Zhou H. Mol Cell Proteomics. 2005;4:1358–1369. doi: 10.1074/mcp.M500115-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Foss EJ. Genetics. 2001;157:567–577. doi: 10.1093/genetics/157.2.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katou Y, Kanoh Y, Bando M, Noguchi H, Tanaka H, Ashikari T, Sugimoto K, Shirahige K. Nature. 2003;424:1078–1083. doi: 10.1038/nature01900. [DOI] [PubMed] [Google Scholar]
- 21.Osborn AJ, Elledge SJ. Genes Dev. 2003;17:1755–1767. doi: 10.1101/gad.1098303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szyjka SJ, Viggiani CJ, Aparicio OM. Mol Cell. 2005;19:691–697. doi: 10.1016/j.molcel.2005.06.037. [DOI] [PubMed] [Google Scholar]
- 23.Tourriere H, Versini G, Cordon-Preciado V, Alabert C, Pasero P. Mol Cell. 2005;19:699–706. doi: 10.1016/j.molcel.2005.07.028. [DOI] [PubMed] [Google Scholar]
- 24.Cobb JA, Bjergbaek L, Shimada K, Frei C, Gasser SM. EMBO J. 2003;22:4325–4336. doi: 10.1093/emboj/cdg391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lopes M, Cotta-Ramusino C, Pellicioli A, Liberi G, Plevani P, Muzi-Falconi M, Newlon CS, Foiani M. Nature. 2001;412:557–561. doi: 10.1038/35087613. [DOI] [PubMed] [Google Scholar]
- 26.Santocanale C, Diffley JF. Nature. 1998;395:615–618. doi: 10.1038/27001. [DOI] [PubMed] [Google Scholar]
- 27.Shirahige K, Hori Y, Shiraishi K, Yamashita M, Takahashi K, Obuse C, Tsurimoto T, Yoshikawa H. Nature. 1998;395:618–621. doi: 10.1038/27007. [DOI] [PubMed] [Google Scholar]
- 28.Tercero JA, Diffley JF. Nature. 2001;412:553–557. doi: 10.1038/35087607. [DOI] [PubMed] [Google Scholar]
- 29.Downs JA, Lowndes NF, Jackson SP. Nature. 2000;408:1001–1004. doi: 10.1038/35050000. [DOI] [PubMed] [Google Scholar]
- 30.Redon C, Pilch DR, Rogakou EP, Orr AH, Lowndes NF, Bonner WM. EMBO Rep. 2003;4:678–684. doi: 10.1038/sj.embor.embor871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pike BL, Tenis N, Heierhorst J. J Biol Chem. 2004;279:39636–39644. doi: 10.1074/jbc.M405080200. [DOI] [PubMed] [Google Scholar]
- 32.Pike BL, Yongkiettrakul S, Tsai M-D, Heierhorst J. J Biol Chem. 2003;278:30421–30424. doi: 10.1074/jbc.C300227200. [DOI] [PubMed] [Google Scholar]
- 33.Schwartz MF, Lee SJ, Duong JK, Eminaga S, Stern DF. Cell Cycle. 2003;2:384–396. [PubMed] [Google Scholar]
- 34.Sweeney FD, Yang F, Chi A, Shabanowitz J, Hunt DF, Durocher D. Curr Biol. 2005;15:1364–1375. doi: 10.1016/j.cub.2005.06.063. [DOI] [PubMed] [Google Scholar]
- 35.Bartek J, Falck J, Lukas J. Nat Rev Mol Cell Biol. 2001;2:877–886. doi: 10.1038/35103059. [DOI] [PubMed] [Google Scholar]
- 36.Cohen PTW, Philp A, Vazquez-Martin C. FEBS Lett. 2005;579:3278–3286. doi: 10.1016/j.febslet.2005.04.070. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.