Abstract
Stromal interaction molecule 1 (STIM1) has recently been identified by our group and others as an endoplasmic reticulum (ER) Ca2+ sensor that responds to ER Ca2+ store depletion and activates Ca2+ channels in the plasma membrane (PM). The molecular mechanism by which STIM1 transduces signals from the ER lumen to the PM is not yet understood. Here we developed a live-cell FRET approach and show that STIM1 forms oligomers within 5 s after Ca2+ store depletion. These oligomers rapidly dissociated when ER Ca2+ stores were refilled. We further show that STIM1 formed oligomers before its translocation within the ER network to ER–PM junctions. A mutant STIM1 lacking the C-terminal polybasic PM-targeting motif oligomerized after Ca2+ store depletion but failed to form puncta at ER–PM junctions. Using fluorescence recovery after photobleaching measurements to monitor STIM1 mobility, we show that STIM1 oligomers translocate on average only 2 μm to reach ER–PM junctions, arguing that STIM1 ER-to-PM signaling is a local process that is suitable for generating cytosolic Ca2+ gradients. Together, our live-cell measurements dissect the STIM1 ER-to-PM signaling relay into four sequential steps: (i) dissociation of Ca2+, (ii) rapid oligomerization, (iii) spatially restricted translocation to nearby ER–PM junctions, and (iv) activation of PM Ca2+ channels.
Keywords: Ca2+ release-activated Ca2+, fluorescence recovery after photobleaching, FRET, store-operated Ca2+ influx
Ca2+ signals are used by cells for transducing receptor or electrical inputs into functional outputs such as gene expression, contraction, and secretion (1). Although endoplasmic reticulum (ER) Ca2+ stores are the main source for inositol trisphosphate-induced transient Ca2+ signals, a necessary step for activation of T lymphocytes, mast cells, and many other cells is a persistent increase in Ca2+ concentration (2). These sustained Ca2+ signals require retrograde signaling from the lumen of the ER to Ca2+ channels in the plasma membrane (PM) in a process called store-operated Ca2+ (SOC) influx [also termed Ca2+ release-activated Ca2+ (CRAC) in immune cells] (3). The Stauderman/Cahalan groups and our group have recently identified the single transmembrane protein stromal interaction molecule 1 (STIM1) as an ER Ca2+ sensor that responds to the depletion of ER Ca2+ and activates SOC/CRAC channels in the PM (4–6). STIM1 senses Ca2+ by an EF hand Ca2+-binding site in the lumen of the ER (7). A recombinant fragment of the luminal region of STIM1 has been shown to form dimers and oligomers in the absence of Ca2+ in vitro (8). Overexpression of STIM1 mutants with a disrupted EF hand Ca2+-binding motif resulted in constitutive activation of Ca2+ influx (5, 6, 9). It was also observed that depletion of ER Ca2+ stores induced STIM1 translocation to punctate structures near the PM that correspond to ER–PM junctions (5, 10). STIM1 colocalizes at these junctions with the CRAC channel Orai1 (also known as CRACM1) and synergistically activates SOC/CRAC influx (11–21). Although these studies suggested possible scenarios for STIM1 signal transduction, the molecular steps by which STIM1 signals from the ER to the PM are not yet understood.
To elucidate the molecular mechanism of this STIM1 ER-to-PM signaling relay, we used FRET and fluorescence recovery after photobleaching live-cell measurements to dynamically monitor how STIM1 is activated by ER Ca2+ store depletion. We found that depletion of ER Ca2+ rapidly induces STIM1 oligomerization and that STIM1 oligomerization is followed by a much slower translocation of the oligomers to ER–PM junctions. These two processes could be separated by a mutant STIM1 without the C-terminal polybasic motif. This mutant still oligomerized but failed to translocate to ER–PM junctions. We also found that the translocation of STIM1 oligomers is spatially restricted, arguing that ER-to-PM signaling by STIM1 is a local process and that SOC influx can be used to generate local Ca2+ signals and Ca2+ gradients in cells. Together, our results provide evidence for a model that STIM1 functions as a four-step ER-to-PM signaling relay that triggers local SOC entry signals by sequential Ca2+ sensing, oligomerization, PM translocation, and SOC/CRAC channel activation.
Results
A Physiological Receptor Stimulus Triggered Both STIM1 Oligomerization and Puncta Formation.
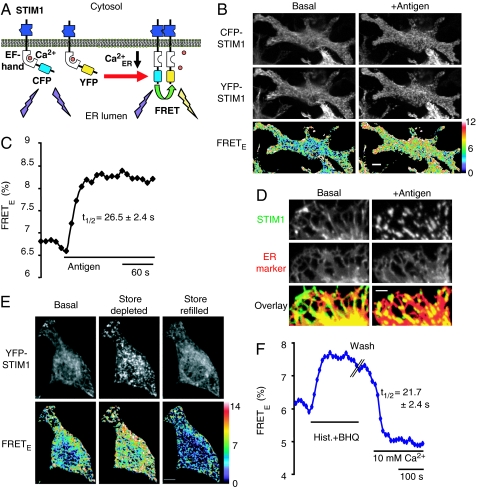
To test whether lowering of ER Ca2+ induces STIM1 oligomerization in live cells, we used FRET imaging (Fig. 1A) to detect the interaction between cyan fluorescent protein (CFP)- and yellow fluorescent protein (YFP)-conjugated STIM1 in RBL cells (a tumor mast cell line) and in HeLa cells. CFP, YFP, and FRET image triplets were taken on a Nipkow confocal microscope using 442- and 514-nm laser excitation. The apparent FRET efficiency (FRETE) between CFP-STIM1 and YFP-STIM1 was calculated on a pixel-by-pixel basis by using a method described previously (22). The FRETE parameter is independent of the CFP-STIM1 concentration and linearly correlates with the fraction of CFP-STIM1 that interacts with YFP-STIM1. Strikingly, antigen stimulation of FcεRI receptors in RBL cells, which induces inositol trisphosphate-mediated Ca2+ release from ER stores, triggered significant increases in STIM1 FRETE signals as well as the formation of STIM1 puncta near the PM (Fig. 1 B and C). We also performed FRET measurements by acceptor photobleaching of YFP-STIM1 and confirmed that stimulus-induced STIM1 FRETE increases indeed reflect an increase in energy transfer [see supporting information (SI) Fig. 5]. Consistent with earlier results (5, 10, 21), the YFP-STIM1 puncta that were formed near the PM overlapped with a luminal ER marker in RBL cells (Fig. 1D). These results with antigen stimulation in RBL cells are the first demonstration that a physiological receptor stimulus is sufficient to trigger STIM1 oligomerization as well as STIM1 puncta formation at ER–PM junctions.
Fig. 1.
ER Ca2+ controls STIM1 oligomerization and puncta formation. (A) Schematic representation of a FRET-based assay for STIM1 oligomerization. (B) STIM1 oligomerization and puncta formation in YFP-STIM1 and CFP-STIM1 cotransfected RBL cells. Confocal images were acquired near the adhesion surface before and 90 s after antigen (2 μg/ml dinitrophenol-BSA) stimulation. (Scale bar, 10 μm.) (C) The average STIM1 FRETE trace of 31 RBL cells. The average and SE of t1/2 are shown. (D) Confocal images of YFP-STIM1 and CFP-ER marker cotransfected RBL cells were acquired near the adhesion surface before and 90 s after antigen stimulation. The overlay images show the colocalization of STIM1 (green) and the ER marker (red). (Scale bar, 5 μm.) (E) YFP-STIM1 and CFP-STIM1 cotransfected HeLa cells were stimulated with 100 μM histamine (Hist.) plus 5 μM BHQ in a Ca2+-free buffer for 4 min. Cells were then washed three times, and 10 mM Ca2+ was added back. Confocal images were acquired near the adhesion surface before (Basal), 75 s after stimulation (Store depleted), and 70 s after Ca2+ readdition (Store refilled). (Scale bar, 10 μm.) (F) The average STIM1 FRETE trace of 10 HeLa cells. The average and SE of t1/2 are shown.
STIM1 Oligomerization and Puncta Formation Are Reversibly Controlled by Depletion of ER Ca2+.
Because antigen receptor stimulation induces not only inositol trisphosphate production but also other signaling events, we used an ER Ca2+ pump inhibitor, thapsigargin, to more selectively deplete ER Ca2+ stores. We then determined whether a reduction in ER Ca2+ is sufficient to trigger STIM1 oligomerization and found similar FRETE increases in thapsigargin-treated HeLa cells and RBL cells (SI Fig. 6). We also used a reversible ER Ca2+ pump inhibitor, 2,5-di-(t-butyl)-1,4-hydroquinone (BHQ), to investigate whether STIM1 oligomerization and puncta formation are reversibly regulated by ER Ca2+ levels. Combined addition of histamine and BHQ triggered a rapid increase in FRETE and STIM1 puncta formation in HeLa cells (Fig. 1E and F). These store-depleted cells were then washed in a Ca2+-free buffer without BHQ and histamine before 10 mM Ca2+ was added to the outside of the cells to force a rapid refilling of Ca2+ stores. The increase in FRETE showed a marked return to below baseline with a half-time of 22 s after the readdition of Ca2+ (Fig. 1F). This decrease in FRETE was paralleled by a loss in STIM1 puncta near the PM (Fig. 1E). We also found that BHQ alone is sufficient to induce a reversible FRETE increase and STIM1 puncta formation in RBL cells (SI Fig. 7). The reversal of the STIM1 FRETE increase and puncta formation during store refilling is consistent with the interpretation that Ca2+ binding to the luminal EF hand of STIM1 rapidly dissociates STIM1 oligomers and at the same time suppresses STIM1 localization to ER–PM junctions. We note that in some cells, such as those shown in SI Fig. 7, a fraction of STIM1 appeared to colocalize not only with the ER but also with microtubule-like structures, but the significance of this localization is not yet known.
STIM1 Oligomerization and Puncta Formation are Sequential Processes.
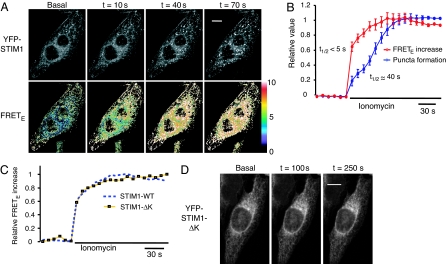
The coregulation of STIM1 oligomerization and STIM1 puncta formation by ER Ca2+ levels prompted us to address the relationship between these two processes. We used 10 μM ionomycin, a Ca2+ ionophore, to more rapidly deplete ER Ca2+ to test whether the STIM1 FRETE increase and puncta formation can be kinetically separated. We found that the time required for a half-maximal FRETE increase after addition of ionomycin was <5 s, but it took ≈40 s to observe significant puncta formation in the same cells (Fig. 2 A and B). Of note, FRETE signals were elevated across the ER and were not restricted to locations where puncta formation subsequently occurred (e.g., t = 10 s panel in Fig. 2A), suggesting that STIM1 first forms oligomers across the ER and that these oligomers subsequently translocate to ER–PM junctions. In these experiments, EGTA was added together with ionomycin to suppress prolonged cytosolic Ca2+ increases and preserve the ER integrity during the experiment (SI Fig. 8).
Fig. 2.
STIM1 oligomerization and translocation to ER–PM junctions are sequential processes. (A) YFP and FRETE images of a YFP-STIM1 and CFP-STIM1 coexpressing HeLa cell acquired near the adhesion surface after 10 μM ionomycin stimulation in a Ca2+-free buffer. (Scale bar, 20 μm.) (B) A kinetic comparison of STIM1 FRETE increases and puncta formation in the same cells. The average FRETE responses of 28 ionomycin-stimulated cells are shown. STIM1 puncta formation was monitored in these cells by measuring the average granule intensity in each cell by using a Gaussian filter (see Materials and Methods). SEs are shown. (C) The average FRETE trace of 29 CFP-STIM1-ΔK and YFP-STIM1-ΔK cotransfected HeLa cells was compared with the wild-type STIM1 FRETE trace shown in B. (D) Confocal images of a YFP-STIM1-ΔK transfected HeLa cell acquired near the adhesion surface after 10 μM ionomycin stimulation in a Ca2+-free buffer. There was no puncta formation in all ionomycin-stimulated YFP-STIM1-ΔK-transfected HeLa cells examined (>100 cells from 30 experiments). (Scale bar, 20 μm.)
A Mutant STIM1 That Lacks the C-Terminal Polybasic Motif Separates the Oligomerization Process from Translocation to ER–PM Junctions.
We further tested whether the oligomerization and translocation processes can be separated by using a mutant STIM1 protein. STIM1 has a polybasic sequence motif at the C terminus, which has been shown in other proteins to function as a PM-targeting motif that binds polyphosphoinositides in the PM (23). This same polybasic region has been shown previously to be necessary for STIM1 function (24). We constructed a mutant STIM1 that lacked this polybasic motif (STIM1-ΔK) and tagged it with CFP and YFP to test whether this region is important for oligomerization and for translocation of STIM1 to ER–PM junctions in response to Ca2+ store depletion. Interestingly, the increase in FRETE after ionomycin addition in cells coexpressing CFP-STIM1-ΔK and YFP-STIM1-ΔK mutants was indistinguishable from that with the wild-type STIM1 pairs (Fig. 2C). In contrast, the STIM1-ΔK mutant failed to form puncta near the PM in all cells after Ca2+ store depletion (>100 cells in 30 experiments) (Fig. 2D). These results demonstrate that the polybasic motif is important for the recruitment of STIM1 to ER–PM junctions but is not required for oligomerization. By uncoupling STIM1 oligomerization from the translocation to ER–PM junctions, the STIM1-ΔK mutant provides direct evidence that oligomerization and translocation are separate steps in STIM1 signal transduction. Furthermore, the STIM1-ΔK mutant also identifies the polybasic region of STIM1 as a motif that targets STIM1 oligomers to ER–PM junctions. By analogy with similar polybasic motifs in other proteins, this peptide may target STIM1 to ER–PM junctions by binding PM polyphosphoinositides.
Consistent with such a role of the polybasic region, a FRETE increase was also observed in HeLa cells coexpressing the wild-type CFP-STIM1 and the YFP-STIM1-ΔK mutant after Ca2+ store depletion (SI Fig. 9A). Notably, when the STIM1-ΔK mutant was coexpressed with the wild-type STIM1, the YFP-STIM1-ΔK mutant now translocated to puncta, albeit to a lesser extent compared with the coexpressed wild-type CFP-STIM1 (SI Fig. 9B). These results indicate that the STIM1-ΔK mutant can oligomerize with the wild-type STIM1 and be recruited to the PM by the polybasic tail of the wild-type STIM1.
STIM1 Signal Transduction Is a Local Process.
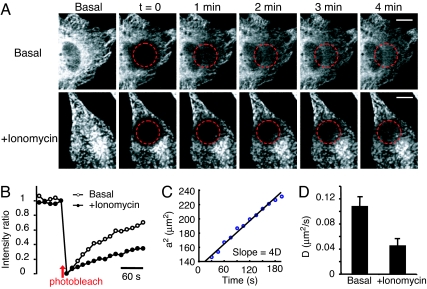
The mechanistic separation of oligomerization and translocation requires that STIM1 oligomers, once formed, diffuse along the ER network to be recruited to ER–PM junctions. We monitored the mobility of YFP-STIM1 by fluorescence recovery after photobleaching measurements and determined how quickly YFP-STIM1 diffuses into a region from which the fluorescence of YFP-STIM1 has been bleached by a brief 514-nm laser pulse. As shown in Fig. 3A, YFP-STIM1 translocation back into the bleached region was relatively slow before Ca2+ store depletion and became even slower after Ca2+ store depletion. This decrease in the diffusion is probably due to the increased size of STIM1 oligomers and/or the interaction of STIM1 with the PM. When we selected two regions 5 μm apart within the bleach profile and calculated the relative intensity equilibration of these two regions, we found that the time required for STIM1 fluorescence recovery either before or after Ca2+ store depletion was >2 min, which is longer than the time required for puncta formation (Fig. 3B). This result indicates that the distance of recruitment for STIM1 puncta formation is <5 μm. To determine the average distance over which STIM1 is recruited to ER–PM junctions, we measured the apparent diffusion coefficient (D) by fitting the two-dimensional relative fluorescence intensity profiles (I) to a Gaussian function, I = exp (−(x2 + y2)/a2) and calculating the increase in the square of the Gaussian peak radius (a2) as YFP-STIM1 diffused back into the bleached area (25). Fig. 3C shows the increase in a2 as a function of time (t) that was used to calculate the diffusion coefficient (slope = 4D) of YFP-STIM1 under the basal condition (25). The average apparent diffusion coefficients of YFP-STIM1 were ≈0.1 and 0.05 μm2/s before and after ER Ca2+ depletion, respectively (Fig. 3D). This diffusion of STIM1 is relatively slow compared with that of other ER membrane proteins (26), which may result from the large size of its cytosolic domain and/or by possible interactions with other proteins. The average distance of recruitment for STIM1 puncta formation was then calculated by using 〈average distance of recruitment〉 = (4Dt/π)1/2 (27) and was found to be between 1.6 and 2.3 μm for the 40-s time period required for STIM1 to accumulate at puncta near the PM. Thus, STIM1 functions as a locally acting ER Ca2+ sensor that transmits signals over an average distance of only 2 μm to a nearby ER–PM junction in HeLa cells. This result suggests that STIM1 can locally regulate SOC influx in response to a local drop in ER Ca2+ concentration. Together with earlier findings of gradients in SOC influx (28), this finding provides a mechanistic explanation of how cells can use STIM1 signaling to create spatially restricted Ca2+ signals and Ca2+ gradients.
Fig. 3.
STIM1 ER-to-PM signaling is a local process. (A) YFP-STIM1 diffusion in basal and ionomycin-treated cells monitored by fluorescence recovery after photobleaching measurements. (Scale bar, 10 μm.) (B) The average YFP-STIM1 intensity ratio of two sites inside and outside of the photobleached region (5 μm apart; n = 8 for each condition). (C) A series of relative Gaussian intensity profiles were fit through images such as those shown in A, and the increase in the square of the Gaussian peak radius (a2) was tracked over time to derive an apparent diffusion coefficient. (D) The average diffusion coefficients of YFP-STIM1 before and after Ca2+ store depletion (n = 5 for each condition; SEs are shown).
Discussion
STIM1 ER-to-PM Signaling Is a New Paradigm for Signal Transduction.
Our results show that STIM1 activation has an intriguing analogy to growth factor and immune cell receptors that are activated by ligand-mediated dimerization and oligomerization (29–31). However, several key aspects of STIM1 activation are different from the activation process of other receptors, arguing that STIM1 activation is a new paradigm for cell signaling. First, Ca2+ sensing occurs in the lumen of the ER and not outside of the cell as for these other receptors. Second, oligomerization is triggered by Ca2+ dissociation and not by ligand binding. Third, a subsequent translocation step is required for STIM1 oligomers to activate effectors at ER–PM junctions. We provide in this study two lines of evidence separating the oligomerization and the translocation processes. We found a significant kinetic difference between oligomerization and translocation to ER–PM junctions (<5 s vs. 40 s), and we were able to show a direct separation of the two processes by using a mutant STIM1 that could still oligomerize but failed to translocate to ER–PM junctions.
Ca2+-Sensing and Rapid STIM1 Oligomerization.
Our study is the first to show that STIM1 rapidly forms oligomers after Ca2+ store depletion in live cells and that this oligomerization process is reversibly regulated by ER Ca2+ levels. These results are supported by a recent in vitro biochemical study, which showed that the luminal portion of STIM1 forms oligomers at low but not at high Ca2+ concentrations (8). Nevertheless, the molecular requirements for STIM1 oligomerization are still controversial. An earlier report has shown that this same luminal region of STIM1 is not necessary for the formation of STIM1 oligomers (32), and a third study argues for a necessary role of the cytosolic coiled-coil region for oligomerization (33). Our method to monitor STIM1 oligomerization by FRET in live cells is useful to track the oligomerization process over time and will be helpful to identify the structural requirements for the oligomerization process.
Signaling from the ER to the PM.
Our study shows that the polybasic motif at the C terminus of STIM1 that has previously been shown to be necessary for activation of SOC influx (24) is not necessary for oligomerization but is required for STIM1 translocation to ER–PM junctions. This result not only separates the oligomerization from the translocation process but strongly argues that puncta formation at ER–PM junctions is an essential step in STIM1 activation. On the basis of these results, we propose that Ca2+ depletion-triggered STIM1 oligomerization exposes or creates a high-affinity PM-targeting motif that includes this polybasic tail, which resembles known polyphosphoinositide binding peptides (34). Through direct interaction of this motif with PM-localized polyphosphoinositides, STIM1 oligomers can then effectively accumulate at ER–PM junctions. Because this polybasic region is conserved in vertebrates (SI Fig. 10) but is missing in Drosophila STIM1, it is plausible that STIM1 interaction with polyphosphoinositides evolved as a mechanism to facilitate PM targeting and thereby enhance interactions with PM-localized effectors. Our study further showed that the rapidly formed STIM1 oligomers diffuse slowly within the ER network with a diffusion constant of 0.05 μm2/s. We estimated that activated STIM1 translocates only to ER–PM junctions that are <2 μm away. Therefore, STIM1 signaling is spatially restricted.
Recent studies showed binding interactions between STIM1 and the PM Ca2+ channel Orai1 (14, 16). Such a direct regulatory role of STIM1 for Orai1 is supported by a >10-fold up-regulation of SOC influx in cells overexpressing both STIM1 and Orai1 (13, 19–21). Two recent reports also showed coclustering of STIM1 and Orai1 at ER–PM junctions (17, 18). Together with the data shown here that the polyphosphoinositide interaction motif from STIM1 is required for translocation, it is likely that oligomerization and translocation of STIM1 to ER–PM junctions both precede the recruitment and activation of the Ca2+ channel Orai1 and possibly other plasma membrane effectors such as TRPC1 (24, 35). This result suggests that the activation process of STIM1 can be separated into a series of distinguishable steps.
The STIM1 ER-to-PM Signaling Relay Has Four Steps.
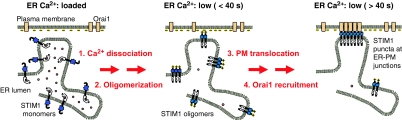
Our study provides evidence for a four-step activation process for STIM1 that can be understood as a “STIM1 ER-to-PM signaling relay” (Fig. 4). These four steps are as follows. (i) Receptor-induced reduction of ER Ca2+ concentration lowers the Ca2+ occupancy of the luminal EF hand of STIM1. (ii) STIM1 without bound Ca2+ rapidly forms oligomers along the ER network. (iii) The oligomerization of STIM1 exposes a C-terminal polybasic PM-targeting motif, which leads to the recruitment of STIM1 oligomers to nearby ER–PM junctions in a diffusion-limited step. This STIM1 recruitment along the ER network is spatially restricted to a 2-μm region surrounding each ER–PM junction. (iv) At ER–PM junctions, STIM1 recruits and activates the Ca2+ channel Orai1 and possibly other PM signaling proteins. Finally, our finding that this four-step STIM1 ER-to-PM signaling relay operates locally provides a mechanistic explanation for how cells may use the SOC influx pathway to generate local Ca2+ signals and cytosolic Ca2+ gradients.
Fig. 4.
Schematic representation of the four steps that define the STIM1 ER-to-PM signaling relay. (Left) When ER stores are loaded with Ca2+, Ca2+ is bound to the EF hand of STIM1, preventing STIM1 oligomerization. (Center) In the first two activation steps, a reduction in luminal ER Ca2+ leads to Ca2+ dissociation from the EF hand, which triggers a rapid oligomerization of STIM1. The oligomerization of STIM1 exposes the C-terminal polybasic PM-targeting motif. (Right) In the next two steps, STIM1 oligomers are recruited via diffusion along the ER network to nearby ER–PM junctions where STIM1 interacts with the PM Ca2+ channel Orai1 and activates SOC influx.
Materials and Methods
Cells, DNA Constructs, and Reagents.
HeLa and RBL cells were purchased from American Type Culture Collection (Manassas, VA). For antigen stimulation, RBL cells were incubated overnight with 0.2 μg/ml anti-dinitrophenol IgE (Sigma, St. Louis, MO) before dinitrophenol–BSA (Sigma) stimulation. YFP-STIM1 and CFP-STIM1 expression constructs have been described (5). CFP-STIM1-ΔK and YFP-STIM1-ΔK mutants were generated by introducing a stop codon at amino acid position 671 of human STIM1. pECFP-ER plasmid was obtained from Clontech. (Mountain View, CA). DNA plasmids were transfected into HeLa or RBL cells by using AMAXA electroporation protocols and solutions (AMAXA Biosystems, Cologne, Germany). BHQ and histamine were acquired from EMD BioSciences (San Diego, CA). Ionomycin was purchased from Sigma, and thapsigargin was purchased from Invitrogen (Carlsbad, CA).
Fluorescence Imaging and Photobleaching.
CFP/YFP/FRET images were acquired with a spinning disk confocal microscope (Nipkow Wallac system; PerkinElmer, Waltham, MA) using a ×40 objective. Cells were imaged in extracellular buffer (5) at room temperature. To better visualize near PM STIM1 puncta, shown images were taken near the adhesion surface of the cells. For photobleaching experiments, the focal plane of a second 100-mW, 514-nm YFP laser was set to create a 19-μm bleach region within the image plane. A 5-s pulse was used to locally photobleach ≈95% of the YFP intensity. Metamorph software (Molecular Devices, Sunnyvale, CA) was used for image analysis.
The time course of puncta formation in Fig. 2 was analyzed by monitoring the increase in average granularity in YFP-STIM1 intensity. Granularity was measured as the average cellular fluorescence intensity following Gaussian filtering (Image Analysis Toolbox for MATLAB; Mathworks, Natick, MA).
FRET Measurements.
CFP, YFP, and FRET image triplets were acquired in CFP-STIM1 and YFP-STIM1 cotransfected cells. The FRET image was taken by using CFP excitation and YFP emission. Intermolecular FRET between CFP-STIM1 and YFP-STIM1 was measured on a pixel-by-pixel basis by using a two-step FRET protocol developed by Zal and Gascoigne (22). First, we corrected for bleed-through and determined a corrected FRET image (Fc): Fc = IDA − dIDD − aIAA. In this equation, IDD, IAA and IDA correspond to the background-subtracted CFP, YFP, and FRET images, respectively. The two microscope-specific bleed-through parameters, d and a, were determined by using cells transfected with CFP or YFP alone. The derived values were d = IDA/IDD = 0.47 ± 0.02 and a = IDA/IAA = 0.09 ± 0.01.
In the second step, we calculated FRETE by using the following algorithm: FRETE = Fc/(Fc + GIDD). FRETE is the “true” FRET efficiency that measures the fraction of CFP exhibiting FRET. The value for the microscope-specific constant G was derived by measuring a CFP fluorescence increase after YFP acceptor photobleaching by using an intramolecular CFP-YFP FRET probe: G = 2.10 ± 0.15. FRETE images were displayed in a color-coded scale. A 3 × 3 low-pass filter was used to remove single-pixel noise in shown FRETE images.
Measurements of FRETE in unstimulated RBL cells was 6.5% as shown in Fig. 1C, probably reflecting a low degree of oligomerization of the overexpressed STIM1 proteins as well as collision FRET between diffusing CFP-STIM1 and YFP-STIM1 proteins. The observed undershot below the baseline in FRETE in Fig. 1F is probably a result of a higher than basal ER Ca2+ load due to the 10 mM Ca2+ refilling protocol used to accelerate ER store filling.
Diffusion Analysis by Fluorescence Recovery After Photobleaching.
Relative intensity equilibration after photobleaching was calculated from the ratio of the intensities of two regions 5 μm apart within the bleach profile. The ratio derived from the image acquired immediately before photobleaching was set to 1, and the ratio derived from the image taken immediately after photobleaching was set to 0. Diffusion coefficients were calculated by using two-dimensional Gaussian fits of the relative intensity in a series of images after the photobleaching pulse. The same fitting approach has been described previously for the case of one dimension (23). Specifically, cells were cotransfected with CFP-STIM1 and YFP-STIM1, and YFP images were first divided by CFP images to obtain normalized YFP intensities. By using the normalized intensities (I), the increasing radius square (a2) of the Gaussian bleach area was calculated by fitting the function I = exp[−(x2 + y2)/a2] through each of the images. The slope of this graph is a direct measure the diffusion coefficient (D) because the square of the Gaussian bleach radius (a2) increases linearly with 4D as a function of time (a2 + a02 = 4Dt, with a0 as the initial bleach radius).
Supplementary Material
Acknowledgments
We thank Angela Hahn and Jia-Yun Chen for carefully reading the manuscript. This work was supported by National Institutes of Health Grant GM030179 and the Sandler Foundation.
Abbreviations
- STIM1
stromal interaction molecule 1
- PM
plasma membrane
- ER
endoplasmic reticulum
- SOC
store-operated Ca2+
- CRAC
Ca2+ release-activated Ca2+
- CFP
cyan fluorescent protein
- YFP
yellow fluorescent protein
- BHQ
2,5-di-(t-butyl)-1,4-hydroquinone.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0702866104/DC1.
References
- 1.Berridge MJ, Lipp P, Bootman MD. Nat Rev Mol Cell Biol. 2000;1:11–21. doi: 10.1038/35036035. [DOI] [PubMed] [Google Scholar]
- 2.Lewis RS. Annu Rev Immunol. 2001;19:497–521. doi: 10.1146/annurev.immunol.19.1.497. [DOI] [PubMed] [Google Scholar]
- 3.Parekh AB, Putney JW., Jr Physiol Rev. 2005;85:757–810. doi: 10.1152/physrev.00057.2003. [DOI] [PubMed] [Google Scholar]
- 4.Roos J, DiGregorio PJ, Yeromin AV, Ohlsen K, Lioudyno M, Zhang S, Safrina O, Kozak JA, Wagner SL, Cahalan MD, et al. J Cell Biol. 2005;169:435–445. doi: 10.1083/jcb.200502019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liou J, Kim ML, Heo WD, Jones JT, Myers JW, Ferrell JE, Jr, Meyer T. Curr Biol. 2005;15:1235–1241. doi: 10.1016/j.cub.2005.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang SL, Yu Y, Roos J, Kozak JA, Deerinck TJ, Ellisman MH, Stauderman KA, Cahalan MD. Nature. 2005;437:902–905. doi: 10.1038/nature04147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Williams RT, Manji SS, Parker NJ, Hancock MS, Van Stekelenburg L, Eid JP, Senior PV, Kazenwadel JS, Shandala T, Saint R, et al. Biochem J. 2001;357:673–685. doi: 10.1042/0264-6021:3570673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stathopulos PB, Li GY, Plevin MJ, Ames JB, Ikura M. J Biol Chem. 2006;281:35855–35862. doi: 10.1074/jbc.M608247200. [DOI] [PubMed] [Google Scholar]
- 9.Spassova MA, Soboloff J, He LP, Xu W, Dziadek MA, Gill DL. Proc Natl Acad Sci USA. 2006;103:4040–4045. doi: 10.1073/pnas.0510050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu MM, Buchanan J, Luik RM, Lewis RS. J Cell Biol. 2006;174:803–813. doi: 10.1083/jcb.200604014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feske S, Gwack Y, Prakriya M, Srikanth S, Puppel SH, Tanasa B, Hogan PG, Lewis RS, Daly M, Rao A. Nature. 2006;441:179–186. doi: 10.1038/nature04702. [DOI] [PubMed] [Google Scholar]
- 12.Vig M, Peinelt C, Beck A, Koomoa DL, Rabah D, Koblan-Huberson M, Kraft S, Turner H, Fleig A, Penner R, et al. Science. 2006;312:1220–1223. doi: 10.1126/science.1127883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang SL, Yeromin AV, Zhang XH, Yu Y, Safrina O, Penna A, Roos J, Stauderman KA, Cahalan MD. Proc Natl Acad Sci USA. 2006;103:9357–9362. doi: 10.1073/pnas.0603161103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yeromin AV, Zhang SL, Jiang W, Yu Y, Safrina O, Cahalan MD. Nature. 2006;443:226–229. doi: 10.1038/nature05108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prakriya M, Feske S, Gwack Y, Srikanth S, Rao A, Hogan PG. Nature. 2006;443:230–233. doi: 10.1038/nature05122. [DOI] [PubMed] [Google Scholar]
- 16.Vig M, Beck A, Billingsley JM, Lis A, Parvez S, Peinelt C, Koomoa DL, Soboloff J, Gill DL, Fleig A, et al. Curr Biol. 2006;16:2073–2079. doi: 10.1016/j.cub.2006.08.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luik RM, Wu MM, Buchanan J, Lewis RS. J Cell Biol. 2006;174:815–825. doi: 10.1083/jcb.200604015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu P, Lu J, Li Z, Yu X, Chen L, Xu T. Biochem Biophys Res Commun. 2006;350:969–976. doi: 10.1016/j.bbrc.2006.09.134. [DOI] [PubMed] [Google Scholar]
- 19.Peinelt C, Vig M, Koomoa DL, Beck A, Nadler MJ, Koblan-Huberson M, Lis A, Fleig A, Penner R, Kinet JP. Nat Cell Biol. 2006;8:771–773. doi: 10.1038/ncb1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soboloff J, Spassova MA, Tang XD, Hewavitharana T, Xu W, Gill DL. J Biol Chem. 2006;281:20661–20665. doi: 10.1074/jbc.C600126200. [DOI] [PubMed] [Google Scholar]
- 21.Mercer JC, Dehaven WI, Smyth JT, Wedel B, Boyles RR, Bird GS, Putney JW., Jr J Biol Chem. 2006;281:24979–24990. doi: 10.1074/jbc.M604589200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zal T, Gascoigne NR. Biophys J. 2004;86:3923–3939. doi: 10.1529/biophysj.103.022087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heo WD, Inoue T, Park WS, Kim ML, Park BO, Wandless TJ, Meyer T. Science. 2006;314:1458–1461. doi: 10.1126/science.1134389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang GN, Zeng W, Kim JY, Yuan JP, Han L, Muallem S, Worley PF. Nat Cell Biol. 2006;8:1003–1010. doi: 10.1038/ncb1454. [DOI] [PubMed] [Google Scholar]
- 25.Oancea E, Teruel MN, Quest AF, Meyer T. J Cell Biol. 1998;140:485–498. doi: 10.1083/jcb.140.3.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lippincott-Schwartz J, Roberts TH, Hirschberg K. Annu Rev Cell Dev Biol. 2000;16:557–589. doi: 10.1146/annurev.cellbio.16.1.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teruel MN, Meyer T. Cell. 2000;103:181–184. doi: 10.1016/s0092-8674(00)00109-4. [DOI] [PubMed] [Google Scholar]
- 28.Petersen OH. Cell Calcium. 2003;33:337–344. doi: 10.1016/s0143-4160(03)00047-2. [DOI] [PubMed] [Google Scholar]
- 29.Schlessinger J. Cell. 2002;110:669–672. doi: 10.1016/s0092-8674(02)00966-2. [DOI] [PubMed] [Google Scholar]
- 30.Metzger M, Eglite S, Haleem-Smith H, Reischl I, Torigoe C. Mol Immunol. 2002;38:1207–1211. doi: 10.1016/s0161-5890(02)00065-2. [DOI] [PubMed] [Google Scholar]
- 31.Bachmann MF, Ohashi PS. Immunol Today. 1999;20:568–576. doi: 10.1016/s0167-5699(99)01543-1. [DOI] [PubMed] [Google Scholar]
- 32.Williams RT, Senior PV, Van Stekelenburg L, Layton JE, Smith PJ, Dziadek MA. Biochim Biophys Acta. 2002;1596:131–137. doi: 10.1016/s0167-4838(02)00211-x. [DOI] [PubMed] [Google Scholar]
- 33.Baba Y, Hayashi K, Fujii Y, Mizushima A, Watarai H, Wakamori M, Numaga T, Mori Y, Iino M, Hikida M, et al. Proc Natl Acad Sci USA. 2006;103:16704–16709. doi: 10.1073/pnas.0608358103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McLaughlin S. Science. 2006;314:1402–1403. doi: 10.1126/science.1136314. [DOI] [PubMed] [Google Scholar]
- 35.Lopez JJ, Salido GM, Pariente JA, Rosado JA. J Biol Chem. 2006;281:28254–28264. doi: 10.1074/jbc.M604272200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.