Abstract
RAS family members are among the most frequently mutated oncogenes in human cancers. Given the utility of zebrafish in both chemical and genetic screens, developing RAS-induced cancer models will make large-scale screens possible to understand further the molecular mechanisms underlying malignancy. We developed a heat shock-inducible Cre/Lox-mediated transgenic approach in which activated human kRASG12D can be conditionally induced within transgenic animals by heat shock treatment. Specifically, double transgenic fish Tg(B-actin-LoxP-EGFP-LoxP-kRASG12D; hsp70-Cre) developed four types of tumors and hyperplasia after heat shock of whole zebrafish embryos, including rhabdomyosarcoma, myeloproliferative disorder, intestinal hyperplasia, and malignant peripheral nerve sheath tumor. Using ex vivo heat shock and transplantation of whole kidney marrow cells from double transgenic animals, we were able to generate specifically kRASG12D-induced myeloproliferative disorder in recipient fish. This heat shock-inducible recombination approach allowed for the generation of multiple types of RAS-induced tumors and hyperplasia without characterizing tissue-specific promoters. Moreover, these tumors and hyperplasia closely resemble human diseases at both the morphologic and molecular levels.
Keywords: myeloproliferative disorder, RAS, rhabdomyosarcoma, intestine, malignant peripheral nerve sheath tumor
RAS genes encode a family of 21-kDa proteins that switch between inactive GDP-bound (RAS-GDP) and active GTP-bound (RAS-GTP) conformations. Once in its activated GTP-bound form, RAS interacts with downstream effectors to modulate diverse cellular responses, including proliferation, differentiation, and survival (1). Point mutations within RAS family members often occur at codon 12, 13, or 61 (2), which abolish RAS-GTP hydrolysis and lead to constitutive activation of downstream signaling pathways. These activating mutations are common in human malignancies; for example, >90% of pancreatic adenocarcinomas, 50% of colorectal cancers, 25–50% of lung cancers, 5–35% of rhabdomyosarcomas (RMS), and 25–50% of myeloid leukemia have mutational activation of RAS family members. Among the three different human RAS genes (H-, N-, and K-RAS), K-RAS is the most frequently mutated member in human tumors (2).
In recent years, mouse models for kRas-induced tumorigenesis have been developed that faithfully recapitulate human disease (3). Such advances have been made possible through use of tissue-specific and inducible expression of oncogenic kRas, mainly achieved through the advent of Cre/Lox technology (4–6). Although possible in mouse models, large-scale, whole-genome, unbiased genetic screens designed to identify genetic modifiers of RAS-induced malignancy are costly and require large number of animals. Moreover, chemical screens with whole animals to discover drugs that affect cancer pathways have not been described in mouse (7). In contrast, live zebrafish can be used in both genetic screens and chemical genetic approaches (8). Combined with the propensity of fish to develop tumors that are similar to human disease (9), zebrafish has become an attractive model system to interrogate cancer biology. Currently, transgenic approaches are beginning to be widely used to generate zebrafish cancer models (10–13), but one of the major challenges in the field is developing conditional gene expression technology (14–16). Additionally, for some organs, tissue-specific promoters have yet to be identified, making transgene delivery unavailable to certain tissue types.
Here, we developed a heat shock-inducible Cre/Lox-mediated transgenic approach, in which activated human kRASG12D can be conditionally induced within transgenic animals by heat shock treatment. Four types of tumors and hyperplasia were generated by using our inducible-transgenic approach. When heat shock treatment was applied to isolated marrow cells ex vivo and transplanted to irradiated recipients, a myeloproliferative disorder (MPD) was specifically induced in recipient fish. Our studies describe a heat shock-inducible cancer model in vertebrates. In addition, we have generated zebrafish disease models of RMS and MPD, both of which have RAS pathway activation in human patients.
Results
Human kRASG12D Expression Can Be Induced in Transgenic Zebrafish by Heat Shock-Induced Cre-Mediated Recombination.
To express the human kRASG12D transgene in various tissue types, a 2.7-kb zebrafish B-actin promoter was used to drive the expression of a floxed transgene cassette containing an enhanced green fluorescent protein (EGFP) followed by a strong transcriptional stop element. This floxed EGFP cassette lies upstream from a constitutively active human kRASG12D transgene (Fig. 1A). Ten stable transgenic zebrafish B-actin-LoxP-EGFP-pA-LoxP-kRASG12D lines were generated (LGL-RAS). All stable lines expressed EGFP as embryos, whereas only four lines had broad EGFP expression as adults (17), suggesting that transgene expression is affected by positional effects [for line 25A, see supporting information (SI) Table 2 and SI Fig. 6]. A heat shock-inducible Cre line (hsp70-Cre) was obtained (generously provided by Andre Quinkertz and the late Jose Campos-Ortega, University of Cologne, Cologne, Germany), and Cre expression was optimized by varying the stage and duration of heat shock at 37°C (SI Fig. 7). In brief, heat shock for 30–120 min at 37°C leads to optimal Cre expression in hsp70-Cre transgenic embryos, and treatment from either 4–5 hours postfertilization (hpf) or 24–25 hpf results in a high level of Cre expression without affecting development or viability. Moreover, Cre RNA is ubiquitously expressed in animals immediately after heat shock, as determined by whole-mount in situ hybridization (SI Fig. 7). By contrast, untreated hsp70-Cre embryos expressed 2.9 ± 2.0% of the Cre transcript levels compared with siblings heat shocked for 1 h (n = 4; heat shock from 24–25 hpf at 37°C and assessed for Cre expression at 28 hpf). Thus, the hsp70 promoter can be induced by heat shock treatment; however, the promoter is active at low levels during development even in the absence of heat shock (18, 19).
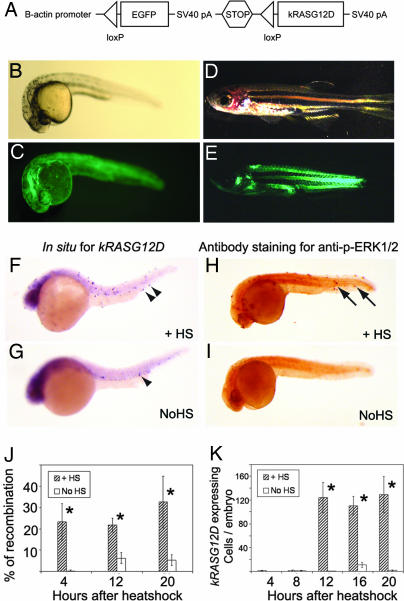
Fig. 1.
Human kRASG12D transgene expression can be induced by Cre-mediated recombination in stable transgenic B-actin-LoxP-EGFP-LoxP-kRASG12D zebrafish. (A) Diagram of B-actin-LoxP-EGFP-LoxP-kRASG12D transgene. (B–E) Stable transgenic animals (line 25A) at 24 hpf (B and C), and 44 dpf (D and E). (B and D) Bright-field. (C and E) EGFP fluorescence. (F and G) Whole-mount in situ hybridization for kRASG12D performed on 24 hpf double transgenic embryos (LGL-RAS; hsp70-Cre) with heat shock (F) or without (G). Arrowheads denote kRASG12D transcript-expressing cells. (H and I) Whole-mount immunostaining with anti-phospho-ERK1/2 antibody performed on 24 hpf double transgenic embryos with heat shock (H) or without (I). Cells with ERK1/2 activation are denoted by arrows. (J) Percentage of recombination at the genomic DNA in single embryos heat shocked from 4 to 5 hpf and analyzed at 8, 16, and 24 hpf. (K) Number of kRASG12D-expressing cells in single embryos heat shocked from 4 to 5 hpf and analyzed at 8, 12, 16, 20, and 24 hpf. Statistic significance (P < 0.001) is indicated by asterisks. +HS, heat shock; NoHS, non-heat shocked.
Each LGL-RAS line was bred to hsp70-Cre fish, heat shocked at 37°C from 4 to 5 hpf, and assessed for genomic DNA recombination and RAS expression at 24 hpf. All tested lines showed recombination at the DNA level (n = 7), whereas two lines expressed detectable kRASG12D as determined by whole-mount in situ hybridization (lines 25A and 48) (Fig. 1F). Whole-mount antibody staining for anti-phospho-ERK1/2 confirmed that the RAS/MAPK signaling pathway was activated in these two lines (Fig. 1H and SI Table 2). However, only the LGL-RAS line 25A developed tumors with short latency after heat shock. Thus, this line was used in all subsequent analysis.
To evaluate the efficiency and dynamics of the heat shock-inducible Cre/Lox recombination approach, a real-time PCR-based assay (20) was designed to access recombination efficiency in line 25A at the genomic DNA level (Fig. 1J and SI Fig. 8). When individual double transgenic embryos (LGL-RAS, line 25A; hsp70-Cre) were heat shocked from 4 to 5 hpf, recombination at genomic level could already be detected 4 h after heat shock (8 hpf, 23.2 ± 8.5% of the transgenes recombined; n = 8 animals), whereas, non-heat-shocked animals did not exhibit recombination (0.3 ± 0.4%; n = 7). By 24 hpf, 32.6 ± 12.2% of the transgenes had recombined in the heat shock cohort (n = 8), whereas non-heat-shocked animals exhibited significantly less recombination (5.3 ± 2.5%, P < 0.001; n = 8). kRASG12D transcripts were first detected by whole-mount in situ hybridization at 16 hpf with heat shock-treated animals having 123 ± 25 kRAS-positive cells per animal (n = 12 animals), whereas expression was absent in non-heat-shocked animals (0.4 ± 0.3 cells per animal, n = 12; Fig. 1K and SI Fig. 9). Consistent with the low-level genomic DNA recombination detected in non-heat-shocked embryos at 24 hpf, these animals exhibited significantly less but detectable kRASG12D expression compared with heat-shocked siblings (129 ± 30 kRAS-positive cells per animal in the heat-shocked group, n = 7 vs. 1.3 ± 0.9 in non-heat-shocked group, P < 0.001; n = 4, Fig. 1 G and K).
Early Activation of kRASG12D Leads to Juvenile Lethality and Four Types of Tumors and Hyperplasia in Adult Transgenic Fish.
A cohort of 300 double transgenic embryos (LGL-RAS; hsp70-Cre) was analyzed to determine whether somatic activation of kRASG12D would lead to tumor formation (21). One hundred eighty animals were heat shocked at 24 hpf for 1 h at 37°C, whereas 120 nontreated animals served as controls. Heat shock-treated, double transgenic animals were smaller in body size [by 43 days postfertilization (dpf), average body length = 0.945 ± 0.367 cm (n = 8) compared with 1.89 ± 0.09 cm in the control group (n = 12), P < 0.0001] and displayed a significant survival disadvantage (P < 0.0001, log rank analysis) (Fig. 2A). To investigate the reasons for juvenile lethality in heat-shocked animals, a second cohort of double transgenic animals was heat shocked and analyzed at 15, 20, and 25 days after fertilization. In situ staining of paraffin-embedded sections revealed that kRASG12D was expressed in multiple types of tissue, including intestinal epithelium (35–62% of juvenile fish at the age of 15–25 days), muscle (18–26%), neural cells (6–13%), and kidney marrow (0–9%) (SI Fig. 10). Among them, the intestinal epithelium is the most prominently affected tissue type, suggesting that gastrointestinal dysfunction may lead to stunted growth and juvenile death in the heat-shocked animals.
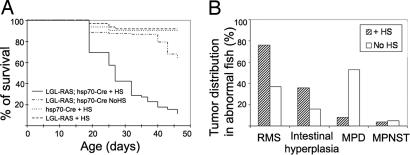
Fig. 2.
Early induction of kRASG12D expression leads to juvenile lethality and four types of tumors and hyperplasia in adult transgenic fish. (A) Kaplan–Meier survival curves. Double transgenic embryos were heat shocked at 24 hpf (LGL-RAS; hsp70-Cre +HS) or were not heat shocked (LGL-RAS; hsp70-Cre NoHS). Single transgenic animals were treated with heat shock (hsp70-Cre + HS or LGL-RAS + HS). (B) Tumor spectrum in diseased fish surviving past 25 days of life (n = 25 of 180 in the heat-shocked group and n = 19 of 120 in the non-heat-shocked group). Four types of lesions were observed: RMS, intestinal hyperplasia, MPD, and MPNST.
Double transgenic animals surviving past 25 days developed external tumor masses, difficulties in breathing and swimming, and/or progressive paleness. Twenty-five heat-shocked animals exhibiting such symptoms (average latency, 34.9 ± 6.2 days) were killed for pathological examination. In contrast, non-heat-shocked animals developed similar symptoms with a longer latency (n = 19, average latency, 65.8 ±21.3 days; P < 0.01). LGL-RAS or hsp70-Cre single transgenic animals that received heat shock did not develop signs of disease by 6 months of age (n = 60 fish per line). Histological examination of double transgenic diseased fish revealed four different types of malignancy, including skeletal muscle tumors (RMS), MPD, intestinal epithelial hyperplasia, and in rare cases, malignant peripheral nerve sheath tumors (MPNST) (SI Table 3). All four abnormalities were observed in both groups, but incidence and latency differed between heat-shocked and nontreated animals (Fig. 2B). The malignancies observed in the non-heat-shocked animals were likely induced by aberrant activation of the hsp70 promoter and subsequently Cre-mediated RAS activation.
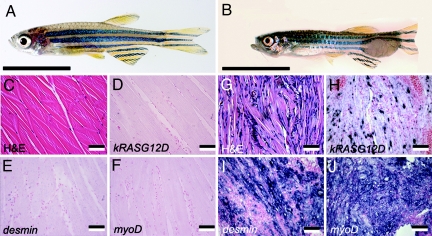
The most common tumor type was RMS, with 76% tumor-bearing fish having skeletal muscle tumors in the heat-shocked group (Fig. 3B). RMS comprised small, round blue cells interspersed with atypical terminally differentiated striated muscle fiber cells (Fig. 3G). RNA in situ hybridization detected kRASG12D expression in the perinuclear area of the multinucleated striated muscle fiber cells and in the mononuclear tumor cells (Fig. 3H). Tumors identified histologically as RMS expressed clinical diagnostic markers of human RMS, including desmin, myoD, and myogenin (Fig. 3 I and J and data not shown). In contrast, only 37% of non-heat-shocked tumor-bearing fish developed RMS. RAS family members are mutationally activated in 5–35% of embryonal rhabdomyosarcomas in human (22), suggesting that RAS pathway activation is required for initiation of this disease in both zebrafish and humans.
Fig. 3.
RMS is the most common tumor type in the heat-shocked group. (A) Normal zebrafish side view. (B) Double transgenic fish (46 dpf) with externally visible tumor mass at the tail region. (C and G) Hematoxylin/eosin (H&E)-stained sections of normal muscle (C) and RMS (G). (D–F) RNA in situ hybridization of normal muscle. (H–J) RNA in situ hybridization of RMS muscle. (C–J) Antisense probes are designated in the lower left corners. [Scale bars: 1 cm (A and B) and 50 μm (C–J).]
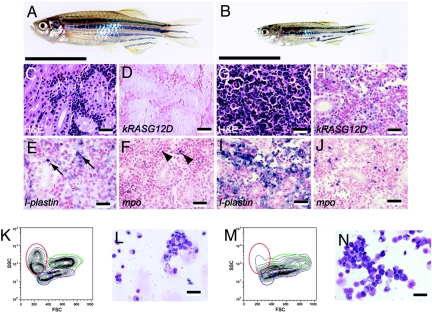
The second most common lesion was MPD (Fig. 4B). Heat-shocked fish developed this disease with a latency of 34 days (n = 2 of 25 fish), whereas non-heat-shocked fish developed MPD with a prolonged onset (mean latency = 66.2 ± 23.1 days, n = 10 of 19 fish, SI Table 3). Histological analysis revealed an expansion of hematopoietic cells in kidneys, comprising predominantly myeloid cells in various differentiation stages (Fig. 4G). In situ staining confirmed the expression of kRASG12D in a subset of kidney marrow cells (Fig. 4H). L-plastin-expressing monocyte/macrophage cells (23) and mpo-expressing granulocytes (24) were increased in diseased fish compared with single transgenic LGL-RAS animals (Fig. 4 I and J), confirming the expansion of the granulocyte and monocyte/macrophage compartments.
Fig. 4.
MPD is common in the non-heat-shocked group. (A) Normal zebrafish side view. (B) Double transgenic adult fish (53 dpf) with MPD. (C and G) Hematoxylin/eosin (H&E)-stained sections of normal (C) and MPD kidney (G). (D–F) RNA in situ hybridization of normal kidneys. (H–J) RNA in situ hybridization of MPD kidneys. Antisense probes are designated within each panel. (E) Arrows denote l-plastin-expressing monocytes in normal kidney. (F) Arrowheads denote mpo-expressing granulocytes in normal kidney. (K and M) FACS analysis of whole kidney marrow cells from normal (K) and MPD fish (M). Erythrocytes are shown in red, lymphocytes in blue, granulocytes and monocytes in green, and blood cell precursors in purple. FSC, Forward scatter; SSC, side scatter. (L and N) May–Grunwald–Giemsa-stained cytospins of normal (L) and MPD (N) kidney marrow cells. [Scale bars: 1 cm (A and B), 25 μm (C–J), and 12.5 μm (L and N).]
To characterize the MPD arising in these animals further, kidney marrow cells were collected and assessed for total cell number, flow cytometric analysis, and differential cell count. Diseased fish had increased numbers of total hematopoietic cells in the kidneys compared with age-matched, single transgenic LGL-RAS fish (3.16 × 106 ± 1.28 × 106 vs. 9.45 × 105 ± 2.31 × 105 cells; n = 4 per group, P = 0.014). Flow cytometric analysis of MPD kidney marrow cells (25) revealed a distinct population of cells lying between myeloid and progenitor populations (Fig. 4M). MPD-affected fish also had severely decreased numbers of mature red blood cells, whereas lymphoid populations remained largely intact. Differential cell counts performed on cytospins of the whole kidney marrow cells (Fig. 4N and Table 1) showed an increase in differentiated myelomonocytes (53.8% ± 4.6% vs. 28.8% ± 4.1%; n = 4 per group, P = 0.0002) and myeloid precursors (14.8% ± 3.3% vs. 6.2% ± 1.3%; P = 0.003) in diseased animals. Erythroid cell development was also perturbed with mature red blood cell numbers in the kidney being severely decreased compared with control animals (7.3% ± 5.6% vs. 41.3% ± 2.6%; P < 10−4), whereas erythroid precursor populations were expanded (12.3% ± 3.3% vs. 3% ± 0.8%; P = 0.002).
Table 1.
Differential cell counts of the normal kidneys, spontaneous MPD kidneys, ex vivo heat shock-induced MPD kidneys, and primary recipient kidneys
| Percentage of cells, % |
|||||
|---|---|---|---|---|---|
| Myelomonocyte | Myelomonocyte precursor | Erythrocyte | Erythroid precursor | Lymphocyte | |
| Normal kidney (n = 4) | 28.8 ± 4.1 | 6.3 ± 1.3 | 41.3 ± 2.6 | 3.0 ± 0.8 | 20.8 ± 5.6 |
| Spontaneous MPD kidney (n = 4) | 53.8 ± 4.6** | 14.8 ± 3.3** | 7.3 ± 5.6** | 12.3 ± 3.3** | 12.0 ± 3.9* |
| Ex vivo heatshock MPD kidney (n = 3) | 40.7 ± 6.4* | 13.3 ± 2.3** | 17.0 ± 3.6** | 12.7 ± 1.2** | 16.3 ± 1.5 |
| Primary recipient kidney (n = 4) | 40.5 ± 5.9* | 13.2 ± 2.5** | 20.0 ± 5.1** | 10.8 ± 2.8** | 15.5 ± 2.6 |
Myelomonocytes include granulocytes, monocytes, and macrophages. Erythroid precursors include cells from proerythroblast to polycharomatophilic erythroblast stages. One hundred cells per kidney were analyzed. Numbers are presented as mean ± 1 SD. Statistical significance is indicated by asterisks.
*, P < 0.05;
**, P < 0.01.
To assess whether zebrafish MPD is transplantable, 3 × 105 whole kidney marrow cells were isolated from diseased individuals and introduced into sublethally irradiated recipient fish. In total, five of six MPDs were transplantable with 14 of 26 primary recipients developing MPD by 2 months after transplantation (SI Table 4). Of those MPDs that were transplantable, secondary transplantation resulted in a striking reduction in transplant efficiency (n = 1 of 26 developed MPD; SI Table 4). Taken together, these data suggest that RAS activation in the zebrafish hematopoietic compartment leads to expansion of myeloid cell populations (26, 27) and ineffective erythropoiesis (28) but does not confer self-renewal potential to progenitor cells.
Intestinal epithelial hyperplasia was also observed in both the heat-shocked and non-heat-shocked groups (36% vs. 16% of histologically examined fish, respectively). In contrast to juvenile fish (day 15–25 dpf; SI Fig. 10), adult fish displaying kRASG12D expression in intestinal epithelial cells had more severely disorganized intestinal epithelial architecture, with some foci forming large outgrowths that protrude into the gastrointestinal cavity (SI Fig. 11). The observation that RAS activation in zebrafish intestinal epithelial cells leads to intestinal hyperplasia is consistent with findings that 50% of human colorectal cancers have RAS mutations (29). Additionally, one fish from each treatment group developed an MPNST (SI Fig. 11), which was morphologically similar to those seen in p53 mutant fish (30). These tumors expressed glial fibrillary acidic protein (GFAP) when assessed by in situ hybridization, a finding present in 30% of human MPNSTs.
MPD Can Be Specifically Induced by ex Vivo Heat Shock of Marrow Cells Followed by Transplantation.
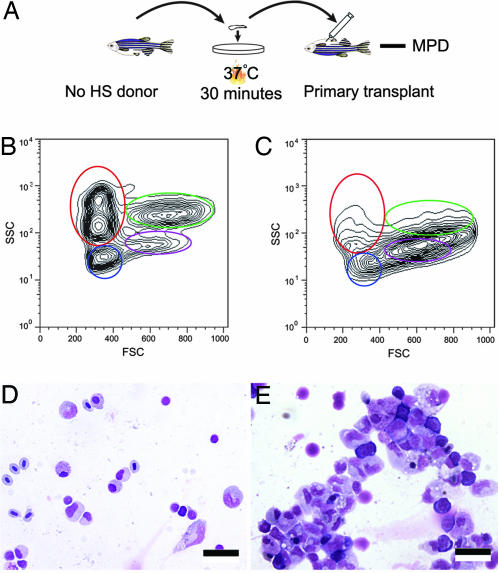
The zebrafish MPD is of particular interest because the disease is remarkably similar to kRASG12D-induced mouse MPD (26, 27), and RAS pathway activation is observed in 30% of human myeloproliferative disorders and leukemia (31). In the heat-shocked group, animals developed MPD with a shorter latency and a lower frequency compared with the non-heat-shocked group, likely because of the severe juvenile lethality and high propensity of animals to develop RMS after heat shock. An ex vivo heat shock strategy was developed to induce kRASG12D expression in hematopoietic cells to induce specifically MPD and circumvent RAS-associated lethality and unwanted tumors induced by the early heat shock (Fig. 5A). Double transgenic fish were raised in the absence of heat shock, and hematopoietic cells from the kidney marrow were isolated from healthy double transgenic fish and analyzed by flow cytometric analysis to confirm that myeloid cell expansion was not already present in these animals (Fig. 5 B and D). Next, half of the kidney marrow cells were heat shocked ex vivo for 30 min at 37°C, and the other half of the marrow cells were left untreated. Both groups of cells were subsequently transplanted into sublethally irradiated recipient fish. At 2 months after transplantation, 8 of 38 recipient fish from the heat-shocked group had developed MPD (Fig. 5 C and E); whereas no fish receiving untreated marrow cells had disease (0 of 38; P = 0.005). MPD arising in these animals was similar to those described above, and no other tumor types were observed (Fig. 5 C and E and Table 1). These results highlight the use of hsp70-Cre and ex vivo heat shock to generate specific tumor types in transgenic animals, even in the absence of tissue-specific gene promoters that drive Cre expression in cell types of interest.
Fig. 5.
MPD can be induced in transplant animals by ex vivo heat shock and cell transplantation strategy. (A) Procedure diagram. No HS donor, non-heat-shocked double transgenic donors. (B) FACS analysis of the donor cells showing that before ex vivo heat shock, the donor fish had normal blood cell numbers within the kidney marrow. (C) FACS analysis showing that primary transplant animals developed MPD 2 months after transplantation. (D and E) May–Grunwald–Giemsa-stained cytospins of the donor cells (D) and primary transplant (E). (Scale bars: 12.5 μm.)
Discussion
Inducible Cre/Lox recombination strategies have been widely used in mammalian genetics to conditionally activate gene expression within targeted tissues of interest (32, 33); however, few studies have been reported with Cre/Lox technology in zebrafish (14–16). Here, we describe a transgenic approach where human kRASG12D can be induced by heat-shocked Cre expression in zebrafish. Two ways of using the system were explored, either heating whole live animals or ex vivo heat shock to cells of interest. Using the first approach, we find that heat shock of whole embryos led to increased tumor incidence, earlier tumor onset, and altered tumor spectrum compared with non-heat-shocked animals. These results demonstrate that the heat shock approach is both inducible and amenable to producing various lesions in double transgenic animals. Moreover, ex vivo heat shock of kidney marrow resulted in increased incidence of MPD in transplanted animals compared with non-heat-shocked controls, providing a robust methodology for inducing specific disease by simply isolating cells from double transgenic animals, ex vivo heat shocking, and transplanting them into irradiated hosts.
Our experiments highlight the use of heat shock-inducible approaches to provide alternatives for conditionally activating genes in whole animals. Compared with other inducible systems, the heat shock-inducible system offers several unique opportunities. First, the recombination can be induced rapidly (4 h after heat shock) and does not require persistent chemical exposure. Second, tissue-specific recombination can be achieved through ex vivo heat shock of cells of interest, a strategy that is especially advantageous in organisms lacking well characterized promoters or established viral transduction, such as Xenopus, medaka, and zebrafish. Moreover, heat shock can be delivered locally by laser activation of individual cells; thus it may be possible to activate a gene of interest selectively in targeted cell types within the normal microenvironment (34). Because the heat shock response is a fundamental cellular mechanism found in most animals and HS-Cre transgenic mice have been reported (35), this heat shock-inducible Cre/Lox approach could be adapted to other model systems.
Our heat shock-inducible Cre/Lox approach can lead to transgene expression in the absence of inducing stimuli, likely caused by the aberrant expression of the hsp70 promoter (18). However, other inducible approaches used to generate tumor models are also leaky. For example, in a kRAS-induced MPD mouse model (27), all double transgenic (LSL-KrasG12D; Mx1-Cre) mice develop MPD regardless of treatment with polyinosinic-polycytidylic acid, although prolonged survival was observed in the untreated group (58 days vs. 35 days). In estrogen-responsive transgenic approaches, 23% of CD2-mycER mice develop lymphoma without treatment compared with the incidence of 62% in the tamoxifen-treated group (36). Thus, similar to other approaches, our heat shock-inducible Cre/Lox strategy is also leaky; however, our strategy does not require continuous chemical exposure to induce conditional gene expression, and it can be delivered to a variety of tissue types. Finally, it is likely our approach can be further optimized by using alternative heat shock Cre lines or promoters. Additionally, raising fish at a lower temperature (26.5°C instead of 28.5°C) may result in decreased heat shock promoter activity and lower tumor incidence (H. Feng, D.M.L., J. A. Madge, A. Quinkertz, A. Gutierrez, D.S.N., J. P. Kanki, and A. T. Look, unpublished data).
To date, we have observed four distinct types of tumors and hyperplasia in this somatic kRASG12D activation model: RMS, MPD, intestinal hyperplasia, and MPNST. These RAS-induced zebrafish diseases are morphologically and molecularly similar to those described in human, providing a powerful tool for future chemical and genetic studies for further understanding of RAS-related tumorigenesis. Compared with a “hit-and-run” KrasG12D mouse model (21), in which lung cancer, thymic lymphoma, and papilloma are the most common lesions, the tumor spectrum found in our zebrafish model is different. This finding may be explained partially by the fact that the B-actin promoter used in this work is not ubiquitously expressed and has different levels of expression in different tissue types. More importantly, our findings support that tumor spectrum differs depending on the species and context. For example, mice deficient in p53 develop predominantly lymphomas (37), whereas p53 mutant zebrafish (30) develop malignant peripheral nerve sheath tumors, and the most common malignancy in Li–Fraumeni patients is osteosarcoma (38). Therefore, use of alternative model systems may provide unique opportunities for modeling human disease that are not readily assessable to other cancer models.
Our studies provide the successful use of heat shock-inducible recombination approaches to model cancer in any vertebrate. This heat shock-inducible recombination technology is especially useful in zebrafish but will also be widely applicable to other model organisms. The RAS-induced zebrafish tumor and hyperplasia models generated herein resemble their associated human malignancies, underscoring the utility of this inducible system to aid in our understanding of conserved mechanisms underlying cancer and will be useful for chemical screening for anticancer agents.
Materials and Methods
Animals and Stable Transgenic Lines.
Zebrafish were maintained in accordance with Animal Research Guidelines at Children's Hospital Boston. The B-actin-LoxP-EGFP-LoxP-kRASG12D construct (SI Methods) was linearized with XhoI, purified, and injected into AB-strain zebrafish embryos to generate stable lines. The hsp70-Cre stable transgenic line was generously provided by Andre Quinkertz from Jose Campos-Ortega Laboratory.
Real-Time PCR to Quantify Recombination Efficiency.
A real-time PCR (SYBR-Green-I based fluorescence, Bio-Rad, Hercules, CA) assay was developed where the threshold cycle number (Ct) is proportional to the logarithm of the initial amount of template used in the reaction (molecules per reaction). Thus, the number of molecules in an unknown sample can be calculated by comparing with a standard curve generated from samples in which the amount of DNA is known (20). First, a RT-PCR primer set 1 (B-actin forward, 5′-GCCTTTTATGGTAATAATGAGAG; and GFP reverse, 5′-GTGAACAGCTCCTCGCCCTTGC) was designed to amplify a 296-bp fragment that detects the nonrecombined B-actin-lox-EGFP-lox-kRASG12D, whereas primer set 2 (B-actin forward, 5′-GAAGTTGACTCCAGATGGTCAC; and RAS reverse, 5′-CTACGCCATCAGCTCCAACTAC) amplifies a 193-bp fragment found only after recombination (SI Fig. 8A). Next, standard curves were obtained by taking plasmids (B-actin-lox-EGFP-lox-kRASG12D and B-actin-kRASG12D) of known concentration to create a log dilution series for each primer set. A linear titration plot (Ct vs. log of the number of template molecules) was generated to correlate the threshold cycle number with the number of template molecules per reaction. A trend line was added and correlated greatly with the individual data points (R2 = 0.9918 and 0.997; SI Fig. 8B). Genomic DNA from individual embryos was extracted, and real-time PCR was performed. Recombination efficiency was defined by the number of recombined molecules (detected by primer set 2) divided by the total molecules of recombined and nonrecombined molecules (detected by primer sets 1 and 2), and then multiplied by 100. This calculation gives the percentage of recombination.
Statistical Analysis.
Survival curves are presented graphically by using the method of Kaplan and Meier. Differences in death rates were assessed by using both log rank and landmark analysis. Student's t tests were used to compare the recombination efficiency, kRAS-expressing cell number, the body length of fish in different treatment groups, and to compare the total number of whole kidney marrow cells and cell numbers of different lineages in MPD-affected fish with controls. The Fisher exact test was used to compare MPD incidence in ex vivo heat shock experiments.
RNA in Situ Hybridization and Immunohistochemistry.
Antisense and sense control probes were made for human kRASG12D, zebrafish desmin, myoD, myogenin, mpo, and l-plastin, and Cre Whole-mount RNA in situ hybridization and antibody staining with anti-phospho-ERK1/2 monoclonal antibody (M9692; Sigma, St. Louis, MO) were performed as described previously (39). In situ hybridization and antibody staining on paraffin-imbedded sections (GFP antibody JL-8; Invitrogen, Carlsbad, CA) were performed as described previously (10).
Transplantation.
Kidney marrow cells were collected and analyzed by FACS analysis as described previously (25). For serial transplantations, 3 × 105 marrow cells from donor fish were introduced into irradiated AB strain adult fish (23 Gy, 2 days before transplantation) by i.p. injection. For ex vivo heat shock transplantations, 1 × 105 marrow cells (with or without heat shock) with 2 × 105 red blood cells (as carrier cells) from AB strain fish were introduced into irradiated AB strain adult fish (23 Gy, 2 days before transplantation) by intracardiac injection. To assess the engraftment of MPD, kidney marrow cells from recipient fish were analyzed by FACS at 4, 6, or 8 weeks after transplantation. Successful engraftment of MPD was defined as having >50% of whole kidney marrow cells falling into the myeloid and precursor gates.
Acknowledgments
We thank Barry Paw, Craig Ceol, Michael Dovey, Xiaoying Bai, Teresa Bowman, Phil Black, and Clemens Graber for critical evaluation of the manuscript; Andre Quinkertz and the late Jose Compos-Ortega for hsp70-Cre transgenic fish and Kenneth Poss (Duke University, Durham, NC) for providing B-actin promoter; Alan Flint, Yu Yang, and Paula Frankel for assistance with flow cytometry, in situ hybridization, and differential cell count analysis, respectively. D.M.L. was supported by a Safra Foundation fellowship from Irvington Institute. L.I.Z. was supported by Howard Hughes Medical Institute and the National Institutes of Health.
Abbreviations
- LGL-RAS
B-actin-LoxP-EGFP-LoxP-kRASG12D
- MPD
myeloproliferative disorder
- MPNST
malignant peripheral nerve sheath tumor
- RMS
rhabdomyosarcoma
- dpf
days postfertilization
- hpf
hours postfertilization.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0611302104/DC1.
References
- 1.Malumbres M, Barbacid M. Nat Rev Cancer. 2003;3:459–465. doi: 10.1038/nrc1097. [DOI] [PubMed] [Google Scholar]
- 2.Bos JL. Cancer Res. 1989;49:4682–4689. [PubMed] [Google Scholar]
- 3.Janssen KP, Abal M, El Marjou F, Louvard D, Robine S. Biochim Biophys Acta. 2005;1756:145–154. doi: 10.1016/j.bbcan.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 4.Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, Jacks T, Tuveson DA. Genes Dev. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tuveson DA, Shaw AT, Willis NA, Silver DP, Jackson EL, Chang S, Mercer KL, Grochow R, Hock H, Crowley D, et al. Cancer Cell. 2004;5:375–387. doi: 10.1016/s1535-6108(04)00085-6. [DOI] [PubMed] [Google Scholar]
- 6.Guerra C, Mijimolle N, Dhawahir A, Dubus P, Barradas M, Serrano M, Campuzano V, Barbacid M. Cancer Cell. 2003;4:111–120. doi: 10.1016/s1535-6108(03)00191-0. [DOI] [PubMed] [Google Scholar]
- 7.Sharpless NE, Depinho RA. Nat Rev Drug Discov. 2006;5:741–754. doi: 10.1038/nrd2110. [DOI] [PubMed] [Google Scholar]
- 8.Stern HM, Murphey RD, Shepard JL, Amatruda JF, Straub CT, Pfaff KL, Weber G, Tallarico JA, King RW, Zon LI. Nat Chem Biol. 2005;1:366–370. doi: 10.1038/nchembio749. [DOI] [PubMed] [Google Scholar]
- 9.Lam SH, Wu YL, Vega VB, Miller LD, Spitsbergen J, Tong Y, Zhan H, Govindarajan KR, Lee S, Mathavan S, et al. Nat Biotechnol. 2006;24:73–75. doi: 10.1038/nbt1169. [DOI] [PubMed] [Google Scholar]
- 10.Langenau DM, Traver D, Ferrando AA, Kutok JL, Aster JC, Kanki JP, Lin S, Prochownik E, Trede NS, Zon LI, Look AT. Science. 2003;299:887–890. doi: 10.1126/science.1080280. [DOI] [PubMed] [Google Scholar]
- 11.Sabaawy H, Azuma M., Embree LJ, Tsai H-J, Starost MF, Hickstein DD. Proc Natl Acad Sci USA. 2006;103:15166–15171. doi: 10.1073/pnas.0603349103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patton EE, Widlund HR, Kutok JL, Kopani KR, Amatruda JF, Murphey RD, Berghmans S, Mayhall EA, Traver D, Fletcher CD, et al. Curr Biol. 2005;15:249–254. doi: 10.1016/j.cub.2005.01.031. [DOI] [PubMed] [Google Scholar]
- 13.Yang HW, Kutok JL, Lee NH, Piao HY, Fletcher CD, Kanki JP, Look AT. Cancer Res. 2004;64:7256–7262. doi: 10.1158/0008-5472.CAN-04-0931. [DOI] [PubMed] [Google Scholar]
- 14.Langenau DM, Feng H, Berghmans S, Kanki JP, Kutok JL, Look AT. Proc Natl Acad Sci USA. 2005;102:6068–6073. doi: 10.1073/pnas.0408708102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thummel R, Burket CT, Brewer JL, Sarras MP, Jr, Li L, Perry M, McDermott JP, Sauer B, Hyde DR, Godwin AR. Dev Dyn. 2005;233:1366–1377. doi: 10.1002/dvdy.20475. [DOI] [PubMed] [Google Scholar]
- 16.Pan X, Wan H, Chia W, Tong Y, Gong Z. Transgenic Res. 2005;14:217–223. doi: 10.1007/s11248-004-5790-z. [DOI] [PubMed] [Google Scholar]
- 17.Higashijima S, Okamoto H, Ueno N, Hotta Y, Eguchi G. Dev Biol. 1997;192:289–299. doi: 10.1006/dbio.1997.8779. [DOI] [PubMed] [Google Scholar]
- 18.Yeh FL, Hsu T. Biosci Biotechnol Biochem. 2000;64:592–595. doi: 10.1271/bbb.64.592. [DOI] [PubMed] [Google Scholar]
- 19.Yeh FL, Hsu T. J Exp Zool. 2002;293:349–359. doi: 10.1002/jez.10093. [DOI] [PubMed] [Google Scholar]
- 20.Ponchel F, Toomes C, Bransfield K, Leong FT, Douglas SH, Field SL, Bell SM, Combaret V, Puisieux A, Mighell AJ, et al. BMC Biotechnol. 2003;3:18. doi: 10.1186/1472-6750-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson L, Mercer K, Greenbaum D, Bronson RT, Crowley D, Tuveson DA, Jacks T. Nature. 2001;410:1111–1116. doi: 10.1038/35074129. [DOI] [PubMed] [Google Scholar]
- 22.Stratton MR, Fisher C, Gusterson BA, Cooper CS. Cancer Res. 1989;49:6324–6327. [PubMed] [Google Scholar]
- 23.Herbomel P, Thisse B, Thisse C. Development (Cambridge, UK) 1999;126:3735–3745. doi: 10.1242/dev.126.17.3735. [DOI] [PubMed] [Google Scholar]
- 24.Bennett CM, Kanki JP, Rhodes J, Liu TX, Paw BH, Kieran MW, Langenau DM, Delahaye-Brown A, Zon LI, Fleming MD, Look AT. Blood. 2001;98:643–651. doi: 10.1182/blood.v98.3.643. [DOI] [PubMed] [Google Scholar]
- 25.Traver D, Paw BH, Poss KD, Penberthy WT, Lin S, Zon LI. Nat Immunol. 2003;4:1238–1246. doi: 10.1038/ni1007. [DOI] [PubMed] [Google Scholar]
- 26.Braun BS, Tuveson DA, Kong N, Le DT, Kogan SC, Rozmus J, Le Beau MM, Jacks TE, Shannon KM. Proc Natl Acad Sci USA. 2004;101:597–602. doi: 10.1073/pnas.0307203101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chan IT, Kutok JL, Williams IR, Cohen S, Kelly L, Shigematsu H, Johnson L, Akashi K, Tuveson DA, Jacks T, Gilliland DG. J Clin Invest. 2004;113:528–538. doi: 10.1172/JCI20476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Braun BS, Archard JA, Van Ziffle JA, Tuveson DA, Jacks TE, Shannon K. Blood. 2006;108:2041–2044. doi: 10.1182/blood-2006-01-013490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bos JL, Fearon ER, Hamilton SR, Verlaan-de Vries M, van Boom JH, van der Eb AJ, Vogelstein B. Nature. 1987;327:293–297. doi: 10.1038/327293a0. [DOI] [PubMed] [Google Scholar]
- 30.Berghmans S, Murphey RD, Wienholds E, Neuberg D, Kutok JL, Fletcher CD, Morris JP, Liu TX, Schulte-Merker S, Kanki JP, et al. Proc Natl Acad Sci USA. 2005;102:407–412. doi: 10.1073/pnas.0406252102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bos JL. Hematol Pathol. 1988;2:55–63. [PubMed] [Google Scholar]
- 32.Branda CS, Dymecki SM. Dev Cell. 2004;6:7–28. doi: 10.1016/s1534-5807(03)00399-x. [DOI] [PubMed] [Google Scholar]
- 33.Maddison K, Clarke AR. J Pathol. 2005;205:181–193. doi: 10.1002/path.1698. [DOI] [PubMed] [Google Scholar]
- 34.Halloran MC, Sato-Maeda M, Warren JT, Su F, Lele Z, Krone PH, Kuwada JY, Shoji W. Development (Cambridge, UK) 2000;127:1953–1960. doi: 10.1242/dev.127.9.1953. [DOI] [PubMed] [Google Scholar]
- 35.Dietrich P, Dragatsis I, Xuan S, Zeitlin S, Efstratiadis A. Mamm Genome. 2000;11:196–205. doi: 10.1007/s003350010037. [DOI] [PubMed] [Google Scholar]
- 36.Blyth K, Stewart M, Bell M, James C, Evan G, Neil JC, Cameron ER. Oncogene. 2000;19:773–782. doi: 10.1038/sj.onc.1203321. [DOI] [PubMed] [Google Scholar]
- 37.Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, Weinberg RA. Curr Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 38.Malkin D, Li FP, Strong LC, Fraumeni JF, Jr, Nelson C. E., Kim DH, Kassel J, Gryka MA, Bischoff FZ, Tainsky MA, et al. Science. 1990;250:1233–1238. doi: 10.1126/science.1978757. [DOI] [PubMed] [Google Scholar]
- 39.Galloway JL, Wingert RA, Thisse C, Thisse B, Zon LI. Dev Cell. 2005;8:109–116. doi: 10.1016/j.devcel.2004.12.001. [DOI] [PubMed] [Google Scholar]