Abstract
The second heart field (SHF), progenitor cells that are initially sequestered outside the heart, migrates into the heart and gives rise to endocardium, myocardium, and smooth muscle. Because of its distinct developmental history, the SHF is likely subjected to different signals from that of the first heart field. Previous experiments revealed that canonical Wnt signaling negatively regulated first heart field specification. We inactivated the obligate canonical Wnt effector β-catenin using a β-catenin conditional null allele and the Mef2c AHF cre driver that directs cre activity specifically in SHF. We also expressed a stabilized form of β-catenin to model continuous Wnt signaling in SHF. Our data indicate that Wnt signaling acts in a positive fashion to promote right ventricular and interventricular myocardial expansion. Cyclin D2 and Tgfβ2 expression was drastically reduced in β-catenin loss-of-function mutants, indicating that Wnt signaling is required for patterning and expansion of SHF derivatives. Our findings reveal that Wnt signaling plays a major positive role in promoting growth and diversification of SHF precursors into right ventricular and interventricular myocardium.
Keywords: conditional genetics, cardiac progenitor, mouse, development
Previous work using retrospective clonal analysis in mouse embryos indicated that the heart developed from two populations of cardiac progenitors. Cells within the first heart field (FHF) uniquely contribute to the left ventricular myocardium whereas the second heart field (SHF) contributes to outflow tract (OFT) and right ventricular (RV) myocardium, endocardium, and smooth muscle of the great vessels (1–5). Each progenitor population expresses distinct and overlapping molecular markers: the Hand1 and Tbx5 transcriptional regulators mark the FHF whereas Fgf10 and the Lim-homeobox gene Isl1 mark the SHF. Recent work indicates that Isl1 may also be transiently expressed in the FHF (6). The Nkx2.5 homeobox gene is expressed in both lineages (7).
Genetic experiments performed in mice revealed that Isl1 had a critical function in SHF. Importantly, although Isl1 is expressed in SHF cardiac progenitors, Isl1 expression is extinguished in the majority of the heart with the exception of a subpopulation of cardiac progenitors that persist in the adult (4, 8). Other experiments, revealing that the Mef2c transcription factor is a direct Isl1 target, uncovered a SHF transcriptional hierarchy (9).
The SHF has been proposed to be a source of adult progenitor cells. Recent work has led to the proposal that Isl1-expressing progenitors provide a source of resident progenitor cells, or cardioblasts, that have the capability to differentiate into mature cardiomyocytes (8). Experiments performed in embryonic stem cells revealed that the Isl1-positive lineage gives rise to endothelial cells, smooth muscle cells, and cardiomyocytes, suggesting the intriguing possibility, as yet unproven, that the adult heart may harbor a stem cell niche (10–13).
Despite the importance of the SHF in cardiac development, a clear picture of the regulatory pathways controlling SHF development and diversification is lacking. An enhancer trap into the Fgf10 locus provided early evidence for the existence of the SHF in mice and indicates, along with conditional loss-of-function experiments, the involvement of Fgf signaling in SHF formation (14–16). Likewise, Bmp signaling has been implicated in SHF development based on data from chick embryos and mouse conditional deletion studies (6, 17, 18).
The Wnt family of secreted glycoproteins has been implicated in numerous events in development and disease (19). β-Catenin, an obligate effector of the canonical Wnt pathway, is stabilized in the presence of Wnt signaling and enters the nucleus, where it interacts with TCF factors, such as Lef1, to regulate gene expression. In the absence of Wnt signaling, β-catenin is targeted for destruction by the APC, Axin, Gsk3b complex that phosphorylates β-catenin and directs it to a destruction pathway (19). β-Catenin also complexes with the cytoplasmic domain of E-cadherin where it plays a fundamental role in promoting cell adhesion.
Previous findings indicated that Wnt signaling from the neural tube inhibits FHF specification (20–22). It is unclear whether Wnt signaling influences the SHF in a similar fashion as the FHF. Because Wnt signaling is known to promote development of trunk skeletal muscle, it was conceivable that Wnt signaling could promote SHF development from splanchnic mesoderm (23).
In this work we investigated β-catenin-dependent Wnt signaling in the SHF. We inactivated β-catenin in the SHF using the Mef2c AHF cre (hereafter referred to as Mef2ccre) that directs cre activity specifically in SHF (9, 24) (kindly provided by B. Black and M. Verzi, University of California, San Francisco, CA). Our findings indicate that β-catenin function is required for expansion of the RV and interventricular septum, as well as patterning of OFT cushions.
Results
Manipulation of β-Catenin Function in Second Cardiac Lineage.
To investigate Wnt signaling in second cardiac lineage, we generated mouse embryos with loss and gain of β-catenin function specifically in SHF. The Mef2ccre transgene directs cre activity specifically in SHF [supporting information (SI) Fig. 7A]. Lineage tracing with Mef2ccre and Rosa26Reporter (R26R) allele labels cells in the dorsal medial aspect of the crescent at 7.5 days postcoitum (dpc) (SI Fig. 7 B and C). As cardiac development progresses, the Mef2ccre lineage contributes to OFT myocardium, pharyngeal mesenchyme, and OFT endocardium (SI Fig. 7 D–I). Activity of the Mef2ccre transgene is excluded from the pharyngeal endoderm, making this an ideal cre driver to investigate gene function in the specified SHF (SI Fig. 7 E, G, and I) (24).
The β-cateninflox allele contains LoxP sites flanking exons 2–6 and has been shown to be a conditional null allele (SI Fig. 7J) (25). We used the Mef2ccre transgenic line to delete β-catenin, an obligatory downstream canonical Wnt effector. β-Catenin deletion in Mef2ccre-expressing cells would render SHF resistant to Wnt signaling. As a complementary experiment, we expressed a stabilized form of β-catenin in the Mef2ccre-expressing SHF to model gain of function or continuous Wnt signaling (26). The β-cateninex3flox allele is a mutant β-catenin allele containing a LoxP-flanked exon 3 that encodes residues that are required for negative regulation of β-catenin (SI Fig. 7K) (26). When β-catenin exon 3 is removed by cre-mediated recombination, the resulting mutant allele encodes a stabilized form of β-catenin that mimics sustained Wnt signaling.
Efficient β-Catenin Deletion by Mef2ccre.
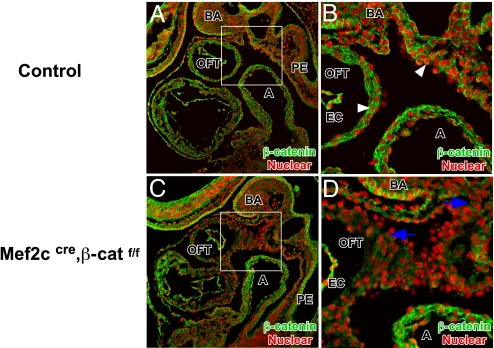
To remove β-catenin function from specified SHF, we crossed the β-cateninflox allele to the Mef2ccre transgenic line (24, 25). We used immunofluorescent microscopy to determine the efficiency of β-catenin deletion in SHF. At 9.5 dpc we detected β-catenin in the pharyngeal mesenchyme and OFT myocardium of control embryos (Fig. 1 A and B). We noted membrane-localized β-catenin in epithelial tissues such as branchial arch ectoderm and pharyngeal endoderm. Moreover, we detected nuclear-localized β-catenin, a hallmark of active Wnt signaling, in the pharyngeal mesenchyme and OFT myocardium (Fig. 1B). In Mef2ccre; β-cateninflox conditional mutants, we found a strong reduction in β-catenin immunostaining in the SHF-derived OFT myocardium and pharyngeal mesenchyme (Fig. 1 C and D), indicating that Mef2ccre efficiently deleted β-catenin in SHF.
Fig. 1.
Mef2ccre induced β-catenin deletion in SHF. Immunofluorescent staining of β-catenin in 9.5 dpc sagittal sections of control (A and B) and Mef2ccre; β-cateninflox mutant (C and D) embryos revealing membrane-localized β-catenin staining in ectoderm and endoderm as well as nuclear β-catenin in OFT myocardium, posterior pericardial mesothelium, and pharyngeal mesenchyme (denoted by white arrowheads in B). In the Mef2ccre; β-cateninflox mutant there is reduced β-catenin expression in pharyngeal mesenchyme and OFT myocardium, but not endocardium, due to mosaic activity of Mef2ccre in OFT endocardium (decreased β-catenin denoted by blue arrows). A, atrium; BA, branchial arch; EC, endocardium; PE, pharyngeal endoderm.
Mef2ccre; β-Cateninflox Conditional Mutants Have a Reduced Right Ventricle.
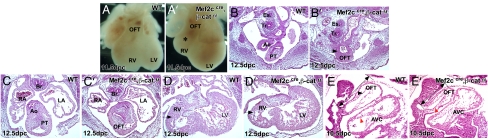
Examination of Mef2ccre; β-catenin conditional null mutant embryos at multiple stages revealed that all β-catenin loss-of-function mutants had defective OFT and RV development. In particular, the RV was drastically reduced when compared with control embryos (Fig. 2 A and A′). Histologic analysis also indicated that, at 12.5 dpc, β-catenin conditional null mutants had defective separation of the distal OFT (Fig. 2 B and B′). More proximal sections revealed that, in the proximal OFT, the pulmonary trunk and aorta were separated (Fig. 2 C and C′). Sections through the RV indicated that the RV was reduced in size with a thin myocardium (Fig. 2 D and D′). The OFT length was also reduced in β-catenin conditional null mutants when compared with control (Fig. 2 E and E′). Together, the phenotypes detected in β-catenin conditional null mutants indicated a major deficiency in SHF derivatives.
Fig. 2.
Phenotype of Mef2ccre; β-catenin mutant embryos. Shown are morphology and histology of 10.5–12.5 dpc β-catenin loss-of-function mutant hearts showing severe RV hypoplasia (∗) (A′) compared with control littermate (A). Transverse section of 12.5 dpc (B, B′, C, C′, D, and D′) and sagittal section of 10.5 dpc (E and E′) control and β-catenin loss-of-function mutant hearts reveal severe cardiac abnormalities: OFT septation defect in the distal OFT (B and B′, arrowheads) but not in more proximal OFT (C and C′), and hypoplasia of RV myocardium (D and D′, arrow) and short OFT (E and E′, double-headed arrows). Endocardium is denoted by arrowheads. PT, pulmonary trunk; LV, left ventricle; RV, right ventricle; RA, right atrium; LA, left atrium; Es, esophagus, Tr, Trachea, Br, bronchi.
Deficiency of SHF Derivatives in β-Catenin Conditional Mutants.
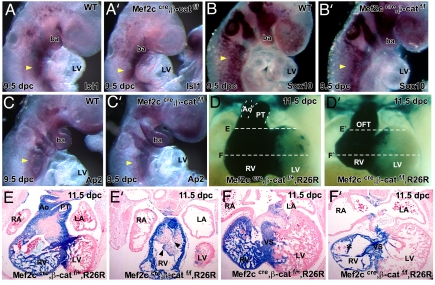
We studied SHF markers to investigate the β-catenin conditional null mutant phenotype in more depth. Expression of Isl1 was intact in the β-catenin conditional null mutants as would be expected because Mef2c is a downstream target of Isl1 in SHF (9) (Fig. 3 A and A′). Moreover, expression of cardiac neural crest (CNC) markers indicated that CNC was specified correctly in β-catenin mutants (Fig. 3 B, B′, C, and C′). Lineage tracing with R26R, to follow the developmental progression of the Mef2ccre lineage, indicated that in β-catenin mutant embryos the SHF was severely defective (Fig. 3 E, E′, F, and F′). Whole-mount LacZ staining showed the reduced size of the RV and defective distal OFT separation in β-catenin mutant embryos (Fig. 3 D and D′). Sections revealed a major deficiency of SHF-derived cells in the RV and interventricular septum (Fig. 3 E, E′, F, and F′). Dysmorphology of the OFT cushions was also evident in these sections, suggesting that a β-catenin target is a signaling molecule that functions to pattern ingressing CNC (Fig. 3 E and E′). These findings indicate that, in contrast to its inhibitory influence on the FHF, Wnt signaling promotes SHF development.
Fig. 3.
Marker analysis of SHF and CNC and Mef2ccre lineage tracing in Mef2ccre; β-cateninflox mutants. In situ hybridizations of Isl1 (A and A′), Sox10 (B and B′), and Ap2 (C and C′) at 9.5 dpc indicated that SHF and CNC were correctly specified in Mef2ccre; β-catenin conditional mutant embryos (yellow arrowheads denote hybridization signal). Mef2ccre lineage tracing (D and D′) and transverse section (E, E′, F, and F′) in wild type and β-catenin mutant revealed a severe deficiency in the Mef2c lineage in Mef2ccre; β-catenin conditional mutant embryos. LV, left ventricle; RV, right ventricle; RA, right atrium; LA, left atrium; AO, aorta; PT, pulmonary trunk; Ba, branchial arch.
Defective Expression of Myocardial Markers in β-Catenin Conditional Mutants.
To address the genetic pathways regulated by Wnt signaling in SHF, we examined molecular markers of the SHF and its derivatives. Cyclin D2, a direct target of β-catenin-dependent Wnt signaling, is expressed in pharyngeal mesenchyme and in proximal OFT (27). At both 9.5 dpc and 10.5 dpc, CyclinD2 was absent in the OFT of β-catenin mutants validating loss of competence to respond to Wnt signaling (Fig. 4 A–D). To determine whether there was a defect in regionalization of the β-catenin mutant OFT, we examined Tgfβ2 expression that marks the most distal OFT myocardium. In β-catenin mutants, Tgfβ2 was severely diminished, indicating that both distal and proximal OFT myocardium was defective in β-catenin mutants (Fig. 4 E and F). In contrast, expression of Smarcd3, which encodes a component of the BAF chromatin remodeling complex and a proposed Wnt target in the heart, was unchanged (Fig. 4 G and H) (28). Absence of Cyclin D2 expression in β-catenin mutants suggested a defect in proliferation. Expression of phospho-Histone H3, which marks mitotic cells, was greatly reduced in β-catenin mutants, supporting the idea that canonical Wnt signaling was required for normal proliferation of OFT and RV myocardium (Fig. 4 I and J). Taken together, these data indicate that patterning and expansion of SHF-derived myocardium are severely disrupted in β-catenin mutant embryos.
Fig. 4.
Marker analysis of OFT myocardium in Mef2ccre; β-cateninflox mutants. In situ hybridization with Cyclin D2 (A–D), Tgfβ2 (E and F), and Smarcd3 (G and H) in control and Mef2ccre; β-cateninflox mutants at 9.5 dpc. Pharyngeal mesenchyme is denoted by yellow arrowheads, and OFT is denoted by black arrowheads. (I and J) Sagittal sections of phospho-histone H3 immunostaining at 9.5 dpc. Arrowheads denote PH3 signal. lv, left ventricle; ba, brachial arch.
Expression of Stabilized β-Catenin Causes Defective Right Ventricle Development.
We used the β-cateninex3flox allele to express stabilized β-catenin in the SHF. This gain-of-function experiment models the effect of continuous, deregulated Wnt signaling on SHF progenitor cells. The RV was severely reduced in Mef2ccre; β-cateninex3flox embryos (Fig. 5 A and B). Sectioning indicated that OFT and RV myocardium was thickened and hypercellular in the Mef2ccre; β-cateninex3flox embryos (Fig. 5 C–F). We also noted that a region of the left ventricular myocardium was thickened (Fig. 5F, arrowhead). This likely reflects the contribution of the SHF to the left ventricular region near the interventricular septum (24). Taken together, these data indicate that both loss and gain of β-catenin function resulted in defective RV development. It is notable that in other systems, such as tooth development, excess and reduced Wnt signaling results in a similar tooth-loss phenotype (29, 30).
Fig. 5.
Phenotype of Mef2ccre; β-cateninEx3flox mutant embryos. Morphology and serial, transverse sections of 9.5 dpc wild-type (A, C, and E) and Mef2ccre; β-cateninEx3flox mutant embryos (B, D, and F) showing short but more dense and hypercellular OFT myocardium (denoted by black arrows). nt, neural tube; ba, branchial arch; LV, left ventricle; ph, pharynx.
Disrupted Bmp4 Expression in β-Catenin Conditional Mutants.
We compared expression of SHF markers in the β-catenin loss- and gain-of-function embryos. Nkx2.5, which plays an early role in SHF specification, was expressed in the pharyngeal mesenchyme and SHF derivatives of both loss- and gain-of-function embryos, supporting the idea that SHF is correctly specified in Mef2ccre; β-catenin flox mutant embryos (Fig. 6 A–C) (6). In contrast, Bmp4 was greatly reduced in the pharyngeal mesenchyme and OFT myocardium of β-catenin loss-of-function embryos (Fig. 6 D and E). β-Catenin gain-of-function embryos expressed Bmp4 in both pharyngeal mesenchyme and OFT myocardium (Fig. 6 D and F). Fgf10 expression in pharyngeal mesenchyme was unchanged in β-catenin loss-of-function embryos and had a considerably reduced expression domain in β-catenin gain-of-function embryos (Fig. 6 G–I). We also noted low levels of Fgf10 in the OFT that was diminished in both β-catenin loss- and gain-of-function embryos. MLC2a, a marker of differentiated myocardium, was reduced in β-catenin gain-of-function embryos, indicating a differentiation defect. In β-catenin loss-of-function mutants, expression of MLC2a was similar to control embryos (Fig. 6 J–L).
Fig. 6.
Marker analysis in β-catenin loss-of-function and gain-of-function mutant embryos. (A–L) Analysis of markers for SHF and differentiated myocardium in 9.5-dpc embryos. Genotypes and markers are labeled. Pharyngeal mesenchyme is denoted by yellow arrowheads, and OFT is denoted by black arrowheads. OFT is outlined by a dotted line in G–I for clarity. (M) Model depicting the proposed role of canonical Wnt signaling in SHF. Based on loss- and gain-of-function experiments, canonical Wnt signaling promotes Bmp4 expression while concurrently acting to restrict Fgf10 in the SHF. The dotted line for Fgf10 repression indicates that this may be a minor function for Wnt signaling in the SHF because the data supporting this inhibitory function were observed only in the gain-of-function experiment. In the SHF-derived myocardium of the OFT, RV, and interventricular septum (IVS), canonical Wnt signaling regulates Bmp4 and promotes cell proliferation through regulation of Cyclin D. ba, branchial arch; RV, right ventricle; lv and LV, left ventricle; oc, otic capsule.
Discussion
Previous experiments revealed an inhibitory role for canonical Wnt signaling in FHF precursors (22, 31, 32). In contrast, canonical Wnt signaling has been shown to promote trunk skeletal muscle development (33, 34). Our data indicate that canonical Wnt signals promote development of SHF progenitors into RV and interventricular myocardium. These findings uncover a striking difference between the two cardiac progenitor populations in the use of Wnt signaling to promote or repress formation of the mature cardiomyocyte phenotype.
Wnt Signaling Is Critical for Development of the Right Ventricle.
We deleted β-catenin using the Mef2ccre driver that is active after Isl1 expression has been initiated in SHF. Therefore, we removed competence to respond to Wnt signaling after SHF specification. Although in situ data indicate that patterning of the proximal and distal OFT is disrupted, the most severely affected cells are the RV and interventricular progenitors. Our findings are consistent with the previous observation that RV and interventricular myocardium derives from SHF (35). One explanation for the stronger RV and interventricular myocardial phenotype may be that the OFT myocardial lineage is specified or expands earlier than RV and interventricular septum. This notion is analogous to data from ES cells in which Nkx2.5 GFP-expressing cells are more restricted than Brachury GFP-expressing cells in their potential for lineage diversification or expansion (36). It is conceivable that other signals, such as Bmp signaling, have a role in specification or expansion of OFT myocardium and endocardial lineages (see below).
Previous work revealed that noncanonical Wnt signaling plays a critical role in cardiac development. The noncanonical Wnt signaling pathway is β-catenin-independent but utilizes Dsh and Rho (37). Wnt11 has been shown to regulate cardiac development through a noncanonical pathway in Xenopus and zebrafish embryos (38, 39). Moreover, Loop-tail (Lp), a naturally occurring mouse mutant that was initially identified based on severe defects in neural tube closure, has cardiac OFT defects. The gene mutated in Lp is Vangl2, a homolog of the Drosophila planar cell polarity gene Strabismus. Recent work has shown that Vangl2 functions in the OFT myocardium to regulate OFT septation (40).
Our work provides insight into the role of Wnt signaling in SHF by showing that the canonical Wnt pathway promotes SHF development. It is notable that Wnt11, highly expressed in OFT myocardium, has also been implicated in canonical Wnt signaling in Xenopus embryos (41). Therefore, it is conceivable that Wnt11 activates the canonical pathway in SHF derivatives, but further experiments will be required to investigate this.
The Intersection of Wnt and Bmp Signaling in SHF and SHF Derivatives.
Bmp signaling has been implicated in the addition of SHF progenitors to the OFT and has been considered to be a prodifferentiation pathway in the SHF. For example, Noggin-coated bead implantation in SHF explants resulted in increased cell proliferation (18). Conditional genetic experiments in mice support this interpretation because Mespcre; Smad1f/f mutants, which delete the Bmp effector Smad1 widely in mesoderm, have elongated OFTs and elevated proliferation in SHF but not OFT myocardium (6). In addition, Nkx2.5cre; Bmp4 flox mutants, which have a broad Bmp4 deletion in SHF, SHF derivatives, and pharyngeal endoderm, show elevated proliferation in OFT myocardium (17). In contrast to what might have been predicted, our data indicate that β-catenin loss-of-function embryos with reduced Bmp4 in pharyngeal mesenchyme and OFT myocardium have less myocardial proliferation.
We also noted that β-catenin loss-of-function embryos had defective patterning of OFT cushions that are partly derived from CNC. Because our strategy to inhibit Wnt signaling competence was limited to the SHF, the defect in CNC patterning indicates that there is a nonautonomous signal acting downstream of β-catenin. Published data reveal that inactivation of Bmp ligands in SHF, and type I Bmp receptors in CNC resulted in defective cardiac cushion development, supporting the notion that Bmp signals from SHF regulate development of the CNC (17, 42–44).
Previous work has uncovered dosage-dependent functions for Bmp signaling in mandibular development (45). This notion of distinct, dosage-dependent functions for Bmp signaling likely also applies to the SHF and SHF derivatives as suggested by the OFT phenotype of the Bmpr2 hypomorph previously reported by Lyons and colleagues (46). Moreover, we have recently found that Bmp2 and Bmp4 function cooperatively in the SHF to regulate distinct events in the forming OFT (L. Ma and J.F.M., unpublished observations). Together our findings suggest that residual Bmp4 expression in β-catenin loss-of-function embryos is sufficient to maintain the antiproliferative Bmp function but fails to support CNC patterning (Fig. 6M). The β-catenin loss-of-function embryos had reduced Fgf10 expression in the OFT myocardium, which also likely contributes to diminished proliferation in the mutant OFT. Fgf signaling has been shown to be an important signal that promotes expansion of SHF derivatives (15, 16).
Stabilized β-Catenin Disrupts SHF Development.
The phenotype we observed in β-catenin gain-of-function embryos, a short hypercellular OFT, likely represents a disruption of the normal balance between progenitor cell expansion and precisely timed differentiation. In support of this idea, we observed reduced Fgf10 expression in the SHF of β-catenin gain-of-function embryos. As noted above, Fgf signaling in SHF is generally considered to promote proliferation; reduced Fgf signaling would likely result in a shorter OFT (15, 16, 18). In contrast, once SHF cells have moved into the OFT canonical Wnt signaling has a more direct role in promoting cellular proliferation and myocardial expansion (Fig. 6M). This would account for the myocardial hypercellularity and reduced MLC2a expression in the β-catenin gain-of-function embryos.
Moreover, these data suggest that elevated Wnt signaling has a negative influence on expansion of undifferentiated SHF in pharyngeal mesenchyme. These data support previous suggestions that Wnt signaling may have a role in delaying SHF from contributing to myocardium until looping stages.
Implications for Signaling in Cardiac Progenitor Cells and Right Ventricular Development.
Our findings show that Wnt signaling plays a critical, positive role in promoting SHF expansion and diversification. This observation may have therapeutic implications because a number of laboratories have identified multipotent cardiac progenitor cells (10–12). An important goal would be to identify the signaling molecules that promote progenitor cell expansion. Our data indicate that Wnt signaling is an excellent candidate to promote growth of cardiac progenitors. Moreover, our findings indicate that Bmp signaling likely has a downstream function in this pathway (Fig. 6M). This is particularly intriguing considering the proposed role for Bmp4 in self-renewal in embryonic stem cells (47).
Materials and Methods
Whole-Mount in Situ Hybridization.
Whole-mount in situ hybridization was performed as previously described (48). Details about the in situ probes will be provided upon request. For all experiments, at least three embryos were used for each probe at each time point examined.
LacZ Staining and Histology.
For histology, embryos were fixed overnight in Bouin's fixative or buffered formalin, dehydrated through graded ethanol, and embedded in paraffin. Sections were cut at 7–10 μm and stained with H&E. Staining for β-galactosidase (LacZ) was as previously described (48).
Mouse Alleles and Transgenes.
The Mef2cAHFcre transgenic line has been previously described (9, 24). Briefly, the Mef2cAHFcre transgenic line uses a 3,970-bp enhancer and promoter element from the Mef2c gene to direct cre recombinase expression in the SHF. The β-catenin conditional null allele has been described previously (25). For genotyping, DNA was extracted from yolk sacs or tails of embryos and adult mice, respectively. PCR was used to determine the genotype. The primers used were described (25). The β-catenin dominant stable allele has also been described previously (26). The genotyping primers used were 5′-AGCTGCTGTGACACCGCT-3′ and 5′-GCTACTTGCTCTTGCGTGAA-3′. The amplified product for the mutant allele is 700 bp.
Immunohistochemical Staining.
Embryos were collected at desired stages and fixed in 4% paraformaldehyde for 30 min. Tissues were washed with PBS twice, dehydrated, and embedded in paraffin as previously described. Immunofluorescence staining was carried out on 5-μm sections as previously described (49). Briefly, sections were first blocked with 2% BSA in PBST and incubated with anti-β-catenin (1:600; Cell Signaling Technology, Beverly, MA) for 1 h. After three 10-min washings, Alexa Fluor 488-conjugated secondary anti-rabbit IgG (1:800; Molecular Probes, Eugene, OR) was added and incubated for 1 h. The secondary antibody was then washed off, and the sections were counterstained with propidium iodide and mounted with Aqua-Mount (Lerner Laboratories, Pittsburgh, PA). Images were captured by an Olympus FluoView confocal microscopy system.
For phospho-histone H3 (PH3) immunostaining antigen retrieval was performed by boiling slides in 10 mM sodium citrate (pH 6.0, 5 min) followed by treatment with 3% H2O2 to quench endogenous peroxidase. Specimens were blocked in 2% normal goat serum (NGS) for 1 h at room temperature and incubated overnight at 4°C with the anti-phospho-Histone H3 (Ser-10) antibody (Upstate Biotechnology, Lake Placid, NY) diluted 1:200 in 2% NGS, followed by incubation in biotinylated anti-rabbit secondary antibody (Vector Laboratories, Burlingame, CA) diluted 1:200 in 2% NGS for 1 h at room temperature. Sections were processed by immunoperoxidase labeling using the Vectastain ABC Kit (Vector Laboratories) and visualized with a DAB kit (Vector Laboratories).
Supplementary Material
Acknowledgments
We thank E. Olson, B. Bruneau, and D. Srivastava for in situ probes, R. Schwartz for discussions, B. Black for the Mef2c AHFcre, and M. M. Taketo for the β-cateninexon3flox allele. This work was supported by National Institutes of Health Grant R01 DE16329-01 (to J.F.M.), National Eye Institute Grant EY11930 (to W.H.K.), and Robert A. Welch Foundation Grant G-0010 (to W.H.K.).
Abbreviations
- FHF
first heart field
- SHF
second heart field
- OFT
outflow tract
- RV
right ventricle
- CNC
cardiac neural crest
- dpc
days postcoitum.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0701212104/DC1.
References
- 1.Meilhac SM, Esner M, Kerszberg M, Moss JE, Buckingham ME. J Cell Biol. 2004;164:97–109. doi: 10.1083/jcb.200309160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meilhac SM, Esner M, Kelly RG, Nicolas JF, Buckingham ME. Dev Cell. 2004;6:685–698. doi: 10.1016/s1534-5807(04)00133-9. [DOI] [PubMed] [Google Scholar]
- 3.Waldo KL, Hutson MR, Ward CC, Zdanowicz M, Stadt HA, Kumiski D, Abu-Issa R, Kirby ML. Dev Biol. 2005;281:78–90. doi: 10.1016/j.ydbio.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 4.Cai CL, Liang X, Shi Y, Chu PH, Pfaff SL, Chen J, Evans S. Dev Cell. 2003;5:877–889. doi: 10.1016/s1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buckingham M, Meilhac S, Zaffran S. Nat Rev Genet. 2005;6:826–835. doi: 10.1038/nrg1710. [DOI] [PubMed] [Google Scholar]
- 6.Prall OW, Menon MK, Solloway MJ, Watanabe Y, Zaffran S, Bajolle F, Biben C, McBride JJ, Robertson BR, Chaulet H, et al. Cell. 2007;128:947–959. doi: 10.1016/j.cell.2007.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwartz RJ, Olson EN. Development (Cambridge, UK) 1999;126:4187–4192. doi: 10.1242/dev.126.19.4187. [DOI] [PubMed] [Google Scholar]
- 8.Laugwitz KL, Moretti A, Lam J, Gruber P, Chen Y, Woodard S, Lin LZ, Cai CL, Lu MM, Reth M, et al. Nature. 2005;433:647–653. doi: 10.1038/nature03215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dodou E, Verzi MP, Anderson JP, Xu SM, Black BL. Development (Cambridge, UK) 2004;131:3931–3942. doi: 10.1242/dev.01256. [DOI] [PubMed] [Google Scholar]
- 10.Moretti A, Caron L, Nakano A, Lam JT, Bernshausen A, Chen Y, Qyang Y, Bu L, Sasaki M, Martin-Puig S, et al. Cell. 2006;127:1151–1165. doi: 10.1016/j.cell.2006.10.029. [DOI] [PubMed] [Google Scholar]
- 11.Wu SM, Fujiwara Y, Cibulsky SM, Clapham DE, Lien CL, Schultheiss TM, Orkin SH. Cell. 2006;127:1137–1150. doi: 10.1016/j.cell.2006.10.028. [DOI] [PubMed] [Google Scholar]
- 12.Kattman SJ, Huber TL, Keller GM. Dev Cell. 2006;11:723–732. doi: 10.1016/j.devcel.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 13.Sun Y, Liang X, Najafi N, Cass M, Lin L, Cai C, Chen J, Evans S. Dev Biol. 2006;304:286–296. doi: 10.1016/j.ydbio.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelly RG, Brown NA, Buckingham ME. Dev Cell. 2001;1:435–440. doi: 10.1016/s1534-5807(01)00040-5. [DOI] [PubMed] [Google Scholar]
- 15.Park EJ, Ogden LA, Talbot A, Evans S, Cai CL, Black BL, Frank DU, Moon AM. Development (Cambridge, UK) 2006;133:2419–2433. doi: 10.1242/dev.02367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ilagan R, Abu-Issa R, Brown D, Yang YP, Jiao K, Schwartz RJ, Klingensmith J, Meyers EN. Development (Cambridge, UK) 2006;133:2435–2445. doi: 10.1242/dev.02408. [DOI] [PubMed] [Google Scholar]
- 17.Liu W, Selever J, Wang D, Lu MF, Moses KA, Schwartz RJ, Martin JF. Proc Natl Acad Sci USA. 2004;101:4489–4494. doi: 10.1073/pnas.0308466101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Waldo KL, Kumiski DH, Wallis KT, Stadt HA, Hutson MR, Platt DH, Kirby ML. Development (Cambridge, UK) 2001;128:3179–3188. doi: 10.1242/dev.128.16.3179. [DOI] [PubMed] [Google Scholar]
- 19.Huelsken J, Birchmeier W. Curr Opin Genet Dev. 2001;11:547–553. doi: 10.1016/s0959-437x(00)00231-8. [DOI] [PubMed] [Google Scholar]
- 20.Lickert H, Kutsch S, Kanzler B, Tamai Y, Taketo MM, Kemler R. Dev Cell. 2002;3:171–181. doi: 10.1016/s1534-5807(02)00206-x. [DOI] [PubMed] [Google Scholar]
- 21.Schneider VA, Mercola M. Genes Dev. 2001;15:304–315. doi: 10.1101/gad.855601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tzahor E, Lassar AB. Genes Dev. 2001;15:255–260. doi: 10.1101/gad.871501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cossu G, Borello U. EMBO J. 1999;18:6867–6872. doi: 10.1093/emboj/18.24.6867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Verzi MP, McCulley DJ, De Val S, Dodou E, Black BL. Dev Biol. 2005;287:134–145. doi: 10.1016/j.ydbio.2005.08.041. [DOI] [PubMed] [Google Scholar]
- 25.Brault V, Moore R, Kutsch S, Ishibashi M, Rowitch DH, McMahon AP, Sommer L, Boussadia O, Kemler R. Development (Cambridge, UK) 2001;128:1253–1264. doi: 10.1242/dev.128.8.1253. [DOI] [PubMed] [Google Scholar]
- 26.Harada N, Tamai Y, Ishikawa T, Sauer B, Takaku K, Oshima M, Taketo MM. EMBO J. 1999;18:5931–5942. doi: 10.1093/emboj/18.21.5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baek SH, Kioussi C, Briata P, Wang D, Nguyen HD, Ohgi KA, Glass CK, Wynshaw-Boris A, Rose DW, Rosenfeld MG. Proc Natl Acad Sci USA. 2003;100:3245–3250. doi: 10.1073/pnas.0330217100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lickert H, Takeuchi JK, Von Both I, Walls JR, McAuliffe F, Adamson SL, Henkelman RM, Wrana JL, Rossant J, Bruneau BG. Nature. 2004;432:107–112. doi: 10.1038/nature03071. [DOI] [PubMed] [Google Scholar]
- 29.Andl T, Reddy ST, Gaddapara T, Millar SE. Dev Cell. 2002;2:643–653. doi: 10.1016/s1534-5807(02)00167-3. [DOI] [PubMed] [Google Scholar]
- 30.Lammi L, Arte S, Somer M, Jarvinen H, Lahermo P, Thesleff I, Pirinen S, Nieminen P. Am J Hum Genet. 2004;74:1043–1050. doi: 10.1086/386293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eisenberg LM, Eisenberg CA. Dev Biol. 2006;293:305–315. doi: 10.1016/j.ydbio.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 32.Marvin MJ, Di Rocco G, Gardiner A, Bush SM, Lassar AB. Genes Dev. 2001;15:316–327. doi: 10.1101/gad.855501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Snider L, Tapscott SJ. Cell. 2003;113:811–812. doi: 10.1016/s0092-8674(03)00474-4. [DOI] [PubMed] [Google Scholar]
- 34.Polesskaya A, Seale P, Rudnicki MA. Cell. 2003;113:841–852. doi: 10.1016/s0092-8674(03)00437-9. [DOI] [PubMed] [Google Scholar]
- 35.Zaffran S, Kelly RG, Meilhac SM, Buckingham ME, Brown NA. Circ Res. 2004;95:261–268. doi: 10.1161/01.RES.0000136815.73623.BE. [DOI] [PubMed] [Google Scholar]
- 36.Garry DJ, Olson EN. Cell. 2006;127:1101–1104. doi: 10.1016/j.cell.2006.11.031. [DOI] [PubMed] [Google Scholar]
- 37.Park M, Moon RT. Nat Cell Biol. 2002;4:20–25. doi: 10.1038/ncb716. [DOI] [PubMed] [Google Scholar]
- 38.Gong Y, Mo C, Fraser SE. Nature. 2004;430:689–693. doi: 10.1038/nature02796. [DOI] [PubMed] [Google Scholar]
- 39.Pandur P, Lasche M, Eisenberg LM, Kuhl M. Nature. 2002;418:636–641. doi: 10.1038/nature00921. [DOI] [PubMed] [Google Scholar]
- 40.Phillips HM, Murdoch JN, Chaudhry B, Copp AJ, Henderson DJ. Circ Res. 2005;96:292–299. doi: 10.1161/01.RES.0000154912.08695.88. [DOI] [PubMed] [Google Scholar]
- 41.Tao Q, Yokota C, Puck H, Kofron M, Birsoy B, Yan D, Asashima M, Wylie CC, Lin X, Heasman J. Cell. 2005;120:857–871. doi: 10.1016/j.cell.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 42.Kaartinen V, Dudas M, Nagy A, Sridurongrit S, Lu MM, Epstein JA. Development (Cambridge, UK) 2004;131:3481–3490. doi: 10.1242/dev.01214. [DOI] [PubMed] [Google Scholar]
- 43.Wang J, Nagy A, Larsson J, Dudas M, Sucov HM, Kaartinen V. BMC Dev Biol. 2006;6:51. doi: 10.1186/1471-213X-6-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stottmann RW, Choi M, Mishina Y, Meyers EN, Klingensmith J. Development (Cambridge, UK) 2004;131:2205–2218. doi: 10.1242/dev.01086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu W, Selever J, Murali D, Sun X, Brugger SM, Ma L, Schwartz RJ, Maxson R, Furuta Y, Martin JF. Dev Biol. 2005;283:282–293. doi: 10.1016/j.ydbio.2005.04.019. [DOI] [PubMed] [Google Scholar]
- 46.Delot EC, Bahamonde ME, Zhao M, Lyons KM. Development (Cambridge, UK) 2003;130:209–220. doi: 10.1242/dev.00181. [DOI] [PubMed] [Google Scholar]
- 47.Ying QL, Nichols J, Chambers I, Smith A. Cell. 2003;115:281–292. doi: 10.1016/s0092-8674(03)00847-x. [DOI] [PubMed] [Google Scholar]
- 48.Lu MF, Pressman C, Dyer R, Johnson RL, Martin JF. Nature. 1999;401:276–278. doi: 10.1038/45797. [DOI] [PubMed] [Google Scholar]
- 49.Fu X, Sun H, Klein WH, Mu X. Dev Biol. 2006;299:424–437. doi: 10.1016/j.ydbio.2006.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.