Abstract
Regulation of chromatin structure is critical in many fundamental cellular processes. Previous studies have suggested that the Rb tumor suppressor may recruit multiple chromatin regulatory proteins to repress E2F, a key regulator of cell proliferation and differentiation. Taking advantage of the evolutionary conservation of the E2F pathway, we have conducted a genome-wide RNAi screen in cultured Drosophila cells for genes required for repression of E2F activity. Among the genes identified are components of the putative Domino chromatin remodeling complex, as well as the Polycomb Group (PcG) protein-like fly tumor suppressor, L3mbt, and the related CG16975/dSfmbt. These factors are recruited to E2F-responsive promoters through physical association with E2F and are required for repression of endogenous E2F target genes. Surprisingly, their inhibitory activities on E2F appear to be independent of Rb. In Drosophila, domino mutation enhances cell proliferation induced by E2F overexpression and suppresses a loss-of-function cyclin E mutation. These findings suggest that potential chromatin regulation mediated by Domino and PcG-like factors plays an important role in controlling E2F activity and cell growth.
Keywords: transcription
Defects in chromatin structure lead to aberrant gene transcription and genetic instability, which contribute to many cancers and developmental defects (1). The E2F and Rb families of transcriptional regulators play crucial roles in cell proliferation and differentiation (2, 3). E2F activity is elevated in a wide variety of human cancers (4). The Rb tumor suppressor physically associates with E2F and restrains it in a transcriptionally inactive state. Modulation of chromatin structure is thought to be an important mechanism underlying Rb-mediated repression primarily due to its interactions with a variety of chromatin-remodeling and -modifying enzymes, including the SWI/SNF ATP-dependent nucleosome remodeling complex, histone deacetylases (HDACs) and methyltransferases (HMTs) (reviewed in refs. 3 and 5). In particular, Rb directs heterochromatin formation through recruitment of the HMTs Suv39h (6) and Suv4–20h (7). Such heterochromatinizing activities may lead to stable silencing of growth-promoting genes and cellular senescence (8), which acts as an important barrier to tumor development (9).
The Rb-E2F network is remarkably conserved in Drosophila. The Drosophila genome encodes two E2F factors (dE2F1 and dE2F2), a single dDP protein as the heterodimeric partner for dE2Fs, and two Rb homologs (Rbf1 and Rbf2). The highly effective RNAi-mediated disruption of gene function in cultured Drosophila cells provides a loss-of-function approach to examining the requirement of potential E2F regulators. However, such a study suggested that the HDACs and SWI/SNF chromatin-remodeling complex were dispensable for E2F inhibition (10), raising questions about the mechanisms underlying actions of Rb, and more generally, repression of E2F, which is important for tumor suppression.
In the present study, we carried out a genome-wide RNAi screen in cultured fly cells (11) to systematically search for essential E2F repressors. Here we describe genes identified from the screen that negatively regulate E2F. These factors are likely to be modulators of chromatin structure, and their mutations cause abnormal cell proliferation in vivo. Interestingly, they apparently confer Rb-independent activities.
Results
A Reporter-Based Genome-Wide RNAi Screen for Negative Regulators of E2F.
To facilitate monitoring of endogenous E2F activity, wegenerated an E2F-responsive firefly luciferase reporter (Fig. 1A). This reporter is driven by a minimal Hsp70 promoter and tandem copies of an E2F-binding site, which can bind E2Fs and Rb/E2F complexes in vitro (12). A Renilla luciferase reporter driven by the same minimal Hsp70 promoter was included as a normalization control. Comparison between the two reporters allowed us to detect E2F-specific activity. Drosophila S2* cells were transiently transfected with both reporter constructs and then cultured in the presence of in vitro synthesized long pieces of dsRNA. S2* cells can take up dsRNAs from the medium and process them into numerous siRNAs to efficiently degrade mRNAs of specific target genes. We tested the reporter system by RNAi of known positive and negative regulators. As expected, RNAi knockdown of dE2F1 or dDP decreased the E2F-dependent reporter activity, whereas RNAi depletion of Rbf1 substantially up-regulated the reporter (Fig. 1B). The level of activation of the E2F reporter is comparable to that of most endogenous E2F target genes (13). Moreover, the ectopic activation that occurred after knockdown of Rbf1 could be efficiently suppressed by RNAi of the downstream positive regulators dE2F1 and dDP (Fig. 1B). These observations support the reporter-based assay as a reliable “sensor” to recapitulate endogenous E2F activity.
Fig. 1.
RNAi analysis of the E2F pathway in Drosophila cell culture. (A) Schematics of the reporter constructs. The E2F-dependent reporter (E2F-Luc) carries four copies of an artificial E2F site (4xE2F), a minimal HSP70 promoter (HSP), and the firefly luciferase gene (Luc). The Renilla control reporter contains the same HSP and the Renilla luciferase gene (RLu). The two constructs share the same plasmid backbone. (B) Effects of RNAi depletion of known E2F pathway components on the E2F-reponsive reporter. Drosophila S2* cells were transiently transfected with E2F-Luc and the control reporter and incubated with dsRNAs in 24-well plates. RNAi of dE2F1/dDP and Rbf1 considerably decreased and increased the E2F-dependent reporter, respectively. RNAi of Rbf2 or dE2F2 had little effect. The reporter responses remain constant during incubation days 4–8. Similar results were also obtained by using several other Drosophila cell lines (e.g., S2, Kc167, Dl2, Cl8).
We next examined a panel of chromatin-modulating proteins, most of which were previously implicated in Rb-E2F regulation. However, RNAi-mediated removal of HMTs, including Suv4–20; Suv3–9; and fly homologs of G9a, ESET, and arginine methyltransferase PRMT5, failed to activate the E2F reporter [supporting information (SI) Fig. 6]. In addition, depletion of HDAC1, or Brm and Mi2, the ATPase subunits of two chromatin-remodeling complexes SWI/SNF and Mi2, respectively, did not increase the reporter activity (SI Fig. 6). Combined RNAi of some of these genes produced similar results (not shown). Therefore, even though Rb represses E2F in this assay, none of these chromatin repressors is required for inhibition of E2F. This conclusion is in agreement with a previous report using endogenous E2F targets as a readout (10).
Although this result might reflect the redundant or context-dependent activities of these regulators, it prompted us to systematically screen the whole Drosophila genome by RNAi for additional chromatin-related factors that are essential for E2F repression. In initial control experiments and pilot screens involving hundreds of genes tested, Rbf1 was detected as the only predominant negative regulator of E2F, and RNAi of the rest of the genes did not display any significant reporter activation (generally a <10% increase) above background. From a high-throughput screen of the Drosophila dsRNA library containing >21,000 dsRNA species and covering >90% of the annotated Drosophila genome (11), only 18 dsRNAs up-regulated the reporter to >50% (SI Table 1), which was set as an arbitrary cutoff. Identification of Rbf1 provided validation for the screen.
Repression of E2F Activity by the Dom and Malignant Brain Tumor (MBT) Proteins.
Among the identified genes are several potential chromatin regulators, including Domino (Dom) (14) and two uncharacterized fly genes, CG14514 and CG4621, all of which are homologous to the yeast SWR1 chromatin-remodeling complex subunits Swr1, Bdf1, and Vps72, respectively (15–17). Dom is a member of the SNF2 family of ATPases (14). The three factors were previously found to be copurified in a fly chromatin-modulating complex (18).
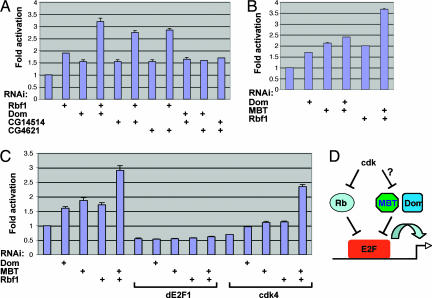
To verify whether these factors repress E2F, we synthesized new dsRNAs from distinct sets of gene-specific primers and repeated the RNAi/reporter assay. Consistent with the original screen, RNAi depletion of Dom, CG14514 or CG4621, resulted in activation of the E2F-reporter (Fig. 2A). Moreover, codepletion of a combination of these genes activated the reporter to the same extent as that induced by RNAi of any gene alone (Fig. 2A). This finding suggests that these factors act in a common pathway consistent with the idea that they exist in the same complex. It is likely they represent three essential subunits of the putative fly SWR1-like chromatin-remodeling complex. As such, ablation of each individual subunit would inactivate the entire complex. Identification of multiple subunits from the same presumed complex also underscores the specificity of the screen. Because other ATP-dependent chromatin-remodeling complexes fail to repress E2F in this assay, the capability to inhibit E2F may be unique to Dom, rather than a general feature of all chromatin remodelers.
Fig. 2.
Regulation of E2F by the Dom complex and MBT proteins. (A) Effects of RNAi of the Dom complex components and their epistatic effect relative to Rbf1. (B) Epistasis analysis of the MBT proteins (L3mbt and CG16975) relative to Dom and Rbf1. (C) Epistasis analysis of known and potential E2F regulators including Rbf1, Dom, and MBTs relative to dE2F1 and Drosophila cdk4. (D) A model for Dom and MBT-mediated regulation of E2F. Dom and MBT confer E2F inhibitory activities that are independent of Rb. They may act downstream of or in parallel to cdk. Absence or small scale of error bar indicates that results from duplicate experiments are nearly identical.
We further attempted to investigate whether the putative fly Dom complex might contribute to Rb-mediated repression. We found, however, that simultaneous disruption of Rbf1 with subunits of this potential complex resulted in strong additive effects on reporter activation as compared with targeting either one alone (Fig. 2A), suggesting that Rbf1 and these factors may independently repress E2F. Such effects appeared to be specific, because codepletion of Rbf1 with Rbf2 or a few other selected genes (SI Fig. 6 and data not shown) or simultaneous removal of any two of Dom, CG14514, and CG4621 (Fig. 2A), did not result in a similar response.
Two related proteins isolated from the screen, the fly tumor suppressor L3mbt (19) and CG16975/dSfmbt (20), share a similar overall architecture to the Polycomb group (PcG) protein Scm (21). These three are the only proteins in Drosophila containing multiple repeats of the so-called MBT domain, which is structurally related to the HP1 chromodomain (22) and can recognize selectively methylated histone tails (20, 23). PcG proteins promote formation of higher-order repressive chromatin structures (24), implicating L3mbt and CG16975 in chromatin regulation. We examined the three MBT factors for their potential role in E2F regulation. RNAi directed at either L3mbt or CG16975 activated the E2F reporter to a similar degree, but RNAi of Scm exhibited little effect (not shown). In all of the subsequent assays, we codepleted L3mbt and CG16975 as a pair, which constantly led to stronger activation of the E2F reporter than did RNAi of Rbf1 (Fig. 2B). Although simultaneous depletion of the MBT proteins with Dom resulted in only a slight increase of reporter activity over RNAi of MBTs alone, codepletion of the MBTs with Rbf1 gave rise to a remarkable additive effect on E2F activation (Fig. 2B), revealing Rbf1-independent activities of the MBT proteins.
The additive effects are unlikely to be due to differences in the RNAi-mediated knockdown of Rbf1. As assessed by Western blot analysis, Rbf1 was depleted to a similar extent under the multiRNAi conditions (SI Fig. 7). Moreover, if addition of other dsRNAs interferes with inactivation of Rbf1, one would expect reduced reporter activation, which is in contrast to our observation.
The repressive activity of these factors is specific to the E2F reporter, because they display no inhibitory effect on the reporters carrying binding sites for p53 or TCF4 (not shown). We further tested whether the reporter responses truly depended on E2F. Indeed, removal of dE2F1 by RNAi completely eliminated reporter activation resulting from RNAi of Rbf1, Dom, or MBTs (Fig. 2C). This requirement for E2F suggests these factors may act upstream of E2F.
We wished to gain further insight into the epistatic relation of these factors with the E2F pathway. It is well established that cyclin-dependent kinases (Cdks) activate E2F by phosphorylating and dissociating Rb-like proteins from E2F. As expected, RNAi of cdk4 resulted in the down-regulation of the E2F reporter activity. Although Rbf1 acts downstream of cdk4, RNAi of Rbf1 only partially reversed the effect caused by RNAi of cdk4 (Fig. 2C), indicating the potential existence of additional cdk4 downstream factors. We found that the reporter response to depletion of cdk4 was also partially overcome by codepletion of Dom or the MBT proteins. Maximal suppression was achieved by simultaneous removal of both Rbf1 and the MBTs (Fig. 2C). These in vitro gene epistasis interactions thus allowed us to tentatively place the Dom and MBT proteins in a hierarchy upstream of E2F and, possibly, downstream of cdk (Fig. 2D).
Association of the Dom and MBT Proteins with E2F.
To explore the mechanism of action of Dom and MBTs, we were interested in determining whether such repression on E2F is direct. Fly cell extracts were immunoprecipitated in parallel with antibodies for Dom, Rbf1, and dE2F1, the latter two serving as positive controls. dE2F1 was readily detected in the immunoprecipitates (Fig. 3A). Similar analyses with cells expressing a Flag-tagged version of L3mbt showed that dE2F1 coprecipitates with Flag-L3mbt (Fig. 3A). We conclude that Dom and MBT proteins associate with endogenous dE2F1. However, Rbf1 was undetectable in the Dom or MBT complexes by anti-Rbf1 Western blot analysis (Fig. 3A), which is consistent with the apparently Rb-independent activities of Dom and MBT. In addition, mammalian orthologs of the presumed Dom complex subunits and MBT proteins interact with E2F as well (SI Fig. 8).
Fig. 3.
The Dom and MBT proteins form complexes with E2F and are present on E2F target gene promoters. (A) The Dom and MBT proteins associate with E2F. Cellular extracts from regular S2* cells or S2* cells transiently transfected with a Flag-L3mbt expression vector were subjected to affinity purification with antibodies for Dom, Rbf1, dE2F1, and Flag and Western blot analysis with anti-dE2F1. No signal was observed in control precipitation in which cellular lysates from untransfected cells were incubated with the anti-Flag antibody. (B) The Dom, H2Av, and MBT proteins are present at the promoters of E2F target genes. Chromatin from cross-linked regular S2* cells was immunoprecipitated with anti-Dom (Dom), anti-H2Av (H2Av), and anti-Flag (control) antibodies. In a parallel experiment (for the right three columns), extracts from S2* cells expressing Flag-L3mbt were precipitated with anti-Flag (F-L3mbt) and nonspecific IgG (control) antibodies. The pelleted chromatin fragments were analyzed by PCR with primers specific for PCNA, DNA polα (Pol), CG14545, CG8399, CG3105 (belonging to the groups A to E, respectively), and RP49 promoters. (C) Recruitment of Dom and MBT proteins to E2F targets depends on E2F. Flag-L3mbt transfected S2* cells were treated with RNAi of dE2Fs (dE2F1, or combined dE2F1 and dE2F2), and then subjected to ChIP with Dom and Flag antibodies, respectively. Several E2F targets were selected for promoter-binding assay. (D) Deposition of H2Av at selected E2F target promoters depends on Dom and dE2F1. S2* cells were incubated with Dom or dE2F1 dsRNAs and subsequently subjected to ChIP assays with an anti-H2Av antibody.
We speculated that Dom and MBT proteins might be recruited to E2F-bound promoters through forming complexes with E2F. Recently, Drosophila genes that are directly regulated by dE2F were identified and categorized into groups A to E (13). Genes of the A and B groups primarily depend on dE2F1 and Rbf1 and are mostly related to cell proliferation, whereas genes of the D and E groups are mainly repressed by dE2F2 and involved in differentiation. We chose E2F-dependent promoters containing E2F-binding sites from each group and analyzed in vivo promoter binding by ChIP. In chromatin immunoprecipitated with antibodies for Dom or Flag (for cells expressing Flag-L3mbt), most E2F target promoters examined were specifically enriched as compared with rp49, a negative control gene whose expression is not subject to E2F regulation (Fig. 3B), suggesting these proteins are normally present at promoter regions with E2F sites. If the selected promoters are representative of these groups in general, then many E2F-controlled genes are likely to be direct targets for Dom and MBT proteins. We further examined the dependence of E2F in the recruitment of these factors. In dE2F1-depleted cells, the presence of Dom at the selected E2F target promoters was greatly reduced (Fig. 3C), suggesting its recruitment is mainly dE2F1-dependent. Similarly, association of L3mbt with these genes was impaired when both dE2F1 and dE2F2 were removed by RNAi (Fig. 3C), pointing out its dependence on either dE2F1 or dE2F2. The molecular mechanism underlying these distinct modes of recruitment remains to be elucidated. These observations indicate that Dom and MBT proteins are brought to E2F targets primarily via interactions with E2F.
Because the yeast SWR1 regulates chromatin structure by incorporating the histone H2A variant, H2AZ, into nucleosomes (15–17), we also looked at the occupancy of the fly histone H2AZ homolog, H2Av, at E2F target promoters using an anti-H2Av antibody (25). The presence of H2Av at those promoters was clearly detected (Fig. 3B), which was both Dom- and dE2F1-dependent (Fig. 3D). This finding supports a potentially conserved function between Dom and SWR1.
However, promoter binding does not fully correlate with gene expression in cultured cells (13, 26). We thus tested the dependence of expression of endogenous E2F targets on the Dom and MBT proteins. Northern blot analysis indicated that depletion of Dom or MBTs failed to relieve repression on S-phase genes, such as cyclin E and PCNA (not shown). We then examined global gene expression by a microarray assay. In cells treated with RNAi of the MBT genes, expression of dE2Fs and Rbfs was not altered. Among the known E2F target genes (13), the overall expression level for many genes of the D and E groups, but not the A and B groups, was up-regulated (SI Table 2). This result was confirmed by Northern blot analysis of several selected genes (Fig. 4A). Because mammalian MBT proteins are detected in a complex with HMTs G9a and EuHMT1 (27), both of which are implicated in dimethylation of histone H3K9 and gene repression, and the MBT domain binds di-methylated H3K9 (20), we wondered whether removal of MBT proteins might cause changes of histone modification at E2F target genes. We found that, at the MBT-dependent E2F target promoters, particularly CG14545, the abundance of methylated H3K9 was decreased, whereas acetylated histone H4, a marker for active chromatin, was increased upon RNAi treatment (Fig. 4B). This result implies that MBT proteins might directly or indirectly contribute to epigenetic status of some E2F target genes. Furthermore, most of the MBT-regulated E2F targets were also derepressed when the Dom complex was depleted (SI Table 2). Therefore, the MBT and Dom factors seem to be rate-limiting for repression of a subset of endogenous E2F target genes. It remains possible that they may regulate some other E2F targets under different contexts. In addition, RNAi assay may be biased in favor of detection of genes whose repressive state is maintained rather than initiated by the Dom and MBT proteins.
Fig. 4.
Changes in gene expression and histone modification induced by depletion of MBT. (A) Northern blot analysis of MBT-repressed E2F target genes. S2* cells were treated with RNAi of indicated E2F regulators. Total RNA was extracted from cells incubated with dsRNAs for 6 days, and the blots were probed for expression of several E2F target genes. The ethidium bromide staining of rRNA showed comparable loadings for each sample. Depletion of MBTs resulted in a strong induction in RNA expression of the selected E2F targets, pointing out their essential role in repression of endogenous E2F-regulated genes. (B) Epigenetic changes at E2F target promoters after RNAi of MBT. S2* cells were treated with RNAi of L3mbt and CG16975 for 6 days and then subjected to ChIP with antibodies recognizing dimethylated H3K9 or acetylated H4. Several E2F targets were chosen for promoter-binding analysis.
Genetic Interactions of Dom with the E2F Pathway.
We further ask whether genes identified from the screen may genetically interact with the E2F pathway in flies. It has been reported that adult flies homozygous for cycEJP, a cycE hypomorphic allele with reduced cyclin E expression, exhibit a small eye phenotype because of impaired cell proliferation during eye development (28) (Fig. 5A). This phenotype is sensitive to E2F activity. Decreasing the dosage of dE2F1 and Rbf1 shows enhancement and suppression of the rough eye phenotype, respectively (28). To assess the effect of altering the dosage of Dom, we generated fly mutants homozygous for cycEJP and heterozygous for dom1, a strong dom allele (14). The eye size and morphology of these flies were essentially restored to normal (Fig. 5B). The penetrance of this genetic suppression is 92% (35 of 38 cases). Thus, halving the dosage of Dom, like Rbf1, is sufficient to suppress cycEJP. This observation is consistent with a role for Dom as a negative E2F regulator possibly downstream of cdk. Because Dom may function through deposition of the histone variant H2Av, we found that mutation in H2Av indeed also suppressed cycEJP (Fig. 5D), although to a lesser penetrance (77%, 53 of 69 cases).
Fig. 5.
dom mutation genetically suppresses cycE and enhances the phenotype of transgenic dE2F1. (A) Homozygotic cycEJP/cycEJP flies display a small eye phenotype because of reduced cell proliferation during eye development. (B) In a dom heterozygotic background, the cycEJP eye phenotype is rescued in the cycEJP, dom1/cycEJP flies. (C) Flies heterozygous for both cycEJP and dom (cycEJP, dom1/+) are normal and do not exhibit any obvious phenotype. (D) Mutation in his2Av also suppresses the cycEJP eye phenotype as shown in the cycEJP/cycEJP; his2Av/+ flies. (E) Eyes of the cycE and his2Av double heterozygous flies (cycEJP/+; his2Av/+) are normal. (F) Transgenic flies (GMR-dE2FdDPp35/+) with overexpression of dE2F1, its dimerization partner dDP, and p35 (a baculovirus antiapoptotic protein) in the eye imaginal disc using the GMR promoter causes an increase in the number of cells in the adult eye and disruption of the regular pattern of ommatidia. (G) Reducing the dosage of dom using the dom14 heterozygotes (GMR-dE2FdDPp35/dom) resulted in strong enhancement of the E2F rough eye phenotype. All pictures were taken at ×100 magnification.
We sought to address genetic interactions of dom directly with E2F. Overexpression of dE2F1 in the developing fly eye induces increased cell proliferation, resulting in mildly overgrown and rough eyes in adults (29) (Fig. 5F). The eye phenotype responds to changes in the level of genes known to affect E2F activity. For example, the severity of rough eye is enhanced by reduction of Rbf1 dosage (29). In an enhancer screen of the entire Bloomington Stock Center's deficiency library collection, only four deficiencies were recovered that enhanced E2F overexpression phenotype, but the responsible genes were not identified (29). Because Dom is located in the region deleted in the deficiency Df(2R)PuD17, we thus propose that Dom is the candidate gene responsible for the E2F enhancement seen with Df(2R)PuD17. Indeed, we found that when the dE2F1 transgene was put into a heterozygous dom background, the degree of disorganization in the arrangement of ommatidia was substantially enhanced, leading to severe eye roughening (Fig. 5G). Therefore, decreasing the level of Dom enhances the phenotype of elevated E2F activity. These genetic interactions support the model in which the Dom complex functions to repress Drosophila E2F in vivo.
Discussion
We have conducted a systematic RNAi screen to seek genes required for repression of E2F transcriptional activity. From the screen, the putative Dom/SWR1 chromatin remodeling complex and the PcG-like MBT domain-containing factors were identified as E2F repressors. We have shown that these proteins are recruited to E2F target promoters through association with E2F and inhibit E2F in an apparently Rb-independent manner. Depletion of these genes resulted in derepression of some endogenous E2F target genes accompanied by changes in histone modification. More importantly, dom genetically interacted with the E2F pathway. These proteins show an extensive degree of evolutionary conservation, indicating the mechanism of E2F regulation provided by these factors may be well conserved.
Regulation of E2F is tightly linked to cell proliferation and differentiation. Existing evidence suggests that perturbation of the Dom and MBT proteins may cause dysregulation of these cellular processes. Apart from the fact that the heterozygous dom mutation modifies cell growth in an E2F-transgenic or a cycE hypomorphic background, fly mutants homozygous for several dom alleles show enlarged lymph glands apparently because of excessive proliferation of prehemocytes (14). In human, the Dom complex subunit YL1 possesses growth suppressive activity (30), and the Dom homolog p400 is an essential target for the viral oncoprotein E1A-mediated transformation (31). Indeed, overexpression of E1A disrupts the association of E2F with the Dom complex in mammalian cells (SI Fig. 8). Furthermore, mutations in the fly tumor suppressor gene l3mbt result in overgrowth of the larval brain lobes and epithelial imaginal discs, and failure of neural differentiation (19). This is intriguing, because in mammalian cells, many E2F-regulated genes are repressed during quiescence and differentiation, and mammalian MBT proteins are found in an inhibitory E2F complex purified from quiescent cells (27).
Although the mechanism of Rb-mediated repression on E2F is complex, our studies indicate that Dom and MBT possess Rb-independent activities. In support of this view, recent studies suggest that the Caenorhabditis elegans Dom and Rb homologs share redundant functions in vulva development, a process controlled by the E2F pathway (32). In addition, these proteins may participate in distinct E2F complexes (Fig. 3A). Mammalian MBT orthologs have been identified from Rb-independent complexes (27), and they can associate with E2F forms lacking the Rb-binding motif, such as E2F6 and a C-terminal truncated E2F3 mutant (SI Fig. 8). Interestingly, L3mbt is shown to interact with dREAM, a dE2F2-Rb complex, even though it is not a stoichiometric subunit (33, 34). But unlike L3mbt, RNAi of dE2F2 and several other components of the core dREAM complex had no effect on the E2F reporter (not shown). This observation may hence indicate the existence of multiple L3mbt-containing complexes or hint at a potential collaboration among different E2F regulatory activities. So far, there is no evidence linking Dom and CG16975 to Rb. It is likely that both Rb-mediated and -independent chromatin modulations play critical roles in E2F regulation and cell proliferation. Future biochemical and genetic studies may shed light on these potentially independent and collaborative relations.
Materials and Methods
Plasmids.
The E2F-dependent luciferase reporter was made by inserting oligos containing four copies of an E2F-binding site (TTTTCGCGCGAAAA) into the pGL3 (Promega, Madison, WI) vector with a minimal Hsp70 promoter. To make the Renilla control reporter, the firefly luciferase coding region in the pGL3-Hsp70 was replaced with the Renilla luciferase cDNA. Fly expression vector for Flag-L3mbt was created by PCR amplification of the l3mbt full-length cDNA with a 5′ primer encoding the Flag tag and cloning of this fragment into pAct5.
Fly Strains.
Fly mutant strains used in this study were described previously: cycEJP (28), dom1 and dom14 (14), GMR-dE2F1 (29), and his2Av05146 (35). The cycEJP dom1/cycEJP flies were made by recombining a second chromosome dom1 line onto a marked cycEJP chromosome (using dp, b, cycEJP, cn, and bw). Individual recombinant candidate chromosomes balanced over CyO were backcrossed to the dom1/CyO and to the homozygous cycEJP stocks to confirm the presence of dom1 and cycEJP, respectively. Adult fly eyes were analyzed by scanning electron microscopy after dehydration of the heads in a series of ethanol washes.
Drosophila Cell Culture and RNAi Treatment.
See SI Text for additional information.
Supplementary Material
Acknowledgments
We thank Susan Armknecht, Nadire Ramadan, and the Harvard Drosophila RNAi Screening Center for expert assistance with the screen; Holger Babbe for FACS analysis; Marie Meister (Institut de Biologie Moleculaire et Cellulaire, Strasbourg, France) for support; and Maki Asano (Duke University Medical Center, Durham, NC), Ramanuj DasGupta, Nick Dyson (Massachusetts General Hospital Cancer Center, Boston, MA), Robert Glaser (Wadsworth Center, New York State Department of Health, New York, NY), Terry Orr-Weaver (Massachusetts Institute of Technology, Cambridge, MA), Helena Richardson (Peter MacCullum Cancer Center, Australia), and Guangchao Sui for kindly providing the fly stocks, antibodies, and plasmids. We thank Steve Elledge, Kevin Freeman, and Robert Weiss (Cornell University, Ithaca, NY) for critically reading the manuscript, and P.L. laboratory members for advice. This study was funded in part by the Howard Hughes Medical Institute. Work at the Drosophila RNAi Screening Center is supported by Grant R01GM067761 from the U.S. National Institute of General Medical Sciences. J.L. was supported by a fellowship from the U.S. Department of Defense Breast Cancer Research Program.
Abbreviations
- HDAC
histone deacetylase
- HMT
histone methyltransferase
- Cdk
cyclin-dependent kinase.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0610279104/DC1.
References
- 1.Lund AH, van Lohuizen M. Genes Dev. 2004;18:2315–2335. doi: 10.1101/gad.1232504. [DOI] [PubMed] [Google Scholar]
- 2.Trimarchi JM, Lees JA. Nat Rev Mol Cell Biol. 2002;3:11–20. doi: 10.1038/nrm714. [DOI] [PubMed] [Google Scholar]
- 3.Classon M, Harlow E. Nat Rev Cancer. 2002;2:910–917. doi: 10.1038/nrc950. [DOI] [PubMed] [Google Scholar]
- 4.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Barrette TR, Ghosh D, Chinnaiyan AM. Nat Genet. 2005;37:579–583. doi: 10.1038/ng1578. [DOI] [PubMed] [Google Scholar]
- 5.Frolov MV, Dyson NJ. J Cell Sci. 2004;117:2173–2181. doi: 10.1242/jcs.01227. [DOI] [PubMed] [Google Scholar]
- 6.Nielsen SJ, Schneider R, Bauer UM, Bannister AJ, Morrison A, O'Carroll D, Firestein R, Cleary M, Jenuwein T, Herrera RE, et al. Nature. 2001;412:561–565. doi: 10.1038/35087620. [DOI] [PubMed] [Google Scholar]
- 7.Gonzalo S, Garcia-Cao M, Fraga MF, Schotta G, Peters AH, Cotter SE, Eguia R, Dean DC, Esteller M, Jenuwein T, et al. Nat Cell Biol. 2005;7:420–428. doi: 10.1038/ncb1235. [DOI] [PubMed] [Google Scholar]
- 8.Narita M, Nunez S, Heard E, Narita M, Lin AW, Hearn SA, Spector DL, Hannon GJ, Lowe SW. Cell. 2003;113:703–716. doi: 10.1016/s0092-8674(03)00401-x. [DOI] [PubMed] [Google Scholar]
- 9.Braig M, Lee S, Loddenkemper C, Rudolph C, Peters AH, Schlegelberger B, Stein H, Dorken B, Jenuwein T, Schmitt CA. Nature. 2005;436:660–665. doi: 10.1038/nature03841. [DOI] [PubMed] [Google Scholar]
- 10.Taylor-Harding B, Binne UK, Korenjak M, Brehm A, Dyson NJ. Mol Cell Biol. 2004;24:9124–9136. doi: 10.1128/MCB.24.20.9124-9136.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Echeverri CJ, Perrimon N. Nat Rev Genet. 2006;7:373–384. doi: 10.1038/nrg1836. [DOI] [PubMed] [Google Scholar]
- 12.Tao Y, Kassatly RF, Cress WD, Horowitz JM. Mol Cell Biol. 1997;17:6994–7007. doi: 10.1128/mcb.17.12.6994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dimova DK, Stevaux O, Frolov MV, Dyson NJ. Genes Dev. 2003;17:2308–2320. doi: 10.1101/gad.1116703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruhf ML, Braun A, Papoulas O, Tamkun JW, Randsholt N, Meister M. Development (Cambridge, UK) 2001;128:1429–1441. doi: 10.1242/dev.128.8.1429. [DOI] [PubMed] [Google Scholar]
- 15.Krogan NJ, Keogh MC, Datta N, Sawa C, Ryan OW, Ding H, Haw RA, Pootoolal J, Tong A, Canadien V, et al. Mol Cell. 2003;12:1565–1576. doi: 10.1016/s1097-2765(03)00497-0. [DOI] [PubMed] [Google Scholar]
- 16.Kobor MS, Venkatasubrahmanyam S, Meneghini MD, Gin JW, Jennings JL, Link AJ, Madhani HD, Rine J. PLoS Biol. 2004;2:E131. doi: 10.1371/journal.pbio.0020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C. Science. 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- 18.Kusch T, Florens L, Macdonald WH, Swanson SK, Glaser RL, Yates JR, III, Abmayr SM, Washburn MP, Workman JL. Science. 2004;306:2084–2087. doi: 10.1126/science.1103455. [DOI] [PubMed] [Google Scholar]
- 19.Wismar J, Loffler T, Habtemichael N, Vef O, Geissen M, Zirwes R, Altmeyer W, Sass H, Gateff E. Mech Dev. 1995;53:141–154. doi: 10.1016/0925-4773(95)00431-9. [DOI] [PubMed] [Google Scholar]
- 20.Klymenko T, Papp B, Fischle W, Kocher T, Schelder M, Fritsch C, Wild B, Wilm M, Muller J. Genes Dev. 2006;20:1110–1122. doi: 10.1101/gad.377406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bornemann D, Miller E, Simon J. Development (Cambridge, UK) 1996;122:1621–1630. doi: 10.1242/dev.122.5.1621. [DOI] [PubMed] [Google Scholar]
- 22.Wang WK, Tereshko V, Boccuni P, MacGrogan D, Nimer SD, Patel DJ. Structure (London) 2003;11:775–789. doi: 10.1016/s0969-2126(03)00127-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim J, Daniel J, Espejo A, Lake A, Krishna M, Xia L, Zhang Y, Bedford MT. EMBO Rep. 2006;7:397–403. doi: 10.1038/sj.embor.7400625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bantignies F, Cavalli G. Curr Opin Cell Biol. 2006;18:275–283. doi: 10.1016/j.ceb.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 25.Leach TJ, Mazzeo M, Chotkowski HL, Madigan JP, Wotring MG, Glaser RL. J Biol Chem. 2000;275:23267–23272. doi: 10.1074/jbc.M910206199. [DOI] [PubMed] [Google Scholar]
- 26.Takahashi Y, Rayman JB, Dynlacht BD. Genes Dev. 2000;14:804–816. [PMC free article] [PubMed] [Google Scholar]
- 27.Ogawa H, Ishiguro K, Gaubatz S, Livingston DM, Nakatani Y. Science. 2002;296:1132–1136. doi: 10.1126/science.1069861. [DOI] [PubMed] [Google Scholar]
- 28.Secombe J, Pispa J, Saint R, Richardson H. Genetics. 1998;149:1867–1882. doi: 10.1093/genetics/149.4.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Staehling-Hampton K, Ciampa PJ, Brook A, Dyson N. Genetics. 1999;153:275–287. doi: 10.1093/genetics/153.1.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horikawa I, Tanaka H, Yuasa Y, Suzuki M, Shimizu M, Oshimura M. Exp Cell Res. 1995;220:11–17. doi: 10.1006/excr.1995.1286. [DOI] [PubMed] [Google Scholar]
- 31.Fuchs M, Gerber J, Drapkin R, Sif S, Ikura T, Ogryzko V, Lane WS, Nakatani Y, Livingston DM. Cell. 2001;106:297–307. doi: 10.1016/s0092-8674(01)00450-0. [DOI] [PubMed] [Google Scholar]
- 32.Ceol CJ, Horvitz HR. Dev Cell. 2004;6:563–576. doi: 10.1016/s1534-5807(04)00065-6. [DOI] [PubMed] [Google Scholar]
- 33.Korenjak M, Taylor-Harding B, Binne UK, Satterlee JS, Stevaux O, Aasland R, White-Cooper H, Dyson N, Brehm A. Cell. 2004;119:181–193. doi: 10.1016/j.cell.2004.09.034. [DOI] [PubMed] [Google Scholar]
- 34.Lewis PW, Beall EL, Fleischer TC, Georlette D, Link AJ, Botchan MR. Genes Dev. 2004;18:2929–2940. doi: 10.1101/gad.1255204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Swaminathan J, Baxter EM, Corces VG. Genes Dev. 2005;19:65–76. doi: 10.1101/gad.1259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.