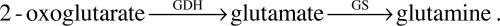Abstract
The central nitrogen metabolic circuit in enteric bacteria consists of three enzymes: glutamine synthetase, glutamate synthase (GOGAT), and glutamate dehydrogenase (GDH). With the carbon skeleton provided by 2-oxoglutarate, ammonia/ammonium (NH4+) is assimilated into two central nitrogen intermediates, glutamate and glutamine. Although both serve as nitrogen donors for all biosynthetic needs, glutamate and glutamine play different roles. Internal glutamine serves as a sensor of external nitrogen availability, and its pool concentration decreases upon nitrogen limitation. A high glutamate pool concentration is required to maintain the internal K+ pool. The configuration of high glutamate and low glutamine pools was disrupted in GOGAT− mutants under low NH4+ conditions: the glutamate pool was low, the difference between glutamate and glutamine was diminished, and growth was defective. When a GOGAT− mutant was cultured in an NH4+-limited chemostat, two sequential spontaneous mutations occurred. Each resulted in a suppressor mutant that outgrew its predecessor in the chemostat. The first suppressor overexpressed GDH, and the second also had a partially impaired glutamine synthetase. The result was a triple mutant in which NH4+ was assimilated by two enzymes instead of the normal three and yet glutamate and glutamine pools and growth were essentially normal. The results indicate preference for the usual ratio of glutamate and glutamine and the resilient and compensatory nature of the circuit on pool control. Analysis of other suppressor mutants selected on solid medium suggests that increased GDH expression is the key for rescue of the growth defect of GOGAT− mutants under low NH4+ conditions.
Keywords: ammonium assimilation, glutamine, metabolic circuit, suppressor
To understand a living organism in a comprehensive way, systems biology has to integrate transcriptomics, proteomics, and metabolomics. Although global metabolite profiling has recently been made possible by advances in analytical technologies, metabolomics and physiology remain to be quantitatively bridged. To fill this deficit, quantitative studies of smaller systems are warranted. The outcome of such studies may reveal novel principles and provide strategic guidance for genome-wide research.
The intensively studied central nitrogen metabolic circuit in enteric bacteria is a subsystem well suited for this purpose (1, 2). The circuit is compact, consisting of only three enzymes and three metabolites (Fig. 1). By assimilating the preferred N-source ammonia/ammonium (NH4+) using 2-oxoglutarate (a metabolite derived from the tricarboxylic acid cycle), the circuit synthesizes the other two metabolites, glutamine and glutamate. Glutamine synthetase (GS, encoded by glnA) catalyzes the only pathway for glutamine biosynthesis. Glutamate can be synthesized by two pathways: through the combined actions of GS and glutamate synthase (GOGAT, encoded by gltBD) forming the so called GS/GOGAT cycle, or by the action of glutamate dehydrogenase (GDH, encoded by gdhA). The GS/GOGAT cycle has a high affinity for NH4+ [Km < 0.2 mM for GS (3)], whereas GDH has a low affinity [Km > 1 mM (3, 4)]. The two central nitrogen intermediates, glutamine and glutamate, provide N for the synthesis of all other N-containing components: ≈88% of cellular N comes from glutamate, and the rest comes from glutamine (5).
Fig. 1.
The central nitrogen metabolic circuit in enteric bacteria. The three-enzyme circuit assimilates NH4+ and produces two central intermediates, glutamine and glutamate. GS catalyzes glutamine synthesis. Glutamate can be synthesized by the action of either GS/GOGAT or GDH, respectively, with high or low affinity for NH4+. The two glutamate molecules shown have no known functional difference.
Beyond their necessity for biosynthesis, glutamine and glutamate play different physiological roles. It has been demonstrated that internal glutamine serves as a sensor of external N availability and may be responsible for modulating growth rate (6). The glutamine pool concentration is lower than that of glutamate under all conditions of N availability and drops as external N becomes scarce. The setting and response fit well with the multicascade regulatory systems governing GS synthesis and activity, for which extensive knowledge has been acquired over decades of research by many groups (2). On the other hand, the internal glutamate pool concentration is relatively high and does not respond to N status (6). Instead, it responds to another environmental stimulus: external osmolarity (7, 8). Internal glutamate accumulates as external osmolarity increases (9–11), and glutamate is the largest anion pool to partially counterbalance the internal pool of K+, the most prevalent osmolyte inside a cell (7). K+ uptake and glutamate accumulation are among the primary cellular responses upon a hyperosmotic shock (9, 10). Moreover, glutamate is also required to maintain the internal K+ pool (12). Manipulated glutamate deficit (at levels apparently still above biosynthetic needs) results in a suboptimal K+ pool concentration and a growth defect. The molecular mechanism for coordinating the pool levels of glutamate and K+ has yet to be understood.
The setting of glutamine and glutamate in the intertwined circuitry needs to be further explored to understand the different roles of these two metabolites. Mutants missing either glutamate biosynthetic enzyme (GOGAT− or GDH−) provide simplified versions to address the system configuration. GOGAT− is particularly useful. It turns the intertwined circuitry (Fig. 1) into a linear pathway (Scheme 1). With only the low-affinity NH4+ assimilatory pathway for glutamate biosynthesis, the metabolic flux through GDH can be limited experimentally under low NH4+ conditions. This results in a growth defect (e.g., the doubling time of a GOGAT− strain was ≈2 h in comparison to ≈1 h of its congenic wild type in a medium of intermediate osmolarity with 1 mM NH4+), a decreased glutamate pool accompanied by a suboptimal K+ pool, and an abnormally increased glutamine pool (12, 13). The ratio of glutamate to glutamine pools of a GOGAT− strain growing in low NH4+ is ≈0.5–1.5, much lower than the ≈5–12 in wild-type strains (13). The relative pool values of glutamate and glutamine are drastically disturbed in this two-enzyme linear pathway.
Scheme 1.
I report here suppressor mutant selections of Salmonella enterica serovar Typhimurium and Escherichia coli GOGAT− strains under low NH4+ conditions by two independent methods. All suppressor mutants showed elevated GDH activities to different degrees that correlated with the extent of growth rescue. Most interesting was the restoration of a high glutamate pool and a low glutamine pool with the two-enzyme linear pathway in these suppressor mutants. I conclude that the central nitrogen metabolic circuit is a resilient and compensatory system, within which the two central nitrogen intermediates, glutamate and glutamine, coordinate with each other to fulfill their diverse roles: K+ pool maintenance for glutamate and N limitation sensor for glutamine.
Results
Isolation of Suppressors of an S. typhimurium GOGAT− Deletion Mutant in an NH4+-Limited Chemostat.
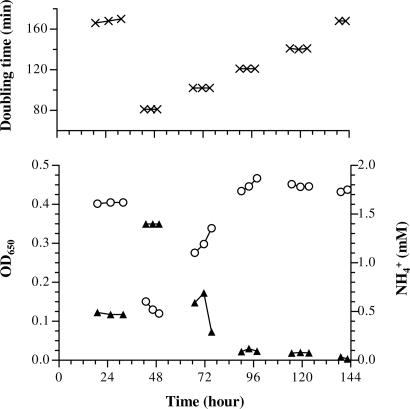
In GOGAT− strains, glutamate biosynthesis depends solely on the GDH pathway (Fig. 1), which has a low affinity for NH4+. The metabolic flux through GDH is titratable with NH4+ concentrations close to or below the Km of GDH (13). I attempted to futher explore this by growing an S. typhimurium GOGAT− deletion strain in an NH4+-limited chemostat (Fig. 2). SK3062 (ΔgltB824) was used to prevent GOGAT+ revertants. A low-osmolarity, diluted minimal medium was used; in this medium, the growth defect of SK3062 is mild. High osmolarity would cause a more severe growth problem (12, 13).
Fig. 2.
GOGAT− SK3062 (ΔgltB824) in an NH4+-limited chemostat. The diluted minimal medium (0.2 × N−C−DB) contained 2 mM NH4+ as the limiting N-source, and 0.2% glycerol as the C-source. Five dilution rates were chosen from 0.52 h−1 (close to maximal growth rate, doubling time of 81 min, day 2) to 0.25 h−1 (about half of the maximal growth rate, doubling time of 168 min, days 1 and 6), with each interval corresponding to a change in doubling time of ≈20 min. After each overnight equilibration, two to three samples were collected several hours apart. (Upper) Cell doubling time calculated from dilution rate measurement after each sample collection. (Lower) Cell density (○) and free NH4+ concentration in the medium (▴).
After an initial equilibration of the chemostat, SK3062 appeared to reach a steady-state growth on day 1 (from ≈18 to 30 h in Fig. 2). The culture showed the signature of GOGAT− growth in an NH4+-limited chemostat. At a slow growth rate (0.25 h−1; doubling time of 168 min), SK3062 failed to exhaust NH4+; ≈0.5 mM NH4+ (of the initial 2 mM) remained in the medium. This was different from wild type or a GDH− strain under similar situations, where free NH4+ dropped below 0.02 mM (6, 13, 14). After the dilution rate was adjusted to approximately maximum, the cell density on day 2 declined. This “washed-out” phase likely resulted from setting the dilution rate beyond the ability of cell growth. Two subsequent decreases of the dilution rate (days 3 and 4) failed to stabilize the culture: the cell density continually increased. The instability was puzzling because equilibration should have been established overnight. During the same period, the unused NH4+ dropped to ≈0.1 mM. One possible explanation was that the cells were evolving to use more of the NH4+. The population stabilized during day 5 with residual NH4+ at ≈0.08 mM. The experiment was stopped after day 6, when the dilution rate was set the same as on day 1. Comparing cultures between day 6 and day 1, the cell density increased while free NH4+ decreased (to <0.02 mM in the last sample). The cells apparently changed into a state that closely resembled wild-type cells in similar chemostat cultures (6, 13).
To investigate whether the original SK3062 was mutated, 10 single colonies were isolated from the first sample of each day. Their abilities to use arginine as the sole N-source were examined (Aut phenotype). Aut− is one of the pleiotropic phenotypes of GOGAT− (15). The isolates were checked by using a four-grade system: from Aut++ of wild-type SK2633 to Aut− of SK3062. The results were as follows: Aut− for all isolates from days 1 to 3; Aut+/− for 19 of 20 isolates from days 4 and 5; and seven Aut+/− and three Aut+ for isolates from day 6. These phenotypes fit well with the chemostat growth (Fig. 2). Aut+/− was replacing Aut− between days 3 and 4, which coincided with increasing cell density and decreasing free NH4+ (from ≈0.5 to ≈0.1 mM). And Aut+ was in the process of outgrowing Aut+/− during the last day, exhausting remaining NH4+ to <0.02 mM. These results suggested that two sequential spontaneous mutational events had occurred during the experiment. One Aut+/− isolate was designated as SK3074 (early suppressor), and one Aut+ was designated as SK3075 (late suppressor).
Growth Characterization and Pools Measurements for the Suppressor Mutants in Batch Cultures.
The growth of suppressor strains SK3074 and SK3075 was analyzed in low NH4+ (2 mM) batch cultures, together with that of parental strain SK3062 (ΔgltB824) and wild-type SK2633 (Table 1). Growth was tested in media of three different osmolarities. During the exponential phase of growth, wild-type cells had stable high glutamate and low glutamine pools, a normal combination. Different external osmolarities caused only small changes in their ratio (5.1–8.5). SK3062 (ΔgltB824) did not grow exponentially in low NH4+ media (growth curves not shown). It had a progressive growth defect, with increasing doubling times as NH4+ was consumed by cell growth (the reason for showing individual rather than average values in Table 1). This was expected because of the decreasing availability of NH4+ to GDH. The glutamate pool of SK3062 (ΔgltB824) was decreasing and lower than that of wild type, and the glutamine pool was increasing and higher. As a result, the ratio of glutamate to glutamine dropped to 0.6–2.7. In contrast to their parent, both suppressors exhibited normal exponential phase of growth. Their growth was almost indistinguishable from that of wild type in low and medium osmolarity media. Their recovered growth corresponded well with the pool values. The pool ratios in the early suppressor SK3074 were 2.8–5.3 and further increased in the late suppressor SK3075 to ≈10–15. Taken together, there was a good correlation between the growth rate and the glutamate pool.
Table 1.
Batch culture doubling times and pool sizes
| Media osmolarity | Strain | Genotype | Doubling time, min | Pools, nmol/ml·OD650 |
Ratio, glutamate:glutamine | |
|---|---|---|---|---|---|---|
| Glutamate | Glutamine | |||||
| Low | SK2633 | WT | 63 | 20 | 4.0 | 5.1 |
| SK3062 | GOGAT− | 81, 140 | 16, 12 | 5.8, 8.5 | 2.7, 1.4 | |
| SK3074 | Chemostat early suppressor | 62 | 18 | 3.4 | 5.3 | |
| SK3075 | Chemostat late suppressor | 65 | 22 | 1.5 | 14.7 | |
| Medium | SK2633 | WT | 67 | 62 | 7.3 | 8.5 |
| SK3062 | GOGAT− | 105, 165 | 26, 16 | 18.7, 22.8 | 1.4, 0.7 | |
| SK3074 | Chemostat early suppressor | 66 | 52 | 10.4 | 5.0 | |
| SK3075 | Chemostat late suppressor | 70 | 55 | 4.5 | 12.2 | |
| High | SK2633 | WT | 77 | 93 | 11.7 | 8.0 |
| SK3062 | GOGAT− | 143, 206 | 33, 20 | 32.6, 34.7 | 1.0, 0.6 | |
| SK3074 | Chemostat early suppressor | 85 | 73 | 26.3 | 2.8 | |
| SK3075 | Chemostat late suppressor | 90 | 85 | 8.2 | 10.4 | |
Media were 0.2× N−C−DB (low osmolarity), N−C− (medium osmolarity), or 0.2× N−C−DB plus 0.3 M NaCl (high osmolarity), with 2 mM NH4+ as N-source, and 0.2% glycerol as the C-source. Cultures were inoculated with overnight cultures grown in the indicated media with 5 mM NH4+ and started with an initial OD650 of 0.05–0.06. Two single samples were taken from each culture at OD650 ≈0.2 and ≈0.4. Pool values of SK2633, SK3074, and SK3075 were expressed as averages (mean coefficient of variation = 8%). Values of SK3062 were given separately (mean coefficient of variation = 21%). The doubling time for each SK3062 sample was calculated based on two OD650 readings sandwiching the sampling point.
Characterization of Suppressor Mutations.
The presence of the ΔgltB824 allele in the suppressors was verified first. A GDH− strain, carrying a null gdh-51 allele and a linked zch-1436::Tn10 marker (90% linkage by P22-mediated transduction), was used as the donor to transduce into SK3074 or SK3075. In both crosses, ≈80% of the Tetr transductants were glutamate auxotrophs (Glu−), confirming that the ΔgltB824 allele was retained.
Reasoning that the growth rescue could be due to enhanced GDH activity either as a result of an altered enzyme with a higher affinity for NH4+ or overexpression, I next looked for linkage of suppressor mutations to the gdhA locus. By using the Glu− strain SK3064 (ΔgltB824 gdh-51 zch-1436::Tn10) as the recipient, glutamate prototrophic colonies (Glu+) were obtained by transducing P22 phage grown on SK3074 or SK3075. In both crosses, all Glu+ transductants were identical to SK3074 but not SK3075. The percentage of Tets among Glu+ transductants resembled the linkage between gdhA and zch-1436::Tn10. This suggested the two suppressors shared one mutation (later designated as gdh-515), either at or closely linked to gdhA, that was solely responsible for the phenotypes of the early suppressor SK3074. With the linked zch-1436::Tn10, gdh-515 was moved into wild-type SK2633. The resulting SK3111 (gdh-515 zch-1436::Tn10) was confirmed by crossing back into SK3062 (ΔgltB824) and obtaining Tetr transductants (ΔgltB824 gdh-515 zch-1436::Tn10) whose phenotypes were identical to those of SK3074 (ΔgltB824 gdh-515).
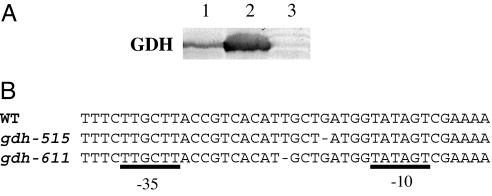
Strains bearing wild-type gdhA, gdh-515, and the null gdh-51 alleles were subjected to Western blot analysis with antibodies against GDH. gdh-515 caused a substantial overexpression of GDH (Fig. 3A). It was ≈5- to 10-fold higher than that of the wild type (estimated from band intensities), regardless of whether cells were grown in rich medium (Fig. 3A) or minimal medium (data not shown). By sequencing the gdhA promoter region, I found that the early suppressor mutation gdh-515 was a 1-bp deletion between the −10 and −35 motifs (Fig. 3B).
Fig. 3.
GDH overexpression with gdhA promoter-up mutations. (A) Isogenic strains SK2633 (wild type, lane 1), SK3111 (gdh-515 zch-1436::Tn10, chemostat suppressor mutation, lane 2), and SK3128 (gdh-51 zch-1436::Tn10, null mutation, lane 3) were grown in nutrient broth. Proteins of lysates from equal amounts of cells were separated by 10% SDS/PAGE and examined by Western blot. (B) Sequences of the −10 and −35 motifs of gdhA promoter in wild type, gdh-515 (from chemostat early suppressor SK3074), and gdh-611 (from plate suppressor FG1143) alleles.
As mentioned above, I expected the late suppressor SK3075 to have a mutation(s) in addition to gdh-515. The glnA locus was of primary interest because the most significant difference between SK3075 and its predecessor SK3074 was the decreased glutamine pool (Table 1). Glutamine auxotroph (Gln−) SK3106 (ΔgltB824 gdh-515 glnA120::Tn10) was transduced to glutamine prototrophy (Gln+) with P22 phage grown on SK3075. Most transductants were phenotypically identical to SK3075, indicating that the additional mutation (later designated as glnA424) was at or closely linked to glnA. By using the linked marker zif-214::Tn10, glnA424 was moved into wild-type SK2633. The resulting SK3112 (glnA424 zif-214::Tn10) was confirmed by backcrossing it into the early suppressor SK3074 and phenotypic examination of the transductants. SK3112 is a “leaky” glutamine auxotroph whose internal glutamine pool concentration is limited (6, 14). The strains carrying glnA424 impair but do not eliminate GS activity and grow slightly more slowly on NH4+ as N-source. The growth defect is relieved if glutamine is provided in the growth medium. Sequencing the glnA promoter and coding regions revealed that the late suppressor mutation glnA424 had a 1-bp substitution (CCT to CTT) in the coding region, resulting in a mutant GSP95L enzyme. The basis for the catalytic defect of GSP95L is unknown.
The identification of the mutations and their associated phenotypes helped to understand the sequential events during the chemostat experiment. Overexpressed GDH, as a result of the early gdhA promoter-up mutation, accelerated NH4+ flux through GDH and primarily refilled the low glutamate pool in GOGAT− cells under low NH4+ conditions (Table 1). Then a partially crippled GS enzyme, caused by the late glnA mutation, further pushed the glutamate pool up by cutting the glutamine pool down slightly below that of the wild type. The two-step evolution provided the cells with the ability to use low to trace amounts of NH4+ (Fig. 1) even without an intact high-affinity GS/GOGAT cycle. It revealed how the cell endowed with the altered linear pathway, which was disadvantageous in low NH4+, could achieve near-optimal growth simply by fine-tuning the pathway. Similarly important is that the bacteria reestablished their preferred relative pool values of high glutamate and low glutamine together with the recovery of growth.
Selection of S. typhimurium and E. coli GOGAT− Suppressor Mutants from Low NH4+ Plates.
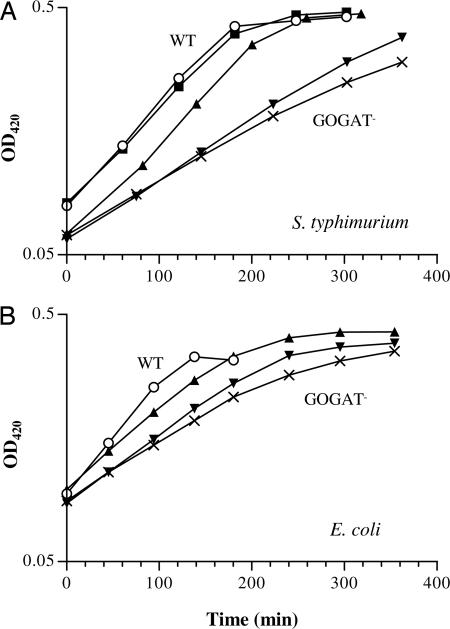
Intrigued by the above outcome, I selected suppressors on agar plates, where they would not be in competition. Full-strength minimal medium plates were used to ensure a significant growth defect (Table 1). Both S. typhimurium and E. coli GOGAT− strains barely grew on 0.1 mM NH4+ plates but did grow with 1 mM NH4+. From SK3062, the same GOGAT− strain used in the chemostat, more than a dozen independent suppressors were isolated from plates with 0.1–0.5 mM NH4+. Their gdhA loci were examined by the same cross described for the chemostat suppressors. Surprisingly, only one isolate (FG1143) carried a gdhA-linked allele that was responsible for the phenotype. Mutation(s) in all others were not linked to gdhA. FG1143 grew as well as the chemostat early suppressor SK3074 with low NH4+ (Fig. 4A). Another representative isolate, FG1158, showed subtle but reproducible growth enhancement over the parent SK3062. Genetic analysis excluded that its glnA locus was mutated (data not shown).
Fig. 4.
Growth of GOGAT− suppressor mutants selected from low NH4+ plates. Cells were grown in N−C− medium with 1 mM NH4+ as N-source and 0.2% glycerol as C-source. Batch cultures were inoculated with overnight cultures grown in the same medium except with 5 mM NH4+. (A) S. typhimurium: SK2633 (wild type, ○), SK3062 (ΔgltB824, ×), the chemostat early suppressor SK3074 (ΔgltB824 gdh-515, ■), and the plate suppressors FG1143 (▴) and FG1158 (▾). (B) E. coli: NCM3722 (wild type, ○), FG1079 (gltD::Tn5KAN-I-SceI, ×), and the plate suppressors FG1126 (▴) and FG1127 (▾).
Similarly, from E. coli GOGAT− FG1079 (gltD::Tn5KAN-I-SceI; gltD encodes GOGAT small subunit), eight independent suppressors were isolated. Their growth rates under low NH4+ conditions were all between wild-type NCM3722 and the parent FG1079 but hardly distinguishable among themselves. Shown in Fig. 4B are two representatives, FG1126 and FG1127. The gltBD, gdhA, and glnA loci in these two suppressors, respectively, were examined genetically. The gltD insertion did not change, proved by analyses of (i) PCR amplifications of the flanking gltD region and (ii) P1 transductants by crossing the Kanr marker back into wild-type NCM3722. By using strains FG1118 (gltD::Tn5KAN-I-SceI ΔgdhA; Glu−) and FG1134 (gltD::Tn5KAN-I-SceI ΔglnA; Gln−), respectively, and similar strategies to those described for S. typhimurium chemostat suppressors, genetic crosses revealed that neither their gdhA nor glnA loci were mutated (data not shown). The mutation(s) responsible for their enhanced growth is unknown.
GDH Activities of GOGAT− Suppressor Mutants.
Because only S. typhimurium FG1143 carried a gdhA mutation, the question remained on the nature of other plate suppressors. Their GDH activities were measured (Table 2). Compared with the S. typhimurium parent SK3062, the chemostat early suppressor SK3074 showed an ≈8-fold GDH activity increase, which agreed well with the Western blot result (Fig. 3A). The fold increase was about the same for FG1143. This was consistent with their similar growth at low NH4+ (Fig. 4A). The other S. typhimurium plate suppressor, FG1158, showed growth only slightly enhanced over SK3062. Its GDH activity was increased ≈30%. For the two E. coli plate suppressors, FG1126 and FG1127, GDH activities both doubled over their parent FG1079 (Table 2). Overall, there was a good correlation between GDH activity and the degree of growth enhancement, regardless of whether a lesion in the gdhA locus was responsible. Interestingly, in both species, GOGAT− strains showed ≈2.5-fold GDH activity increases over their wild-type counterparts (see Discussion).
Table 2.
GDH activities
| Strain | Genotype | Specific activity, nmol NADPH oxidized/min·mg of protein |
|---|---|---|
| S. typhimurium | ||
| SK2633 | WT | 124 |
| SK3128 | GDH− | 0 |
| SK3062 | GOGAT− | 306 |
| SK3074 | Chemostat suppressor | 2,350 |
| FG1143 | Plate suppressor | 2,516 |
| FG1158 | Plate suppressor | 410 |
| E. coli | ||
| NCM3722 | WT | 242 |
| FG1113 | GDH− | 2 |
| FG1079 | GOGAT− | 562 |
| FG1126 | Plate suppressor | 1,132 |
| FG1127 | Plate suppressor | 1,140 |
Strains were grown in N−C− medium with 2 mM NH4+ as N-source and 0.2% glycerol as C-source. Cultures were inoculated with overnight cultures grown in the same medium except with 5 mM NH4+. Samples were collected at similar OD650 (between 0.38 and 0.39 for S. typhimurium and between 0.27 and 0.28 for E. coli) after at least two doublings of growth.
gdhA Promoter-Up Mutations.
The S. typhimurium plate suppressor FG1143 carried a new gdhA mutation, designated as gdh-611. Like gdh-515 in the chemostat suppressors, gdh-611 was also a 1-bp deletion between the −10 and −35 motifs of the promoter (Fig. 3B). The two deletions are not identical (3 bp apart). Their similar GDH expression levels and growth phenotypes suggested a similar molecular mechanism. Both deletions shortened the native gdhA 19-bp spacer between the −35 and −10 motifs to 18 bp, closer to the 17 bp for consensus σ70 promoters (16). This could account for GDH overexpression. However, the gdhA −10 region belongs to an extended form with an immediate upstream TGN (17). The −35 sequence and spacer length in between are more flexible in such σ70 promoters. I cannot rule out the possibility of an unidentified S. typhimurium regulatory protein acting in trans on this region. A different gdhA promoter-up mutation (gdh-3, a base substitution mutation also located in the spacer region) was reported in Klebsiella aerogenes by Bender and coworkers (18). It was identified from a Glu+ revertant of a Glu− strain that lacked GOGAT activity and repressed GDH expression. The authors provided evidence for a likely mechanism of GDH overexpression by this mutated cis element. But they also cautioned that the exact nature could be more complex and was yet to be fully understood (18).
Discussion
Growth Defect of GOGAT− Mutants.
The direct cause of the growth defect of GOGAT− mutants under low NH4+ conditions is insufficient glutamate production through the GDH pathway. However, the intermediate glutamate levels in a GOGAT− strain appear sufficient for cellular biosynthetic needs. For instance, ≈20 mM glutamate, the level present in a wild-type strain at low osmolarity, was able to support maximum growth rate, whereas a GOGAT− strain showed a growth defect at high osmolarity with a similar or even higher pool concentration (Table 1). In terms of biosynthesis, internal glutamate pools of wild type appear to be in excess, especially at high osmolarity. The same is not true for the glutamine pool, which is low especially under N-limiting conditions. The “extra” glutamate is required to maintain and partially balance the K+ pool (12). Other hypotheses to explain the growth defect of GOGAT− mutants have been convincingly disproved (19).
Configuration of Internal Glutamate and Glutamine Pools and Their Relationship.
Pool measurements in a GOGAT− strain and its suppressors also showed an inverse relationship between glutamate and glutamine pools (Table 1). The glutamine pool in the GOGAT− strain was high because of the blockage of GS/GOGAT cycle: GOGAT is the major glutamine-metabolizing enzyme. As NH4+ in the medium was consumed, shrinking NH4+ flux through GDH resulted in a declining glutamate pool. This decrease did not limit GS action because glutamate was still saturating GS [Km ≈ 4 mM (20, 21)]. On the contrary, the glutamine pool climbed even higher. It has been shown in vitro (widely accepted to be true in vivo as well) that 2-oxoglutarate enhances but glutamine limits both GS synthesis and activity as allosteric regulators through the regulatory cascades (2, 22, 23). In both GOGAT− or GDH− strains, the 2-oxoglutarate pool (substrate for both enzymes) was higher than in their parental wild-type strain (unpublished result). The climbing glutamine pool presumably countered the elevated 2-oxoglutarate pool, thus limiting GS action to match the shrinking NH4+ flux through GDH. Otherwise, the low glutamate pool would be further depleted. Indeed, monitoring glnA expression in a GOGAT− strain with a glnA-lacZ fusion showed ≈2-fold repression compared with the wild type (unpublished result).
Pools were reversed in a GOGAT− strain carrying a gdhA promoter-up mutation (gdh-515): this primary suppressor had a largely refilled glutamate pool by an enhanced GDH flux, a growth recovery, and a lower glutamine pool (Table 1). The last agreed with observations of both a decrease of the 2-oxoglutarate pool and glnA-lacZ repression by gdh-515 (unpublished results). The secondary suppressor mutation (glnA424) impaired GS and directly decreased glutamine synthesis, and, as a consequence, levels of the substrate glutamate increased slightly. From GOGAT− through the early suppressor to the late suppressor, the glutamate and glutamine pools showed a series of changes that were inversely coordinated. In the end, the triple mutant (ΔgltB824 gdh-515 glnA424) possessed a combination of high glutamate and low glutamine like the wild type, but with a linear two-enzyme pathway instead of the normal three-enzyme circuit. The two remaining reactions were balanced by enhancing NH4+ flux through GDH and limiting GS action. The system was impressively resilient.
GDH Regulation.
Compared with the vigorous and well studied GS regulation (2), GDH regulation is less understood (1). The only well studied regulatory component is N limitation-induced Nac (the nitrogen assimilation control protein). Nac represses GDH expression in K. aerogenes and E. coli, but not in the nac-free S. typhimurium (24–26). Genetic analyses prompted Gowrishankar and coworkers (27, 28) to suggest that argP (encoding a LysR-type transcriptional regulator) and spoT (encoding ppGpp-3′ pyrophosphohydrolase) were also involved in regulation of GDH synthesis and/or activity. With the requirement for high glutamate, the coordinated pool configuration in the circuit, and the intriguing adaptive responses by the altered GDH–GS pathway, it is conceivable that GDH could be subjected to a multilevel regulation to fine-tune the system. The observed ≈2.5-fold increase of GDH activities in both E. coli and S. typhimurium GOGAT− strains over the wild types (Table 2) suggests that there is a “built-in” GDH regulatory mechanism to combat glutamate deficiency. Others have previously reported an ≈10-fold increase in GDH activity in GOGAT− strains when high NH4+ was replaced with limited glutamine as N-source (19).
The mutations in several S. typhimurium and E. coli GOGAT− suppressor mutants described in this report have not been identified. They all had elevated GDH activities (Table 2) that appeared responsible for the suppressor phenotypes. Nac cannot be the answer for S. typhimurium suppressor mutants and is also unlikely the answer for E. coli GOGAT− suppressor mutants, because Nac presumably is not available in the first place (the Ntr system that activates Nac is repressed in GOGAT− with low NH4+). Identification of the unknown mutations will likely shed light on GDH control and perhaps even on protection of a high glutamate pool.
Materials and Methods
Strains, Media, and Growth Conditions.
All named strains were isogenic of prototrophic S. typhimurium SK2633 [derived from strain LT2 (13)] or E. coli NCM3722 (29). Phages P22 HT105 and P1vir were used for transductions in S. typhimurium and E. coli, respectively. E. coli allele gltD::Tn5KAN-I-SceI was obtained from the Blattner KO collection (30), and alleles ΔgdhA and ΔglnA were derived from the Keio collection (31).
The full-strength minimal medium was Na+-based N−C− medium plus 1 mM KCl, from which other media with different osmolarities (e.g., diluted and then double-buffered 0.2 × N−C−DB) were derived (12). Media were supplemented with NH4Cl as N-source and glycerol as C-source. When necessary, glutamate or glutamine was provided at 10 mM. Arginine (2.5 mM) was used as the sole N-source for Aut examination. Rich media were nutrient or LB broth. When required, antibiotics were added (25 μg/ml kanamycin and 10 μg/ml tetracycline).
Cell growth experiments were carried aerobically at 37°C. Operations of chemostat and batch culture were performed as described (6, 12). Cell density was measured by absorbance at either 420 or 650 nm (1 OD650 ≈ 1.9 OD420). Remaining NH4+ in chemostat samples was determined by a GDH-based colorimetric assay (6).
Measurement of Glutamate and Glutamine Pools.
Metabolites were extracted from cells by using a “no-harvest” protocol, and fluorescent derivatives of glutamate and glutamine were separated and quantified by a HPLC-based method (6, 13, 32). By using an approximate conversion factor of 1 μl of intracellular water per milliliter per OD650 cells (33, 34), pool value in nmol/ml·OD650 can be translated to mM internal concentration.
Selection of GOGAT− Suppressor Mutants from Low NH4+ Plates.
GOGAT− strains were streaked onto full-strength minimal medium plates with 0.2% glycerol as C-source and 0.1–2 mM NH4Cl as N-source. After 4–7 days of incubation, tiny suppressor colonies appeared on cell streaking lines in plates with 0.1–0.5 mM NH4+. They were picked with a sharp-pointed platinum wire under a dissecting microscope, isolated on rich medium plates, and confirmed on minimal medium plates with 0.1 mM NH4+.
GDH Assays.
A standard Western blot protocol was performed for semiquantitative analysis of GDH protein. E. coli GDH antibody was a gift from H. Zalkin (Purdue University, West Lafayette, IN). Assay for GDH activity was mostly carried as described (35). Bacteria were disrupted by passing through a French press cell, and extracts were clarified by centrifugation. The modified assay system contained 50 mM Tris·Cl (pH 7.5), 5 mM 2-oxoglutarate, 5 mM NH4Cl, and 0.1 mM NADPH. The absorbance of NADPH at 340 nm was monitored for 3 min at room temperature after adding an appropriate amount of extract that resulted in a linear decrease of OD340. Protein amounts in extracts were determined by the Bradford method.
PCR Primers and Sequencing.
Primer pair 5′-CGGCAACATTGAGCGCTATATC and 5′-GTTACGCGTCAGGACATCCG, which flank the Tn5KAN-I-SceI insertion site, was used in confirmation of the E. coli gltD::Tn5KAN-I-SceI allele (1,387 bp instead of 148 bp without the insertion). The S. typhimurium gdhA promoter region (352 bp) was amplified with 5′-TTTGCCAGCCCGACCATTGG and 5′-GGTCGCGCTTTTGTACATGGTT, and the glnA promoter plus coding region (1,827 bp) was amplified with 5′-CAACTTTGCCTCAGGCATTAGAA and 5′-CCTTGCAGCAACGCGAAATT. For sequence identification of the gdh-515, gdh-611, and glnA424 alleles, the respective PCR products were amplified from the original mutants and sequenced. One point mutation in each case was identified by sequence alignment with the published S. typhimurium LT2 genome sequence. Then the mutation was confirmed by sequencing the corresponding region in (i) wild-type SK2633 and (ii) an independent SK2633-originated strain carrying the allele obtained through a P22-mediated transduction.
Acknowledgments
I thank Robert Bender, Terence Hwa, and Steven Larsen for detailed and valuable criticisms of the manuscript, and I am grateful to Hal Broxmeyer and Frank Yang for providing laboratory space to complete this research. This work was initiated and partially carried out while I was a postdoctoral fellow with Sydney G. Kustu in the Department of Plant and Microbial Biology, University of California, Berkeley, and was supported by National Institutes of Health Grant GM38361 (to Dr. Kustu).
Abbreviations
- GS
glutamine synthetase
- GOGAT
glutamate synthase
- GDH
glutamate dehydrogenase.
Footnotes
The author declares no conflict of interest.
References
- 1.Reitzer L. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Bock A, Curtiss R III, Kaper JB, Neidhardt FC, Nystrom T, Rudd KE, Squires CL, editors. Washington, DC: Am Soc Microbiol; 2004. [Accessed May 2, 2006]. Chap 3.6.1.3. Available at www.ecosal.org. [Google Scholar]
- 2.Stadtman ER. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Bock A, Curtiss R III, Kaper JB, Neidhardt FC, Nystrom T, Rudd KE, Squires CL, editors. Vol. 30. Washington, DC: Am Soc Microbiol; 2006. [Accessed April 2004]. Chap 3.6.1.6. Available at www.ecosal.org. [Google Scholar]
- 3.Miller RE, Stadtman ER. J Biol Chem. 1972;247:7407–7419. [PubMed] [Google Scholar]
- 4.Sakamoto N, Kotre AM, Savageau MA. J Bacteriol. 1975;124:775–783. doi: 10.1128/jb.124.2.775-783.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wohlheuter RM, Schutt H, Holzer H. In: The Enzymes of Glutamine Metabolism. Prusiner SB, Stadtman ER, editors. New York: Academic; 1973. pp. 45–64. [Google Scholar]
- 6.Ikeda TP, Shauger AE, Kustu S. J Mol Biol. 1996;259:589–607. doi: 10.1006/jmbi.1996.0342. [DOI] [PubMed] [Google Scholar]
- 7.Csonka LN, Hanson AD. Annu Rev Microbiol. 1991;45:569–606. doi: 10.1146/annurev.mi.45.100191.003033. [DOI] [PubMed] [Google Scholar]
- 8.Tempest DW, Meers JL, Brown CM. J Gen Microbiol. 1970;64:171–185. doi: 10.1099/00221287-64-2-171. [DOI] [PubMed] [Google Scholar]
- 9.Dinnbier U, Limpinsel E, Schmid R, Bakker EP. Arch Microbiol. 1988;150:348–357. doi: 10.1007/BF00408306. [DOI] [PubMed] [Google Scholar]
- 10.McLaggan D, Naprstek J, Buurman ET, Epstein W. J Biol Chem. 1994;269:1911–1917. [PubMed] [Google Scholar]
- 11.Richey B, Cayley DS, Mossing MC, Kolka C, Anderson CF, Farrar TC, Record MT., Jr J Biol Chem. 1987;262:7157–7164. [PubMed] [Google Scholar]
- 12.Yan D, Ikeda TP, Shauger AE, Kustu S. Proc Natl Acad Sci USA. 1996;93:6527–6531. doi: 10.1073/pnas.93.13.6527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Csonka LN, Ikeda TP, Fletcher SA, Kustu S. J Bacteriol. 1994;176:6324–6333. doi: 10.1128/jb.176.20.6324-6333.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu P, Leighton T, Ishkhanova G, Kustu S. J Bacteriol. 1999;181:5042–5050. doi: 10.1128/jb.181.16.5042-5050.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brenchley JE, Prival MJ, Magasanik B. J Biol Chem. 1973;248:6122–6128. [PubMed] [Google Scholar]
- 16.Aoyama T, Takanami M, Ohtsuka E, Taniyama Y, Marumoto R, Sato H, Ikehara M. Nucleic Acids Res. 1983;11:5855–5864. doi: 10.1093/nar/11.17.5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mitchell JE, Zheng D, Busby SJ, Minchin SD. Nucleic Acids Res. 2003;31:4689–4695. doi: 10.1093/nar/gkg694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Janes BK, Pomposiello PJ, Perez-Matos A, Najarian DJ, Goss TJ, Bender RA. J Bacteriol. 2001;183:2709–2714. doi: 10.1128/JB.183.8.2709-2714.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goss TJ, Perez-Matos A, Bender RA. J Bacteriol. 2001;183:6607–6619. doi: 10.1128/JB.183.22.6607-6619.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meek TD, Villafranca JJ. Biochemistry. 1980;19:5513–5519. doi: 10.1021/bi00565a008. [DOI] [PubMed] [Google Scholar]
- 21.Timmons RB, Rhee SG, Luterman DL, Chock PB. Biochemistry. 1974;13:4479–4485. doi: 10.1021/bi00719a002. [DOI] [PubMed] [Google Scholar]
- 22.Jiang P, Peliska JA, Ninfa AJ. Biochemistry. 1998;37:12795–12801. doi: 10.1021/bi9802420. [DOI] [PubMed] [Google Scholar]
- 23.Jiang P, Peliska JA, Ninfa AJ. Biochemistry. 1998;37:12802–12810. doi: 10.1021/bi980666u. [DOI] [PubMed] [Google Scholar]
- 24.Bender RA. Mol Microbiol. 1991;5:2575–2580. doi: 10.1111/j.1365-2958.1991.tb01965.x. [DOI] [PubMed] [Google Scholar]
- 25.Goss TJ, Janes BK, Bender RA. J Bacteriol. 2002;184:6966–6975. doi: 10.1128/JB.184.24.6966-6975.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muse WB, Bender RA. J Bacteriol. 1998;180:1166–1173. doi: 10.1128/jb.180.5.1166-1173.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nandineni MR, Laishram RS, Gowrishankar J. J Bacteriol. 2004;186:6391–6399. doi: 10.1128/JB.186.19.6391-6399.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saroja GN, Gowrishankar J. J Bacteriol. 1996;178:4105–4114. doi: 10.1128/jb.178.14.4105-4114.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soupene E, van Heeswijk WC, Plumbridge J, Stewart V, Bertenthal D, Lee H, Prasad G, Paliy O, Charernnoppakul P, Kustu S. J Bacteriol. 2003;185:5611–5626. doi: 10.1128/JB.185.18.5611-5626.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kang Y, Durfee T, Glasner JD, Qiu Y, Frisch D, Winterberg KM, Blattner FR. J Bacteriol. 2004;186:4921–4930. doi: 10.1128/JB.186.15.4921-4930.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Mol Syst Biol. 2006;2:2006–0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kustu S, Hirschman J, Burton D, Jelesko J, Meeks JC. Mol Gen Genet. 1984;197:309–317. doi: 10.1007/BF00330979. [DOI] [PubMed] [Google Scholar]
- 33.Cayley S, Lewis BA, Guttman HJ, Record MT., Jr J Mol Biol. 1991;222:281–300. doi: 10.1016/0022-2836(91)90212-o. [DOI] [PubMed] [Google Scholar]
- 34.Stock JB, Rauch B, Roseman S. J Biol Chem. 1977;252:7850–7861. [PubMed] [Google Scholar]
- 35.Broach J, Neumann C, Kustu S. J Bacteriol. 1976;128:86–98. doi: 10.1128/jb.128.1.86-98.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]