Abstract
Genome-wide linkage studies have defined a broad susceptibility region for late-onset Alzheimer's disease on chromosome 12, which contains the Low-Density Lipoprotein Receptor-Related Protein 6 (LRP6) gene, a coreceptor for Wnt signaling. Here, we report the association between common LRP6 variants and late-onset Alzheimer's disease in a multicenter case-control series as well as in a large family-based series ascertained by the National Institute of Mental Health–National Institute on Aging Genetics Initiative. As shown in the genome-wide linkage studies, our association depends mainly on apolipoprotein E-ε4 (APOE-ε4) carrier status. Haplotype tagging single-nucleotide polymorphisms (SNPs) with a set of seven allelic variants of LRP6 identified a putative risk haplotype, which includes a highly conserved coding sequence SNP: Ile-1062 → Val. Functional analyses revealed that the associated allele Val-1062, an allele previously linked to low bone mass, has decreased β-catenin signaling in HEK293T cells. Our study unveils a genetic relationship between LRP6 and APOE and supports the hypothesis that altered Wnt/β-catenin signaling may be involved in this neurodegenerative disease.
Keywords: neurodegenerative, LRP-6, single-nucleotide polymorphism, APOE, Wnt
Alzheimer's disease (AD) [Online Mendelian Inheritance in Man (OMIM) 104300], the most common form of age-associated dementia, is a progressive neurodegenerative disorder characterized by a deficit in cognitive processes that manifest as alterations in memory, judgment, and reasoning (1). Although the etiology of AD remains to be fully understood, it is well accepted that, along with age, family history is the most prominent risk factor for the development of the disease.
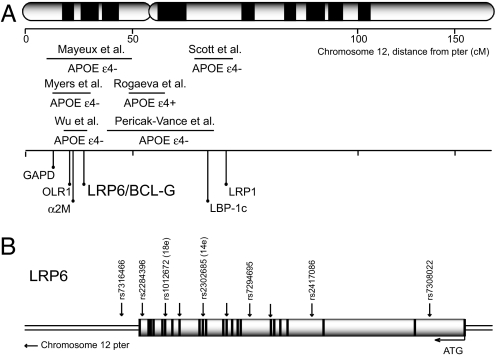
Inheritance of the apolipoprotein E-ε4 (APOE-ε4) allele is a risk factor for AD, including both sporadic and late-onset familial forms of the disease (2, 3). Nevertheless, epidemiological studies estimate that 42–68% of AD patients do not present the APOE-ε4 allele, suggesting that additional genetic or environmental factors could play essential roles in the disease (4). A fact consistent with this observation is that genome-wide screens have identified several regions that show significant linkage to AD, of which the most likely to harbor new risk factors are chromosomes 9, 10, and 12 (5–13). The reported linkage peaks for chromosome 12 show significant association with AD, mainly when samples have been stratified according to APOE carrier status and cluster into two distinct regions (Fig. 1A). One region is located at the p-ter, from ≈6–30 cM, in the vicinity of the Low Density Lipoprotein Receptor (LDLR)-Related Protein 6 (LRP6) (14) gene (at ≈26 cM), whereas the other is pericentromeric, from ≈48–68 cM close to the LRP1 gene (≈68 cM). We and others have proposed that altered function of Wnt signaling components may be involved in AD (15–19), leading us to examine whether the LRP6 gene, which encodes a Wnt coreceptor (20–22), is associated with this disease.
Fig. 1.
Late onset AD and chromosome 12. (A) Summary of reported genome-wide linkage regions according to APOE-ε4 carrier status and location of the LRP6 locus relative to candidate genes showing association to AD (see also www.alzforum.org/res/com/gen/alzgene/default.asp). (B) Diagram of exon–intron boundaries of LRP6 gene and chromosomal position of SNPs (arrows) analyzed in this study.
Results
Genetic Variation in LRP6 Is Associated with Late-Onset AD.
We sought to confirm the existence of single-nucleotide polymorphisms (SNPs) in exons 7, 11, 14, 16, and 18 of LRP6, which were described in the dbSNP database (National Center for Biotechnology Information) (Fig. 1B). We observed that a nonsynonymous coding sequence SNP Ile-1062 → Val in exon 14 (14e, rs2302685; T → C) and a synonymous SNP in exon 18 (18e, rs1012672; C → T) were polymorphic and therefore we continued our analysis with LRP6 SNPs 14e and 18e.
Upon genotyping, 398 Caucasian cases and 339 age- and ethnicity-matched controls coming from the Zurich series (23), the Newcastle Brain Bank (U.K. series) (24), and various brain banks throughout the United States (U.S. series) (24) (see Methods), we noted a trend toward association in single-locus tests for 14e (P = 0.075, Table 1) and significant association for 18e with AD (P = 0.037, Table 1) in the combined multicenter sample (Zurich/U.K./U.S. series). Although 14e was not associated with AD in any of the case-control series analyzed separately, we found that 18e was highly associated in the Zurich series (P = 0.0048, P = 0.011, allelic and genotypic frequencies, respectively), accounting for most of the effect in the combined sample (Table 1). LRP6 18e was also associated in the U.K. series (P = 0.039, genotypic frequency, data not shown).
Table 1.
Association analysis of LRP6 SNPs in the Multicenter case-control series
| Strata | SNP | Sample | MAF Controls | MAF Cases | P | OR (95% CI) |
|---|---|---|---|---|---|---|
| All | 14e | Combined | 18.7 | 15.1 | 0.075 | 1.30 (0.97:1.71) |
| Zurich | 22.9 | 18.5 | 0.24 | 1.31 (0.83:2.07) | ||
| U.K. | 17.1 | 14.2 | 0.43 | 1.25 (0.71:2.20) | ||
| U.S. | 15.1 | 13.5 | 0.61 | 1.14 (0.70:1.86) | ||
| 18e | Combined | 5.9 | 8.9 | 0.037 | 1.54 (1.03:2.31) | |
| Zurich | 5.8 | 13.5 | 0.0048 | 2.54 (1.30:4.94) | ||
| U.K. | 5.5 | 10.1 | 0.111 | 1.93 (0.85:4.41) | ||
| U.S. | 6.3 | 5.4 | 0.66 | 1.81 (0.60:2.35) | ||
| APOE4-ε4-negative | 14e | Combined | 18.2 | 14.8 | 0.24 | 1.29 (0.84:1.97) |
| Zurich | 22.2 | 21.2 | 0.87 | 1.06 (0.53:2.11) | ||
| U.K. | 17 | 13.8 | 0.54 | 1.28 (0.58:2.80) | ||
| U.S. | 14.7 | 10.7 | 0.38 | 1.44 (0.64:3.23) | ||
| 18e | Combined | 4.9 | 10.2 | 0.0075 | 2.20 (1.22:3.98) | |
| Zurich | 6.3 | 15.6 | 0.025 | 2.74 (1.11:6.82) | ||
| U.K. | 2.8 | 12.5 | 0.0083 | 4.91 (1.36:17.8) | ||
| U.S. | 4.7 | 3.4 | 0.76 | 0.72 (0.19:2.76) |
Allelic P values. SNP IDs 14e, rs2302685; 18e, rs1012672; MAF, minimal allele frequency; CI, confidence interval; OR, odds ratio.
Given that the evidence for linkage to chromosome 12 is significant mainly when the analysis has been limited to individuals lacking an APOE-ε4 allele (Fig. 1A), we then stratified the multicenter sample on the basis of whether individuals possessed at least one APOE-ε4 allele (7, 8, 10). The combined sample thus stratified gave 361 individuals corresponding to the ε4-positive stratum (at least one APOE-ε4 allele: 48.9%) and 377 individuals within the ε4-negative subgroup (51.1%). Remarkably, we observed that 18e was strongly associated with AD in the ε4-negative stratum in the combined sample (P = 0.0075, Table 1), as well as in the Zurich and the U.K. series examined separately (P = 0.025 and P = 0.0083, respectively). Confirming widely accepted results, logistic regression analysis in the whole multicenter sample revealed that APOE-ε4 was highly associated with risk in AD [P = <0.0001, supporting information (SI) Table 4]. Likewise, LRP6-18e was strongly associated with disease (P = 0.0092), where individuals carrying at least one copy of the minor allele T had 69–80% greater risk of getting AD compared with individuals being 18e (CC) homozygotes (SI Table 4). We thus conclude that 18e allele T is a previously uncharacterized risk allele for late-onset AD.
Linkage Disequilibrium and Haplotype Analysis Within the LRP6 Gene.
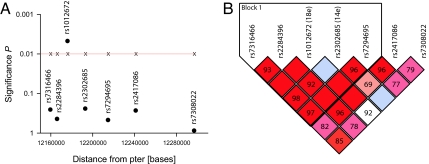
LRP6 is a 23-exon gene covering 150 Kbp in a region with dense genomic content (Fig. 1). To determine whether other variants within LRP6 were responsible for the association signal seen for 18e in the nonstratified Zurich case-control series, we assessed the contribution of the whole gene with a panel of seven haplotype-tagging SNPs (htSNPs): rs7316466, rs2284396, rs1012672 (18e), rs2302685 (14e), rs7294695, rs2417086, and rs7308022 (Fig. 1 and 2). Consistent with what has been determined for the LRP6 gene in the CEPH population (i.e., Caucasians with ancestry from northern and western Europe) by the International HAPMAP Project (25), the region described by our htSNPs had a high degree of linkage disequilibrium (Fig. 2B and SI Fig. 4) and did not extend to other chromosome 12 candidate genes previously associated with AD (SI Fig. 4 and A.P., personal communication on OLR1 and LRP1 in this sample, rs1050286 and rs1799986, respectively). Single locus tests for each marker revealed that 18e was the only LRP6 htSNP showing association with AD (Fig. 2A and Table 2). Nonetheless, haplotype analysis revealed that a 5-htSNP haplotype could be drawn (Block 1: TTTTC; rs7316466, rs2284396, rs1012672 (18e), rs2302685 (14e), and rs7294695, respectively; Fig. 2B) and that this haplotype is strongly associated with AD (P = 0.0069, Table 2). Finally, in agreement with our findings regarding APOE-ε4 stratification, haplotype analysis of the combined multicenter sample revealed that 14eT/18eT was associated in the combined (P = 0.04) and highly significant in the ε4-negative stratum (P = 0.0086) of the multicenter sample (SI Table 5).
Fig. 2.
LRP6 SNP mapping and linkage disequilibrium in the Zurich series. (A) Overall statistical significance of tag-SNPs (dots) covering the whole LRP6 gene (see also Fig. 1B). (B) Plot of the relative D′/LOD values between each of the markers used in this analysis by using the default confidence intervals algorithm automatically generated by the program Haploview (www.broad.mit.edu/mpg/haploview). Note that Block 1 indicates that >95% of informative comparisons between included LRP6 markers are in strong linkage disequilibrium. The standard color D′/LOD scheme was used throughout comparisons: bright red (D′ = 1; LOD ≥2); blue (D′ = 1; LOD <2); shades of pink/red (D′ < 1; LOD ≤2); white (D′ < 1; LOD <2). Numbers in the figure indicate D′ values × 100. Quadrants were left blank if there was perfect linkage disequilibrium (D′ = 1.0).
Table 2.
LRP6 tagging SNPs and haplotype association results in the nonstratified Zurich series
| Stat | SNP ID/haplotype | Allele/frequency | Case, control ratios | χ2 | P |
|---|---|---|---|---|---|
| Single marker | rs7316466 (1) | T, C | 133 : 77, 168 : 120 | 1.27 | 0.26 |
| rs2284396 (2) | T, C | 122 : 90, 157 : 133 | 0.58 | 0.45 | |
| 18e, rs1012672 (3) | C, T | 166 : 26, 243 : 15 | 7.94 | 0.0048 | |
| 14e, rs2302685 (4) | T, C | 163 : 37, 205 : 6 | 1.35 | 0.25 | |
| rs7294695 (5) | C, G | 123 : 89, 159 : 131 | 0.51 | 0.48 | |
| rs2417086 (6) | A, G | 118 : 90, 147 : 137 | 1.19 | 0.28 | |
| rs7308022 (7) | A, G | 192 : 16, 265 : 21 | 0.02 | 0.88 | |
| Haplotype (12345) | TCCTG | 43 | 87.6 : 124.4, 128.3 : 161.7 | 0.44 | 0.51 |
| CTCCC | 20.3 | 39.5 : 172.5, 62.5 : 227.5 | 0.65 | 0.42 | |
| CTCTC | 17.9 | 35.0 : 177.0, 54.7 : 235.3 | 0.47 | 0.5 | |
| TTCTC | 8.6 | 20.4 : 191.6, 22.6 : 267.4 | 0.53 | 0.47 | |
| TTTTC | 8.4 | 26.2 : 185.8, 16.1 : 273.9 | 7.3 | 0.0069 |
LRP6 Variants Are Associated with AD in the Original Linkage Sample to Chromosome 12.
We wanted to determine whether LRP6 association could explain the linkage peak to late-onset AD on chromosome 12, originally described in the affected siblings pairs (ASPs) series from the National Institute of Mental Health (NIMH) and the National Cell Repository for AD (8, 10, 12). We thus genotyped 14e and 18e in 474 families (1,372 individuals) corresponding to our entire NIMH-NIA ASPs sample and analyzed the data by using the Family Based Association Test package FBAT (26, 27). Interestingly, stratification of the ASP sample according to APOE-ε4 carrier status gave 295 nuclear families corresponding to the ε4-positive stratum (76.9%) and 93 nuclear families within the ε4-negative subgroup (23.1%), which is in marked contrast to the combined multicenter case-control series where an equivalent number of individuals is found in both strata (48.9 and 51.1%; ε4-positive/negative, respectively).
FBAT analysis revealed that LRP6 14e was significantly associated in the ε4-negative stratum (P = 0.026), whereas we detected no association in the whole ASPs sample (Table 3) or in the ε4-positive stratum (data not shown). Moreover, and in agreement to the multicenter case-control series, 18e was significantly associated with AD (P = 0.049; Z = 1.96, Table 3). However, because of lack of proper number of “informative families” required by FBAT in the ε4-negative stratum (n ≥ 10; a family is informative when it has a nonzero contribution to the FBAT statistic; i.e., families with members that have all of the same genotypes are not informative) we regard 18e data as suggesting a trend.
Table 3.
Association of LRP6 SNPs 14e and 18e in the NIMH ASP sample
| Stat/strata | Allele | Haplotype | Frequency | Families | Z score | P | p_2side |
|---|---|---|---|---|---|---|---|
| Single marker/all | 14e | C | 20.4 | 80 | — | 0.68 | — |
| 18e | T | 9 | 51 | — | 0.26 | — | |
| APOE-ε4 negative | 14e | C | 21.7 | 14 | — | 0.026 | — |
| 18e | T | 8.5 | 7 | — | 0.049 | — | |
| Haplotype/all | 14e, 18e | TC | 75.4 | 100 | −0.35 | 0.72 | 0.7 |
| CC | 17.9 | 81 | −0.93 | 0.35 | 0.37 | ||
| TT | 6.7 | 53 | 1.64 | 0.1 | 0.078 | ||
| Global | — | — | — | 0.23 | 0.29 | ||
| APOE-ε4 negative | 14e,18e | TC | 73.1 | 17 | −2.96 | 0.003 | 0.0029 |
| CC | 20 | 16 | 1.64 | 0.1 | 0.12 | ||
| TT | 6.9 | 7.7 | 2.19 | 0.029 | 0.031 | ||
| Global | — | — | — | 0.017 | 0.0079 |
Results of FBAT single-locus and HBAT haplotype (univariate) and multihaplotype (global) tests for LRP6 SNPs 14e: rs2302685 and 18e: 1012672. Families, number of informative pedigrees; Z score, FBAT Z statistics. P value, FBAT or HBAT nominal P value. P_2side, Monte Carlo-based two-tailed P value.
The use of haplotypes in family-based association testing is a robust and powerful method especially when the informative nature of individual markers is low (28). Therefore we examined the haplotype contribution of LRP6 14e and 18e SNPs with the aid of the program haplotype-FBAT (HBAT) (28). Interestingly, LRP6 haplotype 14eT/18eC appears to have a protective effect (Z = −2.96) and is highly associated within the ε4-negative stratum of the ASP sample (P = 0.003; Table 3). Likewise, and as observed before in the multicenter sample, haplotype 14eT/18eT confers risk to develop AD (P = 0.029, Z = 2.19; Table 3). Finally, a global (multihaplotype) test between 14e and 18e indicated that both SNPs were strongly associated only in the ε4-negative stratum (P = 0.0079). Therefore, we conclude that LRP6 SNPs are associated with AD in both regular case-control and family-based series and that such association depends mainly on APOE stratification.
LRP6 Is Expressed in the Adult Hippocampus.
Wnts are essential molecules in defining the boundaries of the developing human forebrain, most notably in the hippocampus (29–31). In the mature human brain, the physiological function of Wnts and their receptor complexes containing Frizzled and LRP5/6 is less understood. Therefore, given that preclinical AD is manifested early in the hippocampus, we examined whether the LRP6 gene is expressed in the adult human hippocampus. Quantitative RT-PCR experiments revealed that LRP6 as well as LRP5 transcripts were present in human hippocampal tissue (SI Fig. 5), suggesting that the coreceptor for Wnt signaling may be functional in vivo in the mature brain.
LRP6 Val-1062 Allele Displays Reduced Activation of a β-Catenin Reporter in HEK293T Cells.
Although both LRP6 14e and 18e variants are coding sequence polymorphisms, only 14e results in an amino acid substitution (Ile-1062 → Val). Full sequence alignment of LRP6 and LRP5 from human, mouse, and Arrow (its ortholog in Drosophila) (21, 32) shows that Ile-1062 has been conserved during evolution (Fig. 3A). Likewise, Ile-1062 is also conserved in human and mouse LDLR (Fig. 3A), which constitutes the signature for this family of transmembrane proteins (33), suggesting that this residue may be important for proper protein function. Structurally, Ile-1062 is located after the second YWTD tetrarepeat within the fourth EGF homology domain of LRP6 and appears to be buried inside the hydrophobic core of the β-propeller (SI Fig. 6). Most mutations in the LDLR and LRP5 genes that are responsible for different syndromes reside within β-propeller domains, which are regions involved in proper folding and ligand recognition (32, 33).
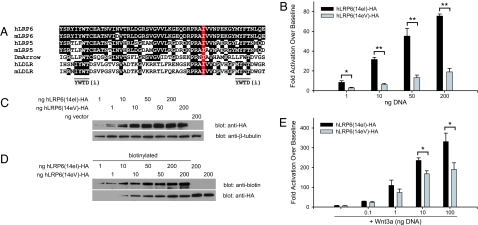
Fig. 3.
Wnt signaling activity of LRP6 alleles 14eI and 14eV in HEK293T cells. (A) ClustalW multiple sequence alignment showing hLRP6 Ile-1062 as a highly conserved amino acid among LDLR-family members in humans (h), mice (m), and Drosophila melanogaster (Dm). (B) Dose-dependent activation by alleles 14eI and 14eV on Wnt/β-catenin signaling activity in HEK293T/STF cells. Values represent averages (n = 3) of fold activation over control plasmid (pCS2p+) from three independent experiments. Errors bars represent standard deviation. ∗, P < 0.01; ∗∗, P < 0.001. (C) Representative Western blot showing equivalent total expression of each allele at each dose from the whole-cell lysates measured in B. (D) Representative Western blot showing equivalent plasma membrane expression of each allele from samples in B. (E) Dose-dependent activation by alleles 14eI and 14eV on Wnt/β-catenin signaling activity in HEK293T/STF cells in the presence of Wnt3a-conditioned media. Values represent averages (n = 3) of fold activation over control plasmid (pCS2p+). Errors bars represent standard deviation. ∗, P < 0.01.
It is well established that LRP6 is a coreceptor in the canonical Wnt/β-catenin signaling pathway, and its overexpression in cells results in ligand-independent activation leading to an increase in cytosolic levels of β-catenin, which then binds TCF/LEF (T cell factor/Lymphoid enhancing factor) and activates transcription (20–22). To determine whether the Ile-1062 → Val substitution had any effect on β-catenin signaling, we constructed COOH-terminal HA-tagged wild-type (Ile-1062, 14eI) and associated SNP (Val-1062, 14eV) alleles and examined their activity in HEK293T cells. As a readout of β-catenin signaling, we used HEK293T/sTF cells that stably express a modified version of the luciferase reporter construct superTOPFLASH (34), which contains 12 copies of the TCF-binding sites and is directly activated by the β-catenin/TCF complex.
Remarkably, despite the conserved nature of the amino acid substitution (i.e., Ile-1062 → Val), LRP6 alleles 14eI and 14eV displayed a nearly 5-fold differential activation of the β-catenin luciferase reporter superTOPFLASH, when overexpressed in HEK293T/sTF cells and assayed 48 h later (Fig. 3B). This diminished activation by 14eV could not be explained by lower expression because Western blot analysis revealed that the alleles were expressed at similar levels (Fig. 3C). To determine whether the diminished activity could be explained by defects in trafficking to the plasma membrane, we next biotinylated proteins at the cell surface. As seen in Fig. 3D, plasma membrane expression of each allele was similar. Interestingly, in the presence of a low dose of Wnt3a-conditioned media, the 14eV allele displayed a significantly decreased activation of the reporter only at the two highest doses of transfected DNA (Fig. 3E). These data are consistent with a reduced efficacy of LRP6 14eV to activate β-catenin signaling.
Although it has been established that HEK293 cells are of neuronal origin (35), the preceding functional analyses were also carried out in the HT22 neuronal cell line. Overexpression of the 14e alleles in this cell line showed a trend toward decreased signaling capability for 14eV as compared with the 14eI, although this effect was not significant (SI Fig. 7A). In the presence of a low dose of Wnt3a-conditioned media, the same trend was observed (SI Fig. 7A). Finally, expression analysis revealed that both alleles were equally expressed (SI Fig. 7B) and trafficked to the membrane similarly (SI Fig. 7C), although, relative to HEK293T cells, there was considerably less plasma membrane-associated LRP6.
Discussion
Besides linkage to various diseases including cancer and osteoporosis (18, 36), it has been proposed that Wnt signaling may underlie neurodevelopmental as well as neurodegenerative disorders such as autism, schizophrenia, and AD (15–19, 37–39). Regarding AD, several studies have shown that the Wnt signaling components β-catenin and the glycogen synthase kinase 3β (GSK3β) form multiprotein complexes with the early-onset familial AD-linked presenilin proteins (40–42). β-Catenin levels are significantly reduced in AD individuals bearing presenilin mutations (43) and active GSK3β accumulates in vivo in AD brains (44), thus affecting τ phosphorylation (45). Likewise, Wnt signaling may have an essential role in the processing of the amyloid precursor protein (46–48), as well as in the neurotoxicity of its derivative the amyloid β-peptide (45, 49–52). Therefore, the Wnt signaling cascade is a candidate for genetic studies aimed at understanding AD etiology. As a consequence, several candidate gene association studies have recently focused on Wnt signaling components, including GSK3β in chromosome 3 (53), Disheveled 1 in chromosome 1 (54), and α-T-catenin in the long arm of chromosome 10 (55–57). Nevertheless, it is not clear whether those genes represent risk factors for AD.
Our results regarding the putative association of common variants within the LRP6 gene in the combined multicenter series, as well as in the ASP sample, which was originally used to describe the genetic linkage of chromosome 12 with late-onset AD, support the involvement of Wnt signaling components in AD (15–19). Indeed, although the synonymous LRP6–18eT variant was found markedly overrepresented in late-onset AD individuals throughout this study, LRP6–14e SNP reached significance in the ASP sample and only in non-APOE-ε4 carriers (Table 2). Furthermore, haplotype analysis of LRP6 variants showed a consistently strong association of the haplotype 14eT/18eT, both in the case-control and the family-based samples, mainly in APOE-ε4-negative individuals (Tables 2 and 3 and SI Table 5), a finding that agrees well with whole-genome scans reporting a linkage peak for chromosome 12 in non-APOE-ε4 carriers (Fig. 1). Interestingly, it has been recently reported that LRP6 variants are associated with increased fracture risk in men (58) and also with age-related macular degeneration (59); both conditions are frequently found in the elderly and may share common pathological mechanisms with AD (60).
Our functional analysis of the LRP6 Val-1062 allele revealed that it displays a reduced activation of β-catenin signaling (Fig. 3). This reduced signaling is consistent with the recent report linking the Val-1062 allele to increased fracture risk (58). Specifically active β-catenin signaling is required for mesenchymal progenitor differentiation to osteoblasts, and many studies in mice and humans show that reduced β-catenin signaling leads to reduced bone mass, consistent with the increased fracture risk observed for this allele (58). Thus it is important to note that the same allele we link to AD has been linked to another clinical condition, which is normally observed with reduced β-catenin signaling.
Another recent report has identified an inherited mutation in LRP6 (R611C) that is linked to early coronary artery disease (61). Carriers of this mutation presented with high LDL, high triglycerides, hypertension, diabetes, and low bone density (increased fracture risk). Interestingly, amino acid 611 is also a highly conserved residue in one of the EGF homology domains. Functional analysis of this mutation yielded an essentially identical biochemical profile as the Val-1062 allele. The Val-1062 allele and the R611C mutant each had decreased ligand-independent activation of β-catenin signaling, a minimal decrease in ligand-dependent activation, and they both expressed and trafficked at similar levels as the wild-type/unassociated LRP6. These examples provide strong evidence that highly conserved EGF homology domain residues such as Ile-1062 are important for the proper biochemical function of LRP6.
Currently, variation in the APOE gene is a widely accepted risk factor for AD. APOE is a component of several classes of secreted lipoproteins that mediate ligand-receptor presentation/endocytosis through the LDLR family of single transmembrane proteins, notably LRP1 (62, 63). However, LRP1 levels decline normally in the aging population and are drastically reduced in AD brains (64), suggesting that, once LRP1 is absent, other receptors that belong to this family might become important in modulating APOE effects.
In this context, it has been proposed that LRP5/6 may recognize and be involved in APOE catabolism of plasma lipoproteins (65, 66). Likewise, it was shown that apolipophorins, the ortholog for APOE proteins in Drosophila, copurify and act as vehicles for the movement of lipid-linked morphogens including the Wnt ortholog Wingless (67). Moreover, recent evidence indicates that there may be an isoform-dependent effect of APOE on Wnt signaling activity, where APOE-ε4 had a strong inhibitory effect on the activity of the signaling cascade (68).
In summary, our study unveils a genetic relationship between LRP6 and APOE and supports the hypothesis that altered Wnt signaling may be central in the onset of this neurodegenerative disease. Given that the Wnt/β–catenin signaling pathway can be modulated pharmacologically (15, 18, 19), it is anticipated that further research in this area may contribute to therapeutic approaches to prevent or to treat AD.
Methods
Study Populations.
The Zurich case-control series, the Newcastle Brain Bank (U.K. series), and various brain banks throughout the United States (U.S. series) have been recently described (23, 24). The affected sibling pairs (ASPs) sample was provided by the National Institute of Mental Health (NIMH), the National Cell Repository for AD (Grant U24 AG21886) and the United Kingdom. The ASPs data set includes many of the families used in our Stage I and II late-onset AD genomic screens (8, 10) and more recently ascertained families. Cases used for analysis had to have an age of onset ≥65 years and a diagnosis of definite or probable AD according to National Institute of Neurological and Communication Disorders and Stroke-Alzheimer's Disease and Related Disorders Association diagnostic criteria. DNA was extracted from blood or mouth swab samples, and informed consent was obtained for all patient samples used in the study according to procedures approved by local and national ethics committees.
Markers.
The National Center for Biotechnology Information Single Nucleotide Polymorphism (SNP) Consortium Database (www.ncbi.nlm.nih.gov/SNP) was used to identify SNPs within coding regions of the LRP6 gene. Sequencing and Pyrosequencing (Biotage, Uppsala, Sweden) conditions are provided in SI Methods.
Statistical Analysis.
In the combined as well as in each case-control series, markers were tested for deviation from Hardy–Weinberg equilibrium by using the Finetti program (http://ihg.gsf.de/cgi-bin/hw/hwa1.pl). Allelic association was then examined on nonstratified samples and samples stratified according to APOE-ε4 carrier status by χ2 tests. Odds ratios (OR) and the 95% confidence intervals (CI) for the comparison between groups were also estimated. Likewise, statistical tools provided by Haploview (69) (www.broad.mit.edu/mpg/haploview) were used throughout the combined multicenter case-control series. In the ASPs sample, we tested 14e and 18e for association with AD by using the Family-Based Association Test (FBAT; version 1.5.1) package (26, 27), which includes also haplotype-FBAT (HBAT) (28). We analyzed the sample by using an additive model as a whole and two strata based on APOE-ε4 carrier status. Given that there have been several reports of linkage in this region on chromosome 12 (5–11), we used the empirical variance option in FBAT (FBAT–e).
Cell and Molecular Biology.
Detailed information for expression vectors, cell culture, protein biotinylation, Western blotting, luciferase reporter assays, and Quantitative RT-PCR conditions is provided in SI Methods.
Supplementary Material
Acknowledgments
We especially thank all of the families that allowed the collection of DNA samples. Many data and biomaterials were collected in three projects that participated in the National Institute of Mental Health (NIMH) Alzheimer's Disease Genetics Initiative. From 1991 to 1998, the principal investigators and coinvestigators were as follows: Massachusetts General Hospital, Boston, MA, U01 MH46281, Marilyn S. Albert and Deborah Blacker; John Hopkins University, Baltimore, MD, U01 MH46290, Susan Bassett, Gary A. Chase, and Marshal F. Folstein; and University of Alabama, Birmingham, AL, U01 MH46372, Rodney C. P. Go and Lindy E. Harrell. R.T.M. thanks the Howard Hughes Medical Institute and the T. L. L. Temple Award from the Alzheimer's Association for support and for encouraging the pursuit of new research directions. F.W.D.-V., A.M. and J.H. are supported by the National Institute on Aging/National Institutes of Health Intramural Program. A.P. was supported by the Swiss National Science Foundation (PP00B-68859), the Helmut Horten Stiftung, and the European Union APOPIS program (Contract LSHM-CT-2003-503330). A.M. is a resident research associate of the National Academy of Sciences. A.M. and J.H. thank the Verum Foundation. G.V.D.F. thanks the PEW Latin American Program in the Biomedical Sciences and was also supported by a Pilot Award from University of Washington Alzheimer's Disease Research Center, Fondo Nacional de Desarrollo Científico y Tecnológico Grant 1050097, and Programa Bicentenario de Ciencia y Tecnología Grant ACT-04.
Abbreviations
- AD
Alzheimer's disease
- SNP
single-nucleotide polymorphism
- htSNP
haplotype-tagging SNP
- LDLR
Low-Density Lipoprotein Receptor
- LRP6
LDLR-Related Protein 6
- APOE
apolipoprotein E.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0603523104/DC1.
References
- 1.Hardy J, Selkoe DJ. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 2.Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 3.Strittmatter WJ, Saunders AM, Schmechel D, Pericak-Vance M, Enghild J, Salvesen GS, Roses AD. Proc Natl Acad Sci USA. 1993;90:1977–1981. doi: 10.1073/pnas.90.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Warwick Daw E, Payami H, Nemens EJ, Nochlin D, Bird TD, Schellenberg GD, Wijsman EM. Am J Hum Genet. 2000;66:196–204. doi: 10.1086/302710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pericak-Vance MA, Bass MP, Yamaoka LH, Gaskell PC, Scott WK, Terwedow HA, Menold MM, Conneally PM, Small GW, Vance JM, et al. J Am Med Assoc. 1997;278:1237–1241. [PubMed] [Google Scholar]
- 6.Rogaeva E, Premkumar S, Song Y, Sorbi S, Brindle N, Paterson A, Duara R, Levesque G, Yu G, Nishimura M, et al. J Am Med Assoc. 1998;280:614–618. doi: 10.1001/jama.280.7.614. [DOI] [PubMed] [Google Scholar]
- 7.Wu WS, Holmans P, Wavrant-DeVrieze F, Shears S, Kehoe P, Crook R, Booth J, Williams N, Perez-Tur J, Roehl K, et al. J Am Med Assoc. 1998;280:619–22. doi: 10.1001/jama.280.7.619. [DOI] [PubMed] [Google Scholar]
- 8.Kehoe P, Wavrant-De Vrieze F, Crook R, Wu WS, Holmans P, Fenton I, Spurlock G, Norton N, Williams H, Williams N, et al. Hum Mol Genet. 1999;8:237–245. doi: 10.1093/hmg/8.2.237. [DOI] [PubMed] [Google Scholar]
- 9.Scott WK, Grubber JM, Conneally PM, Small GW, Hulette CM, Rosenberg CK, Saunders AM, Roses AD, Haines JL, Pericak-Vance MA. Am J Hum Genet. 2000;66:922–932. doi: 10.1086/302828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Myers A, Wavrant De-Vrieze F, Holmans P, Hamshere M, Crook R, Compton D, Marshall H, Meyer D, Shears S, Booth J, et al. Am J Med Genet. 2002;114:235–244. doi: 10.1002/ajmg.10183. [DOI] [PubMed] [Google Scholar]
- 11.Mayeux R, Lee JH, Romas SN, Mayo D, Santana V, Williamson J, Ciappa A, Rondon HZ, Estevez P, Lantigua R, et al. Am J Hum Genet. 2002;70:237–243. doi: 10.1086/324773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blacker D, Bertram L, Saunders AJ, Moscarillo TJ, Albert MS, Wiener H, Perry RT, Collins JS, Harrell LE, Go RC, et al. Hum Mol Genet. 2003;12:23–32. doi: 10.1093/hmg/ddg007. [DOI] [PubMed] [Google Scholar]
- 13.Saunders AJ, Bertram L, Mullin K, Sampson AJ, Latifzai K, Basu S, Jones J, Kinney D, MacKenzie-Ingano L, Yu S, et al. Hum Mol Genet. 2003;12:2765–2776. doi: 10.1093/hmg/ddg310. [DOI] [PubMed] [Google Scholar]
- 14.Brown SD, Twells RC, Hey PJ, Cox RD, Levy ER, Soderman AR, Metzker ML, Caskey CT, Todd JA, Hess JF. Biochem Biophys Res Commun. 1998;248:879–888. doi: 10.1006/bbrc.1998.9061. [DOI] [PubMed] [Google Scholar]
- 15.De Ferrari GV, Inestrosa NC. Brain Res Brain Res Rev. 2000;33:1–12. doi: 10.1016/s0165-0173(00)00021-7. [DOI] [PubMed] [Google Scholar]
- 16.Mudher A, Lovestone S. Trends Neurosci. 2002;25:22–26. doi: 10.1016/s0166-2236(00)02031-2. [DOI] [PubMed] [Google Scholar]
- 17.Caricasole A, Copani A, Caruso A, Caraci F, Iacovelli L, Sortino MA, Terstappen GC, Nicoletti F. Trends Pharmacol Sci. 2003;24:233–238. doi: 10.1016/s0165-6147(03)00100-7. [DOI] [PubMed] [Google Scholar]
- 18.Moon RT, Kohn AD, De Ferrari GV, Kaykas A. Nat Rev Genet. 2004;5:691–701. doi: 10.1038/nrg1427. [DOI] [PubMed] [Google Scholar]
- 19.De Ferrari GV, Moon RT. Oncogene. 2006;25:7545–7553. doi: 10.1038/sj.onc.1210064. [DOI] [PubMed] [Google Scholar]
- 20.Tamai K, Semenov M, Kato Y, Spokony R, Liu C, Katsuyama Y, Hess F, Saint-Jeannet JP, He X. Nature. 2000;407:530–535. doi: 10.1038/35035117. [DOI] [PubMed] [Google Scholar]
- 21.Wehrli M, Dougan ST, Caldwell K, O'Keefe L, Schwartz S, Vaizel-Ohayon D, Schejter E, Tomlinson A, DiNardo S. Nature. 2000;407:527–530. doi: 10.1038/35035110. [DOI] [PubMed] [Google Scholar]
- 22.Pinson KI, Brennan J, Monkley S, Avery BJ, Skarnes WC. Nature. 2000;407:535–538. doi: 10.1038/35035124. [DOI] [PubMed] [Google Scholar]
- 23.Papassotiropoulos A, Wollmer MA, Tsolaki M, Brunner F, Molyva D, Lutjohann D, Nitsch RM, Hock C. J Clin Psychiatry. 2005;66:940–947. [PubMed] [Google Scholar]
- 24.Myers AJ, Kaleem M, Marlowe L, Pittman AM, Lees AJ, Fung HC, Duckworth J, Leung D, Gibson A, Morris CM, et al. Hum Mol Genet. 2005;14:2399–2404. doi: 10.1093/hmg/ddi241. [DOI] [PubMed] [Google Scholar]
- 25.The International HapMap Project Consortium. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 26.Lake SL, Blacker D, Laird NM. Am J Hum Genet. 2000;67:1515–1525. doi: 10.1086/316895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rabinowitz D, Laird N. Hum Hered. 2000;50:211–223. doi: 10.1159/000022918. [DOI] [PubMed] [Google Scholar]
- 28.Horvath S, Xu X, Lake SL, Silverman EK, Weiss ST, Laird NM. Genet Epidemiol. 2004;26:61–69. doi: 10.1002/gepi.10295. [DOI] [PubMed] [Google Scholar]
- 29.Abu-Khalil A, Fu L, Grove EA, Zecevic N, Geschwind DH. J Comp Neurol. 2004;474:276–288. doi: 10.1002/cne.20112. [DOI] [PubMed] [Google Scholar]
- 30.Maretto S, Cordenonsi M, Dupont S, Braghetta P, Broccoli V, Hassan AB, Volpin D, Bressan GM, Piccolo S. Proc Natl Acad Sci USA. 2003;100:3299–3304. doi: 10.1073/pnas.0434590100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lako M, Lindsay S, Bullen P, Wilson DI, Robson SC, Strachan T. Hum Mol Genet. 1998;7:813–822. doi: 10.1093/hmg/7.5.813. [DOI] [PubMed] [Google Scholar]
- 32.He X, Semenov M, Tamai K, Zeng X. Development (Cambridge, UK) 2004;131:1663–1677. doi: 10.1242/dev.01117. [DOI] [PubMed] [Google Scholar]
- 33.Jeon H, Meng W, Takagi J, Eck MJ, Springer TA, Blacklow SC. Nat Struct Biol. 2001;8:499–504. doi: 10.1038/88556. [DOI] [PubMed] [Google Scholar]
- 34.Robitaille J, MacDonald ML, Kaykas A, Sheldahl LC, Zeisler J, Dube MP, Zhang LH, Singaraja RR, Guernsey DL, Zheng B, et al. Nat Genet. 2002;32:326–330. doi: 10.1038/ng957. [DOI] [PubMed] [Google Scholar]
- 35.Shaw G, Morse S, Ararat M, Graham FL. FASEB J. 2002;16:869–871. doi: 10.1096/fj.01-0995fje. [DOI] [PubMed] [Google Scholar]
- 36.Nusse R. Cell Res. 2005;15:28–32. doi: 10.1038/sj.cr.7290260. [DOI] [PubMed] [Google Scholar]
- 37.Kozlovsky N, Belmaker RH, Agam G. Eur Neuropsychopharmacol. 2002;12:13–25. doi: 10.1016/s0924-977x(01)00131-6. [DOI] [PubMed] [Google Scholar]
- 38.Miyaoka T, Seno H, Ishino H. Schizophr Res. 1999;38:1–6. doi: 10.1016/s0920-9964(98)00179-0. [DOI] [PubMed] [Google Scholar]
- 39.Katsu T, Ujike H, Nakano T, Tanaka Y, Nomura A, Nakata K, Takaki M, Sakai A, Uchida N, Imamura T, Kuroda S. Neurosci Lett. 2003;353:53–56. doi: 10.1016/j.neulet.2003.09.017. [DOI] [PubMed] [Google Scholar]
- 40.Kang DE, Soriano S, Xia X, Eberhart CG, De Strooper B, Zheng H, Koo EH. Cell. 2002;110:751–762. doi: 10.1016/s0092-8674(02)00970-4. [DOI] [PubMed] [Google Scholar]
- 41.Takashima A, Murayama M, Murayama O, Kohno T, Honda T, Yasutake K, Nihonmatsu N, Mercken M, Yamaguchi H, Sugihara S, Wolozin B. Proc Natl Acad Sci USA. 1998;95:9637–9641. doi: 10.1073/pnas.95.16.9637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou J, Liyanage U, Medina M, Ho C, Simmons AD, Lovett M, Kosik KS. NeuroReport. 1997;8:2085–2090. doi: 10.1097/00001756-199705260-00054. [DOI] [PubMed] [Google Scholar]
- 43.Zhang Z, Hartmann H, Do VM, Abramowski D, Sturchler-Pierrat C, Staufenbiel M, Sommer B, van de Wetering M, Clevers H, Saftig P, et al. Nature. 1998;395:698–702. doi: 10.1038/27208. [DOI] [PubMed] [Google Scholar]
- 44.Pei JJ, Braak E, Braak H, Grundke-Iqbal I, Iqbal K, Winblad B, Cowburn RF. J Neuropathol Exp Neurol. 1999;58:1010–1019. doi: 10.1097/00005072-199909000-00011. [DOI] [PubMed] [Google Scholar]
- 45.Alvarez G, Munoz-Montano JR, Satrustegui J, Avila J, Bogonez E, Diaz-Nido J. Bipolar Disord. 2002;4:153–165. doi: 10.1034/j.1399-5618.2002.01150.x. [DOI] [PubMed] [Google Scholar]
- 46.Phiel CJ, Wilson CA, Lee VM, Klein PS. Nature. 2003;423:435–439. doi: 10.1038/nature01640. [DOI] [PubMed] [Google Scholar]
- 47.Mudher A, Chapman S, Richardson J, Asuni A, Gibb G, Pollard C, Killick R, Iqbal T, Raymond L, Varndell I, et al. J Neurosci. 2001;21:4987–4995. doi: 10.1523/JNEUROSCI.21-14-04987.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun X, Sato S, Murayama O, Murayama M, Park JM, Yamaguchi H, Takashima A. Neurosci Lett. 2002;321:61–64. doi: 10.1016/s0304-3940(01)02583-6. [DOI] [PubMed] [Google Scholar]
- 49.Alvarez AR, Godoy JA, Mullendorff K, Olivares GH, Bronfman M, Inestrosa NC. Exp Cell Res. 2004;297:186–196. doi: 10.1016/j.yexcr.2004.02.028. [DOI] [PubMed] [Google Scholar]
- 50.De Ferrari GV, Chacon MA, Barria MI, Garrido JL, Godoy JA, Olivares G, Reyes AE, Alvarez A, Bronfman M, Inestrosa NC. Mol Psychiatry. 2003;8:195–208. doi: 10.1038/sj.mp.4001208. [DOI] [PubMed] [Google Scholar]
- 51.Caricasole A, Copani A, Caraci F, Aronica E, Rozemuller AJ, Caruso A, Storto M, Gaviraghi G, Terstappen GC, Nicoletti F. J Neurosci. 2004;24:6021–6027. doi: 10.1523/JNEUROSCI.1381-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takashima A, Honda T, Yasutake K, Michel G, Murayama O, Murayama M, Ishiguro K, Yamaguchi H. Neurosci Res. 1998;31:317–323. doi: 10.1016/s0168-0102(98)00061-3. [DOI] [PubMed] [Google Scholar]
- 53.Russ C, Lovestone S, Powell JF. Mol Psychiatry. 2001;6:320–324. doi: 10.1038/sj.mp.4000852. [DOI] [PubMed] [Google Scholar]
- 54.Russ C, Lovestone S, Powell JF. Mol Psychiatry. 2002;7:104–109. doi: 10.1038/sj.mp.4000941. [DOI] [PubMed] [Google Scholar]
- 55.Ertekin-Taner N, Ronald J, Asahara H, Younkin L, Hella M, Jain S, Gnida E, Younkin S, Fadale D, Ohyagi Y, et al. Hum Mol Genet. 2003;12:3133–3143. doi: 10.1093/hmg/ddg343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Blomqvist ME, Andreasen N, Bogdanovic N, Blennow K, Brookes AJ, Prince JA. Neurosci Lett. 2004;358:220–222. doi: 10.1016/j.neulet.2004.01.032. [DOI] [PubMed] [Google Scholar]
- 57.Busby V, Goossens S, Nowotny P, Hamilton G, Smemo S, Harold D, Turic D, Jehu L, Myers A, Womick M, et al. Neuromol Med. 2004;5:133–146. doi: 10.1385/NMM:5:2:133. [DOI] [PubMed] [Google Scholar]
- 58.van Meurs JB, Rivadeneira F, Jhamai M, Hugens W, Hofman A, van Leeuwen JP, Pols HA, Uitterlinden AG. J Bone Miner Res. 2006;21:141–150. doi: 10.1359/JBMR.050904. [DOI] [PubMed] [Google Scholar]
- 59.Haines JL, Schnetz-Boutaud N, Schmidt S, Scott WK, Agarwal A, Postel EA, Olson L, Kenealy SJ, Hauser M, Gilbert JR, Pericak-Vance MA. Invest Ophthalmol Vis Sci. 2006;47:329–335. doi: 10.1167/iovs.05-0116. [DOI] [PubMed] [Google Scholar]
- 60.Malek G, Johnson LV, Mace BE, Saloupis P, Schmechel DE, Rickman DW, Toth CA, Sullivan PM, Bowes Rickman C. Proc Natl Acad Sci USA. 2005;102:11900–11905. doi: 10.1073/pnas.0503015102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mani A, Radhakrishnan J, Wang H, Mani MA, Nelson-Williams C, Carew KS, Mane S, Najmabadi H, Wu D, Lifton RP. Science. 2007;315:1278–1282. doi: 10.1126/science.1136370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Herz J. Neuron. 2001;29:571–581. doi: 10.1016/s0896-6273(01)00234-3. [DOI] [PubMed] [Google Scholar]
- 63.Li Y, Cam J, Bu G. Mol Neurobiol. 2001;23:53–67. doi: 10.1385/MN:23:1:53. [DOI] [PubMed] [Google Scholar]
- 64.Kang DE, Pietrzik CU, Baum L, Chevallier N, Merriam DE, Kounnas MZ, Wagner SL, Troncoso JC, Kawas CH, Katzman R, Koo EH. J Clin Invest. 2000;106:1159–1166. doi: 10.1172/JCI11013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim DH, Inagaki Y, Suzuki T, Ioka RX, Yoshioka SZ, Magoori K, Kang MJ, Cho Y, Nakano AZ, Liu Q, et al. J Biochem (Tokyo) 1998;124:1072–1076. doi: 10.1093/oxfordjournals.jbchem.a022223. [DOI] [PubMed] [Google Scholar]
- 66.Magoori K, Kang MJ, Ito MR, Kakuuchi H, Ioka RX, Kamataki A, Kim DH, Asaba H, Iwasaki S, Takei YA, et al. J Biol Chem. 2003;278:11331–11336. doi: 10.1074/jbc.M211987200. [DOI] [PubMed] [Google Scholar]
- 67.Panakova D, Sprong H, Marois E, Thiele C, Eaton S. Nature. 2005;435:58–65. doi: 10.1038/nature03504. [DOI] [PubMed] [Google Scholar]
- 68.Caruso A, Motolese M, Iacovelli L, Caraci F, Copani A, Nicoletti F, Terstappen GC, Gaviraghi G, Caricasole A. J Neurochem. 2006;98:364–371. doi: 10.1111/j.1471-4159.2006.03867.x. [DOI] [PubMed] [Google Scholar]
- 69.Barrett JC, Fry B, Maller J, Daly MJ. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.