Abstract
Alzheimer's disease (AD) is an age-dependent neurodegenerative disease that causes progressive cognitive impairment. The initiation and progression of AD has been linked to cholesterol metabolism and inflammation, processes that can be modulated by liver x receptors (LXRs). We show here that endogenous LXR signaling impacts the development of AD-related pathology. Genetic loss of either Lxrα or Lxrβ in APP/PS1 transgenic mice results in increased amyloid plaque load. LXRs regulate basal and inducible expression of key cholesterol homeostatic genes in the brain and act as potent inhibitors of inflammatory gene expression. Ligand activation of LXRs attenuates the inflammatory response of primary mixed glial cultures to fibrillar amyloid β peptide (fAβ) in a receptor-dependent manner. Furthermore, LXRs promote the capacity of microglia to maintain fAβ-stimulated phagocytosis in the setting of inflammation. These results identify endogenous LXR signaling as an important determinant of AD pathogenesis in mice. We propose that LXRs may be tractable targets for the treatment of AD due to their ability to modulate both lipid metabolic and inflammatory gene expression in the brain.
Keywords: nuclear receptor, inflammation, macrophage, microglia, cholesterol metabolism
The liver x receptors α and β (LXRα/NR1H3 and LXRβ/NR1H2, respectively) are oxysterol-activated nuclear receptors that play an important role in the control of cellular and whole-body cholesterol homeostasis (1–4). LXRs are also potent inhibitors of inflammatory responses in macrophages, pointing to an additional function for LXR signaling in immune regulation (5–11). The function of LXRs in the brain is not well understood. Ligand activation of LXRs promotes cholesterol efflux from glia (12, 13) and primary neurons (14), whereas mice deficient in expression of Lxrα and Lxrβ develop marked accumulation of neutral lipids in the brain (15). Functionally, loss of LXRβ leads to adult-onset motor neuron degeneration by the age of 7 months (16). The role of LXRs in human neurodegenerative diseases, however, remains largely unknown.
Alzheimer's disease (AD) is an age-dependent neurodegenerative disease typified by progressive neuronal loss and cognitive impairment. AD is characterized by extraneuronal deposits of β-amyloid (Aβ) fibrils (fAβ) (17) and intraneuronal tangles of hyperphosphorylated τ (18). The identification of rare mutations in the amyloid precursor protein (APP) and in presenilin genes (PS) in human AD is consistent with a central role for Aβ in AD pathogenesis (19). AD is a multifactorial disease, and inflammatory processes and cholesterol metabolism are among several factors that have been linked to its etiology.
The AD brain exhibits prominent activation of innate immune responses. For example, elevated levels of inflammatory mediators can be measured in AD brains, and these are postulated to contribute to neuronal loss. The local inflammatory response is mediated by activated microglia and reactive astrocytes that surround the senile plaques (20, 21). Interaction of microglia with fAβ leads to their activation and the release of an array of inflammatory mediators. Despite decorating amyloid plaques, microglia in the AD brain are poor phagocytes of deposited amyloid. This phenomenon has been proposed to be due in part to inhibition of their phagocytic capacity by the chronic inflammatory milieu (22). Interestingly, antiinflammatory drugs are able to reverse this defect in phagocytosis, suggesting that modulating microglial activation may promote amyloid clearance.
In addition to ongoing inflammatory processes, the role of cholesterol metabolism and specifically of apolipoprotein E (apoE) in AD pathogenesis is undisputed (23, 24). The APOE4 allele, with a prevalence of 10–15% in most populations, remains the major genetic susceptibility allele for AD. The discovery that SNPs in genes involved in cholesterol metabolism such as Abca1, Abca2, and Cyp46 are associated with increased risk for AD further strengthens this notion (25–27). Several groups have examined effects of LXR on APP processing (14, 28, 29), which is sensitive to cellular sterol levels (23). These studies support a possible role for ATP-binding cassette A1 (ABCA1), a transporter for cholesterol and phospholipids and a direct transcriptional target of LXR, in AD. Recent studies have shown that AD transgenic (Tg) mice lacking Abca1 develop increased Aβ levels and plaque pathology in the absence of changes in APP processing (30–33).
Owing to their ability to regulate cholesterol metabolism and inflammatory signaling, LXRs have emerged as potential targets for metabolic diseases (34). However, the ability of endogenous LXR signaling to modulate neuroinflammation and AD pathology in vivo has not been addressed. We show here that genetic loss of Lxrα or Lxrβ in APP/PS1 AD-Tg mice exacerbates senile plaque pathology. We further suggest that this effect may be due to defects in brain cholesterol metabolism as well as the ability of LXRs to inhibit the inflammatory response of glial cells to fAβ. These results support the further investigation of synthetic LXR agonists as potential therapy for AD.
Results
LXRs Regulate Lipid-Inducible Gene Expression in the Brain.
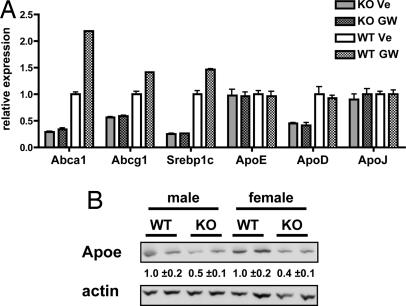
We examined the influence of LXRs on lipid-inducible gene expression in the brain by treating WT and Lxrαβ−/− mice with the synthetic LXR ligand GW3695. Transcriptional profiling revealed that LXR expression was important for both basal and inducible expression levels of several established LXR targets, including Abca1 and Abcg1 (Fig. 1A). These changes were also confirmed in a second experiment using a different LXR ligand, T0901317 [supporting information (SI) Table 1]. Interestingly, Apoe mRNA levels in the LXR-null mice were unchanged, and the transcript was not induced by ligand treatment, despite Apoe being a direct target of LXRs (35). However, Apoe protein levels were substantially decreased in male and female Lxrαβ−/− mice (Fig. 1B). Importantly, in these experiments, we did not detect significant changes in the expression of genes involved in APP processing, including Ps1, Ps2, App, Bace1, Bace2, Adam10, Adam11, and Ncstrn1 (data not shown).
Fig. 1.
Analysis of LXR-dependent gene expression in the brain. (A) LXRαβ−/− [knockout (KO)] and WT mice (n = six mice per group) were gavaged for 3 days with vehicle (Ve) or GW3695 (GW) (20 mg/kg). Brain RNA was analyzed individually for gene expression by quantitative PCR. Results are expressed as mean ± SD relative to vehicle-treated WT mice. (B) Immunoblot analysis of apoE in whole brain homogenates from male and female WT and LXRαβ−/− mice (n = two mice per group). The relative protein level was quantitated by densitometry.
Loss of LXRs Exacerbates Senile Plaque Pathology.
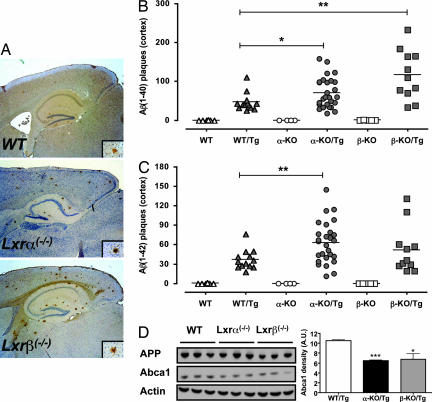
To directly test whether endogenous LXR signaling modulates the progression of AD-associated pathology, we generated mice lacking Lxrα or Lxrβ and that were also hemizygous for a transgene carrying the APP Swedish mutation and PS1 lacking exon 9 (36). This double-Tg model of AD has the advantage that lesions develop at a significantly earlier age compared with single-gene models. Amyloid plaques could be detected as early as 24 weeks of age in our model (data not shown). We chose to analyze the mice for AD pathology at an age of 32 weeks. At this age, amyloid pathology was predominantly cortical (Fig. 2A), consistent with previous studies of this Tg model (37). Remarkably, loss of either LXR isoform resulted in a significant increase in cortical the Aβ1–40 plaque number (Fig. 2B). Similarly, loss of Lxrα led to a significant increase in the Aβ1–42 plaque number (Fig. 2C). The cortical Aβ1–42 plaque count in Lxrβ mice showed a trend toward increased burden relative to their WT/Tg counterparts, but did not reach statistical significance. Of note, in both Lxrα- and Lxrβ-null mice, plaques were characteristically larger (Fig. 2A). It is therefore likely that our results, which are based on plaque counts, underestimate the amyloid burden in LXR-null mice. Loss of Lxrα or Lxrβ had no effect on APP levels or processing as assessed by immunoblotting for C-terminal fragments (CTFs) (Fig. 2D and data not shown). Unexpectedly, loss of either Lxrα or Lxrβ resulted in an ≈40% decrease in Abca1 levels in the brains of 12-week-old Tg mice. At this age, no amyloid plaques can be detected, suggesting that regulation of ABCA1 by LXRs may constitute an early factor that modifies AD progression.
Fig. 2.
Increased amyloid deposition in LXR-null mice. (A) Representative sagittal sections from 32-week-old WT/Tg, Lxrα−/−/Tg, and Lxrβ−/−/Tg mice stained for Aβ1–40. (Inset) A representative amyloid plaque. (Magnification: ×40.) (B and C) Amyloid Aβ1–40 (B) and Aβ1–42 (C) were quantified in 32-week-old mice. Each point represents an individual mouse, with the group average plotted as a bar. The number of mice and male/female distribution within the groups was WT (n = 6; 4/2), WT/Tg (n = 13; 9/4), α-KO (n = 4; 4/0), α-KO/Tg (n = 27; 15/12), β-KO (n = 4; 2/2), and β-KO/Tg (n = 11; 8/3). (D) Immunoblot analysis of APP and ABCA1 levels in whole-brain lysates of 12-week-old male mice of the indicated Tg genotype. Each lane represents an individual mouse. ∗, P < 0.05; ∗∗, P < 0.01.
LXRs Are Repressors of the Microglial Inflammatory Response.
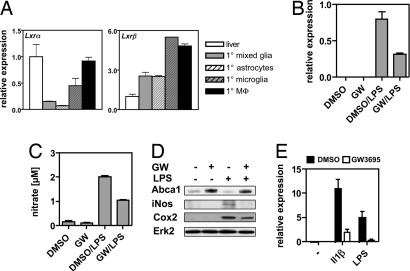
Previous work has established that LXRs attenuate inflammatory responses in macrophages (5). Innate immune responses are linked to AD pathogenesis, but whether LXRs can impact these processes in the context of AD is unknown. Together with astrocytes, resident microglia make up the innate immune system within the CNS. Analysis of the expression patterns of Lxrα and Lxrβ in primary glia cells (Fig. 3A) revealed that primary microglia express Lxrα and Lxrβ at a level comparable with that of peritoneal macrophages, whereas astrocytes express Lxrβ predominantly. As an initial approach to examine the function of LXRs in glial cells we tested whether LXR agonists could modify the response of the microglial-derived cell line BV2 to lipopolysaccharide (LPS). These cells respond to the synthetic LXR ligand GW3695 and to the brain-specific oxysterol 24S-hydroxycholesterol (SI Fig. 7). LPS robustly induced iNos mRNA, nitrate secretion into the medium, and iNOS and Cox2 protein in BV2 cells. In the presence of GW3695, however, this response was severely attenuated (Fig. 3 B–E). Similarly, activated LXRs inhibited the LPS- and Il-1β-induced expression of iNos in primary microglia (Fig. 3E) and in primary astrocytes (data not shown). These results are in line with a recent study by Kim et al. (38) showing that T0901317 represses the microglial response to LPS.
Fig. 3.
Activated LXRs inhibit the inflammatory response of BV2 cells and primary microglia toward LPS and Il1β. (A) Relative expression of Lxrα and Lxrβ in primary mixed glia cultures, astrocytes, microglia, peritoneal macrophages, and liver. (B and C) BV2 cells were treated as indicated, and iNos expression (B) and nitrate release into the medium (C) were determined. (D) Immunoblot analysis of Abca1, iNos, and Cox2 levels in BV2 cells treated as indicated. Erk2 was used to control for loading. (E) Primary microglia were pretreated with DMSO or 1 μM GW3695 for 18 h and subsequently challenged with 15 ng/ml Il1β or 5 ng/ml LPS for 6 h, after which iNos expression was determined. Values shown are the average ± SD.
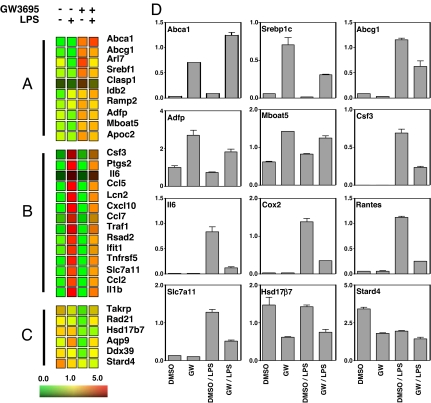
To gain further insight into the roles of LXRs in glial cells, we performed transcriptional profiling on BV2 cells in the presence or absence of GW3695 and LPS. Microarray analysis allowed us to distinguish three major groups of LXR-regulated genes (Fig. 4). The first group consisted of genes induced by LXR activation (Fig. 4A). The majority of these genes are involved in lipid metabolism and transport (e.g., Abca1, Abcg1, Srebp-1c). The second group contained a battery of inflammatory genes that were strongly induced by LPS and potently repressed by LXRs, such as Il1β, Traf1, Ccl5, Ccl2, and Cxcl10 (Fig. 4B). The third group included genes that were repressed by LXR activation (Fig. 4C). These genes could not be readily assigned to a common functional group. Notably, Hsd17β7 and Stard4 are involved in synthesis (39) and intracellular transport of cholesterol (40), respectively, and are known to be repressed by elevated sterol levels (41, 42).
Fig. 4.
Transcriptional profiling of BV2 cells. BV2 cells were treated as indicated, and RNA was isolated and subjected to transcriptional profiling on Affymetrix arrays. (A–C) Heat diagrams of genes induced by GW3695 (A), genes induced by LPS (>5-fold) and repressed by GW3695 (>2-fold repression) (B), and genes repressed (>2-fold repression) by GW3695 (C). (D) Quantitative PCR analysis was used to confirm the changes in expression of the indicated genes in RNA samples from BV2 cells. Values shown are the average ± SD.
LXRs Inhibit the Inflammatory Response of Primary Glial Cells to Fibrillar Aβ Peptide.
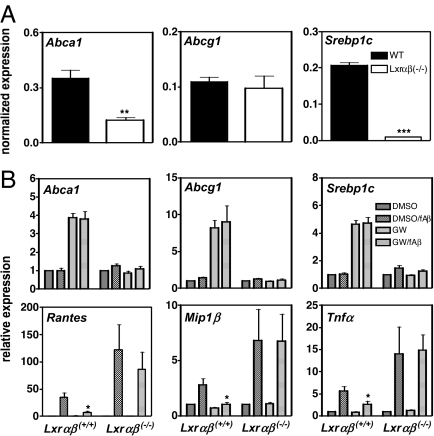
To specifically test whether LXRs could inhibit inflammatory responses in the setting of AD we isolated primary cultures of mixed glia cells from WT and Lxrαβ−/− mice. Loss of LXRs resulted in severely impaired basal expression of Abca1 and Srebp1c, whereas Abcg1 expression remained unchanged (Fig. 5A). Cells from WT mice expressed Lxrα and Lxrβ, and treatment of these cells with an LXR ligand resulted in the induction of LXR target genes in a receptor-dependent manner (Fig. 5B). We also challenged mixed glial cultures with fAβ1–40. Compared with the response to the strong inflammatory stimulus LPS, the response to amyloid fibrils was substantially weaker. Nevertheless, we observed a significant response to a 24-h exposure to 10 μM fAβ1–40. Importantly, under similar conditions, fAβ40–1 did not elicit an inflammatory response nor was there a substantial response to nonfibrillar forms of Aβ1–40 (data not shown). We analyzed expression of a panel of inflammatory genes known to be responsive to fAβ in glial WT and Lxrαβ−/− cells. In line with the results in BV-2 cells, synthetic LXR agonist blunted the induction of Rantes, Tnfα, Mip1β, and Il1β expression by fAβ1–40 in a receptor-dependent manner (Fig. 5B and data not shown). Moreover, despite similar low-level basal expression of these genes, their induction in response to fAβ1–40 was enhanced in cells lacking LXRs. These results strongly suggest that LXRs are endogenous inhibitors of the innate CNS response elicited by fAβ.
Fig. 5.
Analysis of gene expression in primary mixed glia. (A) Primary mixed glial cells were isolated from WT and Lxrαβ−/− mice, and the basal expression level of LXR target genes was analyzed. (B) Primary mixed glial cells were treated as indicated with DMSO or 1 μM GW3695 and in the presence or absence of 10 μM fAβ for 24 h. Transcript levels of the indicated genes were analyzed by quantitative PCR and are normalized to the DMSO-treated sample in each genotype. Values represent the mean ± SD of independent preparations made from WT (n = 4) and Lxrαβ−/− (n = 4) pups. ∗, P < 0.05.
LXRs Couple Their Antiinflammatory Activity to Increased Phagocytic Capacity.
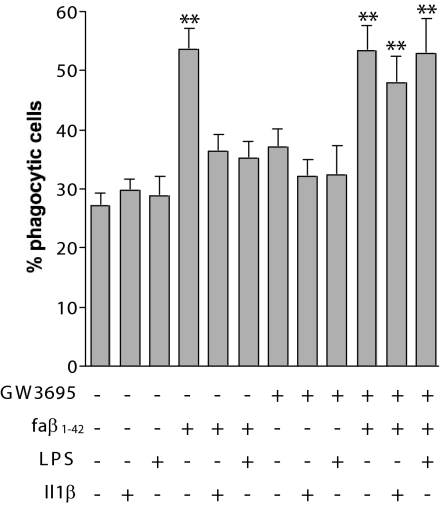
Microglia are competent phagocytes and normally clear Aβ from the brain (22, 43). However, inflammatory cytokines, which are present at elevated levels in the AD brain, suppress the phagocytic activity of these cells. We tested whether LXR-mediated suppression of inflammatory gene expression would increase microglial phagocytosis. Indeed, treatment of microglia with GW3965 reversed the Il1β- and LPS-mediated inhibition of fAβ-stimulated phagocytosis (Fig. 6). These findings suggest that the antiinflammatory activity of LXR agonists facilitates the phagocytic clearance of fAβ by microglia.
Fig. 6.
LXRs regulate the phagocytic response of BV2 cells. BV2 cells were pretreated with DMSO or 1 μM GW3965 for 18 h and treated with or without 15 ng/ml Il1β or 100 ng/ml LPS for an additional 18 h. On the day of phagocytosis assay, the cells were treated with fAβ1–42 for 30 min and incubated with 1 μm of fluorescent microspheres for another 30 min. Phagocytosis of the fluorescent microspheres was quantified as percent of cell phagocytosing. Data represent the mean ± SE from three independent experiments. ∗∗, P < 0.001.
Discussion
Accumulating evidence points to an association between cholesterol metabolism in the CNS and AD (23, 24). The role of endogenous LXR signaling in the development of AD, however, has not been explored. We have shown here that loss of either LXRα or LXRβ expression exacerbates AD-related pathology in APP/PS1 Tg mice. Our data suggest that dysregulated expression of key metabolic genes and increased inflammatory responses toward fibrillar amyloid forms both contribute to this phenotype. The ability of LXRs to regulate inflammatory and metabolic factors relevant to AD pathogenesis supports further exploration of nuclear receptor-signaling pathways as targets for AD therapeutics.
When cellular cholesterol levels rise, activation of LXRs stimulates cholesterol efflux from peripheral tissues and reverse cholesterol transport, thereby promoting whole-body cholesterol homeostasis (2). The ability of cells to unload excess cholesterol is dramatically reduced in mice lacking LXRs, leading to cholesterol accumulation in multiple tissues (44–46). The role of LXRs in the CNS is less well understood. Our findings indicate that LXRs control both basal and inducible expression of a subset of genes important for cellular cholesterol homeostasis in the brain, in agreement with the findings of Wang et al. (15). We have further shown that these genes are controlled in a receptor-dependent manner in primary glial cultures. In contrast to macrophages and the intestine, where loss of LXRs results in derepression of basal Abca1 expression (47), we found that Abca1 expression was reduced in brain samples and primary glial cells from mice lacking one or both LXRs. Cumulatively, these observations support the idea of tissue-specific regulation of Abca1 that may involve differential interaction of LXRs with corepressor proteins (47).
LXR-null AD Tg mice exhibit decreased ABCA1 protein levels, and this may be an early predisposing factor for the development of AD pathology. Three groups have recently reported that loss of Abca1 expression reduces soluble apoE levels in the brain, concomitant with an increase in insoluble apoE (30–32). In contrast to AD mice lacking apoE (48), ABCA1-null mice showed increased amyloid burden. One potential explanation for this is that lipidation of apoE may be ABCA1-dependent (49). Decreased lipidation of apoE-containing lipoproteins has been proposed to underlie the decreased stability of apoE and its deposition in ABCA1-null mice. Despite the fact that the Apoe gene is a target for LXR (35), we observed no effect of LXR on Apoe gene expression in whole brain, consistent with previous work (12, 15). It is plausible, however, that LXR activation may alter the posttranslational stability of apoE by regulating its ABCA1-dependent lipidation. This idea is consistent with our observation that Apoe protein is decreased in LXR-null brains. Currently, we cannot exclude the possibility that LXRs may modulate the lipoprotein receptor-related protein (LRP)-dependent clearance of Aβ from the brain into the periphery (50). The preferential increase of Aβ1–40 deposition in mice lacking LXRβ may hint at this possibility. Further studies are also needed to address whether other metabolic targets of LXR, such as ABCG1 and apoD, may also impact AD-related pathology.
We have previously shown that LXR activation inhibits inflammatory responses (5, 7, 8). In the current work, we show that LXRs are also potent inhibitors of glial cell responses to inflammatory stimuli, including fAβ. The role of inflammation in AD remains a topic of intense debate (21). Most but not all studies suggest that inflammation is not a causative factor in AD but is secondary to amyloid deposition. It has been proposed that blocking inflammation with nonsteroidal antiinflammatory drugs (NSAIDS) may be beneficial in limiting AD progression (51, 52). Additional evidence for detrimental effects of inflammatory signaling in AD comes from experiments with genetically modified mice. Neuronal or astroglial overexpression of Cox2 (53) or Mcp1 (54), respectively, results in increased amyloid burden. Conversely, ablation of iNos (54), Cd40l (55), or Ep2 (54) decreases amyloid burden. We have shown here that activated LXRs are potent inhibitors of Cox2, Mcp1, and iNos expression in glial cells. LXRs also repress Cd40 (Tnfrsf5) and Slc7a11 expression in BV2 cells in response to LPS. Inhibition of the Cd40l-Cd40 axis would be predicted to reduce amyloid deposition and neurotoxicity (55, 56), and repression of Slc7a11 would be predicted to decrease efflux of neurotoxic glutamate (57).
One potential mechanism by which inhibition of neuroinflammation by LXRs may decrease amyloid burden is through modulation of microglial phagocytosis. Microglia normally act to clear fAβ through phagocytosis of amyloid fibrils and large Aβ aggregates (43). Exposure of microglia to fAβ induces a distinct form of phagocytosis mediated by a β1-integrin-dependent process (58), and this process is suppressed in the presence of inflammatory cytokines (22). In our study, LXR agonist treatment resulted in the restoration of fAβ-stimulated phagocytosis, most likely through inhibition of NFκB activity (5). On the basis of this result, activation of LXRs in the brain by endogenous ligands would be predicted to facilitate the clearance of fAβ in an inflammatory milieu. Conversely, loss of this beneficial effect on fAβ clearance may contribute to the elevated plaque burden in APP Tg mice in which the LXR genes have been inactivated.
Recent studies have begun to explore the potential utility of LXR agonists in the therapy of AD. Initial work focused primarily on the ability of LXRs to decrease amyloidogenic processing of APP (28, 59). More recently, Koldamova et al. (32) extended these in vitro observations to show that short-term treatment of 11-week-old APP23 mice with T0901317 resulted in a marked decrease in soluble levels of Aβ40 and Aβ42. Burns et al. (33) further demonstrated that T0901317 reduced the level of endogenous Aβ in WT mice. Here we have advanced these studies to show that endogenous LXR signaling impacts the development of AD-like pathology and attenuates the inflammatory response to deposited Aβ. The antiinflammatory facet of LXR signaling in the brain may be pertinent to other neurodegenerative diseases as well. For example, LXR agonist was recently shown to reduce the severity of experimental autoimmune encephalomyelitis in mice (60). Remarkably, this outcome was not due to an intrinsic effect on T cell activity but was associated with decreased migration of T cells into the CNS. This result is consistent with our observation that LXRs potently attenuate chemokine expression in glial cells.
In summary, we have shown that loss of endogenous LXR signaling in APP/PS1 Tg mice results in increased senile plaque accumulation. Our study also demonstrates that LXRs are potent inhibitors of glial cell response to inflammatory stimuli, including fAβ. The antiinflammatory action of LXRs may promote the ability of microglia to phagocytose fAβ in a chronic inflammatory environment. Cumulatively, these findings support further investigation of the LXR pathway as a potential target for the treatment of AD.
Materials and Methods
Reagents.
GW3695 was provided by Jon Collins (GlaxoSmithKline, Research Triangle Park, NC). 24-hydroxycholesterol was from the American Radiochemical Company (St. Louis, MO). LPS was from Sigma (St. Louis, MO), and cytokines were from PeproTech (Rocky Hill, NJ). Amyloid peptides (Bachem, Torrence, CA) were dissolved in sterile water at a concentration of 1 mM and stored at −80°C. To induce fibril formation, aliquots were incubated at 37°C for 7 days. We cannot rule out the presence of oligomers in the Aβ preparations. All other reagents were from Sigma.
Animals.
Lxrα−/−, Lxrβ−/−, and Lxrαβ−/− mice (Sv129 and C57BL/6 mixed background) (61) were mated with Tg APPswe/PS1ΔE9 mice (36) (Jackson Laboratory, Bar Harbor, ME). Animals were maintained as hemizygotes for the transgene and were genotyped by PCR. At 32 weeks of age, littermates were killed, the left hemibrain was fixed in 4% parafomaldehyde, and the right hemibrain was snap frozen and stored at −80°C. In some experiments, mice were gavaged with vehicle, GW3695 (20 mg/kg), or T0901317 (50 mg/kg) once daily for 3 days. All studies were conducted in accordance with institutional guidelines for animal care and use.
Cell Culture and Phagocytosis Assays.
BV2 cells (62) were maintained in DMEM (Gibco, Carlsbad, CA) containing 10% FBS. For gene expression studies cells were pretreated with 1 μM GW3695 or DMSO for 16 h in 0.5% FBS and then treated for 6 h with 100 ng/ml LPS. NO production was determined as described in ref. 5. In some experiments, cells were sterol-depleted by treatment with 5 μM simvastatin and 100 μM mevalonic acid.
Mixed primary glial cells were isolated as described in ref. 63. For treatment with Aβ peptides, cells were washed extensively and incubated in DMEM/F12 containing no serum and 1 μM GW3695 or DMSO for 18 h. Subsequently, Aβ peptides were briefly sonicated and added to the cultures for an additional 24 h. Primary microglia were isolated following the procedure of Saura et al. (64).
Phagocytosis assays were performed as described in ref. 22. In each experiment, 100 cells per well per condition were counted from two independent wells.
RNA Analysis and DNA Microarray Analysis.
Total RNA was isolated using TRIzol (Life Technologies, Grand Island, NY) and was reverse-transcribed into cDNA by using a reverse transcription kit (Roche, Indianapolis, IN). Real-time quantitative PCR assays were performed using TaqMan or SYBRgreen-based chemistry. Samples were analyzed simultaneously for 36B4 expression as the internal control, and quantitative expression values were extrapolated from separate standard curves. Each sample was assayed in duplicate and normalized to 36B4. Sequences for qPCR primers are available upon request. For cDNA microarray analysis, BV2 cells were treated as indicated above with either DMSO or 1 μM GW3695 in the presence or absence of 100 ng/ml LPS. For each condition, six independent samples were processed. Transcriptional profiling was performed at the University of California, Los Angeles, microarray core facility by using Affymetrix (Santa Clara, CA) 430 2.0 microarrays. Duplicate arrays were hybridized using pooled RNA (three samples per array). Data were analyzed using GeneSpring software (Stratagene, La Jolla, CA).
Immunohistochemistry.
Immunohistochemistry was performed on 5-μm paraffin sections. Antigen retrieval was optimized using an 88% formic acid solution for Aβ-stained slides. Slides were washed in PBS and covered in 5% normal goat serum (Vector Laboratories, Burlingame, CA) diluted in PBS to block nonspecific binding. Slides were then incubated at 4°C overnight with antibodies to Aβ1–40 and Aβ1–42 (Chemicon, Temecula, CA). Sections were washed in PBS and incubated at room temperature with biotinylated goat anti-rabbit Ig, followed by avidin–biotin–peroxidase complex and 3,3- diaminobenzidine tetrahydrochloride as the chromagen. Aβ plaques were quantified manually by two blinded individuals.
Immunoblotting of whole-brain homogenates or BV2 cell lysates was done following standard procedures. The following primary antibodies were used: anti-Abca1, NB 400-105 (Novus Biologicals, Littleton, CO), Anti-APP, 6E10 (Signet, Dedham, MA), anti-Cox2, BD-610203 (BD Biosciences, Franklin Lakes, NJ), anti-iNos, SC-7271 (Santa Cruz Biotechnology, Santa Cruz, CA), and anti-Actin (Sigma).
Statistical Analysis.
Differences between groups were evaluated with Student's t test.
Supplementary Material
Acknowledgments
N.Z. is supported by a long-term Human Frontier Science Program Postdoctoral Fellowship. H.V.V. is supported in part by National Institute on Aging Grant P50 AG16570 and the Daljit S. and Elaine Sarkaria Chair in Diagnostic Medicine. P.T. is an Investigator of the Howard Hughes Medical Institute. This work was supported by National Institutes of Health Grants HL30568 and HL66088.
Abbreviations
- ABCA1
ATP-binding cassette A1
- Aβ
amyloid β peptide
- AD
Alzheimer disease
- apoN
apolipoprotein N
- APP
amyloid precursor protein
- fAβ
fibrillar Aβ
- KO
knockout
- LPS
lipopolysaccharide(s)
- LXR
liver x receptor
- Tg
transgenic.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0701096104/DC1.
References
- 1.Tontonoz P, Mangelsdorf DJ. Mol Endocrinol. 2003;17:985–993. doi: 10.1210/me.2003-0061. [DOI] [PubMed] [Google Scholar]
- 2.Zelcer N, Tontonoz P. J Clin Invest. 2006;116:607–614. doi: 10.1172/JCI27883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Janowski BA, Willy PJ, Devi TR, Falck JR, Mangelsdorf DJ. Nature. 1996;383:728–731. doi: 10.1038/383728a0. [DOI] [PubMed] [Google Scholar]
- 4.Repa JJ, Mangelsdorf DJ. Annu Rev Cell Dev Biol. 2000;16:459–481. doi: 10.1146/annurev.cellbio.16.1.459. [DOI] [PubMed] [Google Scholar]
- 5.Joseph SB, Castrillo A, Laffitte BA, Mangelsdorf DJ, Tontonoz P. Nat Med. 2003;9:213–219. doi: 10.1038/nm820. [DOI] [PubMed] [Google Scholar]
- 6.Ogawa S, Lozach J, Benner C, Pascual G, Tangirala RK, Westin S, Hoffmann A, Subramaniam S, David M, Rosenfeld MG, Glass CK. Cell. 2005;122:707–721. doi: 10.1016/j.cell.2005.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fowler AJ, Sheu MY, Schmuth M, Kao J, Fluhr JW, Rhein L, Collins JL, Willson TM, Mangelsdorf DJ, Elias PM, Feingold KR. J Invest Dermatol. 2003;120:246–255. doi: 10.1046/j.1523-1747.2003.12033.x. [DOI] [PubMed] [Google Scholar]
- 8.Ogawa D, Stone JF, Takata Y, Blaschke F, Chu VH, Towler DA, Law RE, Hsueh WA, Bruemmer D. Circ Res. 2005;96:e59–e67. doi: 10.1161/01.RES.0000163630.86796.17. [DOI] [PubMed] [Google Scholar]
- 9.Terasaka N, Hiroshima A, Ariga A, Honzumi S, Koieyama T, Inaba T, Fujiwara T. FEBS J. 2005;272:1546–1556. doi: 10.1111/j.1742-4658.2005.04599.x. [DOI] [PubMed] [Google Scholar]
- 10.Valledor AF, Hsu LC, Ogawa S, Sawka-Verhelle D, Karin M, Glass CK. Proc Natl Acad Sci USA. 2004;101:17813–17818. doi: 10.1073/pnas.0407749101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joseph SB, Bradley MN, Castrillo A, Bruhn KW, Mak PA, Pei L, Hogenesch J, O'Connell RM, Cheng G, Saez E, Miller JF, Tontonoz P. Cell. 2004;119:299–309. doi: 10.1016/j.cell.2004.09.032. [DOI] [PubMed] [Google Scholar]
- 12.Whitney KD, Watson MA, Collins JL, Benson WG, Stone TM, Numerick MJ, Tippin TK, Wilson JG, Winegar DA, Kliewer SA. Mol Endocrinol. 2002;16:1378–1385. doi: 10.1210/mend.16.6.0835. [DOI] [PubMed] [Google Scholar]
- 13.Abildayeva K, Jansen PJ, Hirsch-Reinshagen V, Bloks VW, Bakker AH, Ramaekers FC, de Vente J, Groen AK, Wellington CL, Kuipers F, Mulder M. J Biol Chem. 2006;281:12799–12808. doi: 10.1074/jbc.M601019200. [DOI] [PubMed] [Google Scholar]
- 14.Koldamova RP, Lefterov IM, Ikonomovic MD, Skoko J, Lefterov PI, Isanski BA, DeKosky ST, Lazo JS. J Biol Chem. 2003;278:13244–13256. doi: 10.1074/jbc.M300044200. [DOI] [PubMed] [Google Scholar]
- 15.Wang L, Schuster GU, Hultenby K, Zhang Q, Andersson S, Gustafsson JA. Proc Natl Acad Sci USA. 2002;99:13878–13883. doi: 10.1073/pnas.172510899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andersson S, Gustafsson N, Warner M, Gustafsson JA. Proc Natl Acad Sci USA. 2005;102:3857–3862. doi: 10.1073/pnas.0500634102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glenner GG, Wong CW. Biochem Biophys Res Commun. 1984;120:885–890. doi: 10.1016/s0006-291x(84)80190-4. [DOI] [PubMed] [Google Scholar]
- 18.Grundke-Iqbal I, Iqbal K, Tung YC, Quinlan M, Wisniewski HM, Binder LI. Proc Natl Acad Sci USA. 1986;83:4913–4917. doi: 10.1073/pnas.83.13.4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hardy J, Selkoe DJ. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 20.Akiyama H, Barger S, Barnum S, Bradt B, Bauer J, Cole GM, Cooper NR, Eikelenboom P, Emmerling M, Fiebich BL, et al. Neurobiol Aging. 2000;21:383–421. doi: 10.1016/s0197-4580(00)00124-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wyss-Coray T. Nat Med. 2006;12:1005–1015. doi: 10.1038/nm1484. [DOI] [PubMed] [Google Scholar]
- 22.Koenigsknecht-Talboo J, Landreth GE. J Neurosci. 2005;25:8240–8249. doi: 10.1523/JNEUROSCI.1808-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wolozin B. Neuron. 2004;41:7–10. doi: 10.1016/s0896-6273(03)00840-7. [DOI] [PubMed] [Google Scholar]
- 24.Mahley RW, Rall SC., Jr Annu Rev Genomics Hum Genet. 2000;1:507–537. doi: 10.1146/annurev.genom.1.1.507. [DOI] [PubMed] [Google Scholar]
- 25.Mace S, Cousin E, Ricard S, Genin E, Spanakis E, Lafargue-Soubigou C, Genin B, Fournel R, Roche S, Haussy G, et al. Neurobiol Dis. 2005;18:119–125. doi: 10.1016/j.nbd.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 26.Katzov H, Chalmers K, Palmgren J, Andreasen N, Johansson B, Cairns NJ, Gatz M, Wilcock GK, Love S, Pedersen NL, et al. Hum Mutat. 2004;23:358–367. doi: 10.1002/humu.20012. [DOI] [PubMed] [Google Scholar]
- 27.Kolsch H, Lutjohann D, Ludwig M, Schulte A, Ptok U, Jessen F, von Bergmann K, Rao ML, Maier W, Heun R. Mol Psychiatry. 2002;7:899–902. doi: 10.1038/sj.mp.4001109. [DOI] [PubMed] [Google Scholar]
- 28.Sun Y, Yao J, Kim TW, Tall AR. J Biol Chem. 2003;278:27688–27694. doi: 10.1074/jbc.M300760200. [DOI] [PubMed] [Google Scholar]
- 29.Fukumoto H, Deng A, Irizarry MC, Fitzgerald ML, Rebeck GW. J Biol Chem. 2002;277:48508–48513. doi: 10.1074/jbc.M209085200. [DOI] [PubMed] [Google Scholar]
- 30.Hirsch-Reinshagen V, Maia LF, Burgess BL, Blain JF, Naus KE, McIsaac SA, Parkinson PF, Chan JY, Tansley GH, Hayden MR, et al. J Biol Chem. 2005;280:43243–43256. doi: 10.1074/jbc.M508781200. [DOI] [PubMed] [Google Scholar]
- 31.Wahrle SE, Jiang H, Parsadanian M, Hartman RE, Bales KR, Paul SM, Holtzman DM. J Biol Chem. 2005;280:43236–43242. doi: 10.1074/jbc.M508780200. [DOI] [PubMed] [Google Scholar]
- 32.Koldamova R, Staufenbiel M, Lefterov I. J Biol Chem. 2005;280:43224–43235. doi: 10.1074/jbc.M504513200. [DOI] [PubMed] [Google Scholar]
- 33.Burns MP, Vardanian L, Pajoohesh-Ganji A, Wang L, Cooper M, Harris DC, Duff K, Rebeck GW. J Neurochem. 2006;98:792–800. doi: 10.1111/j.1471-4159.2006.03925.x. [DOI] [PubMed] [Google Scholar]
- 34.Collins JL. Curr Opin Drug Discov Devel. 2004;7:692–702. [PubMed] [Google Scholar]
- 35.Laffitte BA, Repa JJ, Joseph SB, Wilpitz DC, Kast HR, Mangelsdorf DJ, Tontonoz P. Proc Natl Acad Sci USA. 2001;98:507–512. doi: 10.1073/pnas.021488798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jankowsky JL, Slunt HH, Ratovitski T, Jenkins NA, Copeland NG, Borchelt DR. Biomol Eng. 2001;17:157–165. doi: 10.1016/s1389-0344(01)00067-3. [DOI] [PubMed] [Google Scholar]
- 37.Garcia-Alloza M, Robbins EM, Zhang-Nunes SX, Purcell SM, Betensky RA, Raju S, Prada C, Greenberg SM, Bacskai BJ, Frosch MP. Neurobiol Dis. 2006;24:516–524. doi: 10.1016/j.nbd.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 38.Kim OS, Lee CS, Joe EH, Jou I. Biochem Biophys Res Commun. 2006;342:9–18. doi: 10.1016/j.bbrc.2006.01.107. [DOI] [PubMed] [Google Scholar]
- 39.Marijanovic Z, Laubner D, Moller G, Gege C, Husen B, Adamski J, Breitling R. Mol Endocrinol. 2003;17:1715–1725. doi: 10.1210/me.2002-0436. [DOI] [PubMed] [Google Scholar]
- 40.Soccio RE, Adams RM, Romanowski MJ, Sehayek E, Burley SK, Breslow JL. Proc Natl Acad Sci USA. 2002;99:6943–6948. doi: 10.1073/pnas.052143799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ohnesorg T, Adamski J. Mol Cell Endocrinol. 2006;248:164–167. doi: 10.1016/j.mce.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 42.Soccio RE, Adams RM, Maxwell KN, Breslow JL. J Biol Chem. 2005;280:19410–19418. doi: 10.1074/jbc.M501778200. [DOI] [PubMed] [Google Scholar]
- 43.Rogers J, Strohmeyer R, Kovelowski CJ, Li R. Glia. 2002;40:260–269. doi: 10.1002/glia.10153. [DOI] [PubMed] [Google Scholar]
- 44.Repa JJ, Turley SD, Lobaccaro JA, Medina J, Li L, Lustig K, Shan B, Heyman RA, Dietschy JM, Mangelsdorf DJ. Science. 2000;289:1524–1529. doi: 10.1126/science.289.5484.1524. [DOI] [PubMed] [Google Scholar]
- 45.Tangirala RK, Bischoff ED, Joseph SB, Wagner BL, Walczak R, Laffitte BA, Daige CL, Thomas D, Heyman RA, Mangelsdorf DJ, et al. Proc Natl Acad Sci USA. 2002;99:11896–11901. doi: 10.1073/pnas.182199799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schuster GU, Parini P, Wang L, Alberti S, Steffensen KR, Hansson GK, Angelin B, Gustafsson JA. Circulation. 2002;106:1147–1153. doi: 10.1161/01.cir.0000026802.79202.96. [DOI] [PubMed] [Google Scholar]
- 47.Wagner BL, Valledor AF, Shao G, Daige CL, Bischoff ED, Petrowski M, Jepsen K, Baek SH, Heyman RA, Rosenfeld MG, et al. Mol Cell Biol. 2003;23:5780–5789. doi: 10.1128/MCB.23.16.5780-5789.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bales KR, Verina T, Dodel RC, Du Y, Altstiel L, Bender M, Hyslop P, Johnstone EM, Little SP, Cummins DJ, et al. Nat Genet. 1997;17:263–264. doi: 10.1038/ng1197-263. [DOI] [PubMed] [Google Scholar]
- 49.Wahrle SE, Jiang H, Parsadanian M, Legleiter J, Han X, Fryer JD, Kowalewski T, Holtzman DM. J Biol Chem. 2004;279:40987–40993. doi: 10.1074/jbc.M407963200. [DOI] [PubMed] [Google Scholar]
- 50.Zlokovic BV. J Neurochem. 2004;89:807–811. doi: 10.1111/j.1471-4159.2004.02385.x. [DOI] [PubMed] [Google Scholar]
- 51.Morgan D, Gordon MN, Tan J, Wilcock D, Rojiani AM. J Neuropathol Exp Neurol. 2005;64:743–753. doi: 10.1097/01.jnen.0000178444.33972.e0. [DOI] [PubMed] [Google Scholar]
- 52.Hoozemans JJ, O'Banion MK. Curr Drug Targets CNS Neurol Disord. 2005;4:307–315. doi: 10.2174/1568007054038201. [DOI] [PubMed] [Google Scholar]
- 53.Xiang Z, Ho L, Yemul S, Zhao Z, Qing W, Pompl P, Kelley K, Dang A, Qing W, Teplow D, Pasinetti GM. Gene Expr. 2002;10:271–278. doi: 10.3727/000000002783992352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yamamoto M, Horiba M, Buescher JL, Huang D, Gendelman HE, Ransohoff RM, Ikezu T. Am J Pathol. 2005;166:1475–1485. doi: 10.1016/s0002-9440(10)62364-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tan J, Town T, Crawford F, Mori T, DelleDonne A, Crescentini R, Obregon D, Flavell RA, Mullan MJ. Nat Neurosci. 2002;5:1288–1293. doi: 10.1038/nn968. [DOI] [PubMed] [Google Scholar]
- 56.Tan J, Town T, Paris D, Mori T, Suo Z, Crawford F, Mattson MP, Flavell RA, Mullan M. Science. 1999;286:2352–2355. doi: 10.1126/science.286.5448.2352. [DOI] [PubMed] [Google Scholar]
- 57.Qin S, Colin C, Hinners I, Gervais A, Cheret C, Mallat M. J Neurosci. 2006;26:3345–3356. doi: 10.1523/JNEUROSCI.5186-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koenigsknecht J, Landreth G. J Neurosci. 2004;24:9838–9846. doi: 10.1523/JNEUROSCI.2557-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Koldamova RP, Lefterov IM, Staufenbiel M, Wolfe D, Huang S, Glorioso JC, Walter M, Roth MG, Lazo JS. J Biol Chem. 2005;280:4079–4088. doi: 10.1074/jbc.M411420200. [DOI] [PubMed] [Google Scholar]
- 60.Hindinger C, Hinton DR, Kirwin SJ, Atkinson RD, Burnett ME, Bergmann CC, Stohlman SA. J Neurosci Res. 2006;84:1225–1234. doi: 10.1002/jnr.21038. [DOI] [PubMed] [Google Scholar]
- 61.Peet DJ, Turley SD, Ma W, Janowski BA, Lobaccaro JM, Hammer RE, Mangelsdorf DJ. Cell. 1998;93:693–704. doi: 10.1016/s0092-8674(00)81432-4. [DOI] [PubMed] [Google Scholar]
- 62.Blasi E, Barluzzi R, Bocchini V, Mazzolla R, Bistoni F. J Neuroimmunol. 1990;27:229–237. doi: 10.1016/0165-5728(90)90073-v. [DOI] [PubMed] [Google Scholar]
- 63.McDonald DR, Brunden KR, Landreth GE. J Neurosci. 1997;17:2284–2294. doi: 10.1523/JNEUROSCI.17-07-02284.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Saura J, Tusell JM, Serratosa J. Glia. 2003;44:183–189. doi: 10.1002/glia.10274. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.