Abstract
Chromosomes are compacted hundreds of times to fit in the cell, packaged into dynamic folds whose structures are largely unknown. Here, we examine patterns in gene locations to infer large-scale features of bacterial chromosomes. Specifically, we analyzed >100 genomes and identified thousands of gene pairs that display two types of evolutionary correlations: a tendency to co-occur and a tendency to be located close together in many genomes. We then analyzed the detailed distribution of these pairs in Escherichia coli and found that genes in a pair tend to be separated by integral multiples of 117 kb along the genome and to be positioned in a 117-kb grid of genomic locations. In addition, the most pair-dense locations coincide with regions of intense transcriptional activity and the positions of top transcribed and conserved genes. These patterns suggest that the E. coli chromosome may be organized into a 117-kb helix-like topology that localizes a subset of the most essential and highly transcribed genes along a specific face of this structure. Our approach indicates an evolutionarily maintained preference in the spacing of genes along the chromosome and offers a general comparative genomics framework for studying chromosome structure, broadly applicable to other organisms.
Keywords: chromosome structure, computational genomics, nucleoid, spatial organization
The interplay between structure and function of chromosomes is a critical aspect of spatial organization in the cell, intimately involved in transcription (1–3), recombination (4), and replication (1, 2). Despite this importance, the detailed structure of a chromosome in any organism is unknown. Bacteria offer appealing systems in which to study the fundamental factors governing chromosome structure, because they exhibit exquisite spatial organization and contain functional homologs of many eukaryotic DNA-associated proteins, yet they have a small genome generally packaged in a single circular chromosome (2, 5, 6).
Bacterial chromosomes must be compacted 1,000-fold to fit within the cell. The resulting structures could therefore be highly disordered; for example, 10 kb of uncompacted DNA (1/400th of the genome) could span the entire cell. However, in vivo the chromosome exhibits a high degree of order. At a local level, it is wound into ≈10-kb supercoiled domains that topologically isolate different regions of the genome from each other (7, 8). At larger scales, certain regions of the genome are physically inaccessible to each other, suggesting that loci undergo limited diffusion (9). More recently, fluorescence microscopy has shown that loci are not randomly positioned in the cell but occupy reproducible 3D positions that undergo specific cell-cycle movements (10–14). In Escherichia coli and Caulobacter crescentus, evidence suggests furthermore that the positions of loci in the cell are linearly correlated with their coordinate along the genome, with the origin and terminus at opposite cell poles (11, 12). In E. coli, recent data confirm this linear correlation but suggest the origin is located at midcell, with the two arcs of the chromosome in two different (longitudinal) halves of the cell (13, 14). At a finer scale, below the resolution of current confocal microscopy, positional correlations and periodicities in sequence (15), expression levels (16–19), and transcription factor-binding sites (17) suggest a functionally important, possibly regular chromosome conformation. However, beyond coarse high-level outlines, the structure has been largely inaccessible to experiment and remains largely unknown.
Here, we approach the problem of chromosome structure from an evolutionary perspective. Our method, based on comparative genomics, is similar to statistical coupling analysis in proteins (20). In proteins, the 3D arrangement of specific residues is critical for function; for example, the WW protein domain contains a small 3D network of residues that is crucial for both folding and function (20). Maintaining this arrangement constrains the identities of the amino acids at the involved residues and causes them to coevolve, generating statistical correlations in a multiple sequence alignment (20).
In the chromosome, we reasoned analogously that if a particular 3D arrangement of genes is critical for function, such genes would tend to occupy genomic locations where they can achieve this arrangement in the folded chromosome (Fig. 1); for example, a regulatory region may tend to occupy positions where it can be folded close to the gene it regulates, as in the β-globin locus control region (21). We reasoned that the coevolution of genes to locations compatible with such 3D arrangements would create statistical correlations in gene locations, analogous to the observed correlations in protein residues. Given the dynamical and fluid nature of chromosome organization, we expected such constraints to be less rigid than those found in protein residues, yet potentially significant enough to create correlations detectable in a multiple genome comparison.
Fig. 1.
We identify SC pairs of genes based on correlations in their positions (tendency to be close together in many genomes) and phylogenetic co-occurrence over many genomes (22, 38). (a) Example of identification of such a pair (red and blue rectangles). The pair tends to occur at short distance along the chromosome (genomes S1, S2, and S4) and also with high phylogenetic co-occurrence [both genes either present (genome S1, S2, S4, S5) or absent (genome S3)]. Note that, despite this tendency, the same genes may be far apart on a specific genome (S5). (b) This process yields a large set of SC pairs, selected based on the statistical significance of their correlations. (c) Statistical patterns in the distributions of the distances between the paired genes and of their positions along a specific genome (here a significant distance preference D, as seen in S5) may reflect substructures of the chromosome fold, for example regions folded into spatial proximity. (d) The contour length of the loop (which may have a complex internal structure) reflects the preferred distance D.
In our analysis, we explore this hypothesis by analyzing a large set of statistically correlated (SC) gene pairs. We first select a set of correlated pairs. We then investigate patterns in the position and distance distributions of these pairs along the genome of E. coli. We next explore the functional basis of these distributions by analyzing the levels of conservation and transcription along the genome. Finally, we discuss possible implications of these distributions in terms of geometrical features of the E. coli chromosome fold.
Results
To begin, we identified a large set of strongly correlated gene pairs. Specifically (Fig. 1a), we searched across many genomes for pairs of genes and their orthologs (which we will simply refer to as genes) that exhibit two specific types of correlations: a tendency to be located close together in many genomes, and phylogenetic co-occurrence (22) (where one gene tends to be present in a particular genome only if the other gene is also present). Genes in such pairs are known to often share function and transcriptional regulation and have gene products that physically associate (22, 23). Consequently, we reasoned, the 3D chromosomal arrangement of these pairs may be important for function.
Based on these criteria, we searched across 10 million gene pairs in >100 genomes and selected 22,500 strongly SC pairs [supporting information (SI) Methods and SI Table 1; Fig. 1b]. In any given genome, genes belonging to these SC pairs will occupy a specific set of genomic locations: for many SC pairs, the two genes will be located close together along the genome. However, in this same genome, the genes in other SC pairs may be far apart (Fig. 1a). In addition, many of the genes may be concentrated in particular regions of the genome. We reasoned that if the correlated pairs are constrained by chromosome structure, their distributions along a given genome (Fig. 1c) might reveal structural features of the chromosome fold, for example, regions that are folded into spatial proximity (Fig. 1d) or are constrained to particular subspaces of the nucleoid or cell.
We first investigated properties of the SC pairs across all organisms and found that the genes in a pair exhibit a strong preference for positions that are symmetric about the origin of replication (SI Fig. 6). This symmetry is consistent with fluorescence microscopy in C. crescentus (10–12) and with observations of symmetry in genome alignments and gene order (24–26).
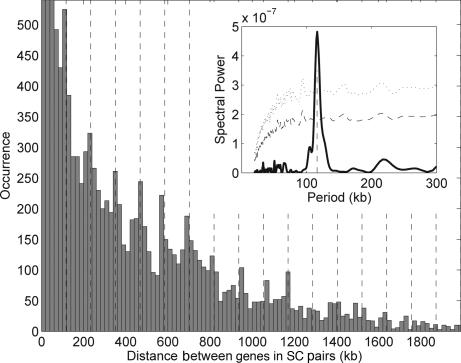
We next examined the detailed genomic organization of the SC pairs in a single organism, E. coli. First, we analyzed the distribution of distances, i.e., the number of times a particular distance separates genes in an SC pair along each chromosome arc (defined by the origin and terminus of replication; see SI Fig. 6a). Because the SC genes were chosen based on their tendency for closeness in bacterial genomes, the expectation (under a null hypothesis of otherwise randomly positioned genes) is that most distances will be close to zero, and that distances larger than zero will taper smoothly to zero (see SI Fig. 7a). In E. coli, however, we observe a markedly different pattern (Fig. 2). Distances near zero are indeed overrepresented, but there is also a series of significant peaks at 117, 234, 351 kb, and n × 117 kb (n integer), out to 1.4 Mb. Using a Fourier transform, we confirmed a strong and highly significant periodicity (P < 0.002; see Fig. 2 Inset and Methods). The SC pairs are therefore not randomly spaced along the genome but prefer specific genomic intervals of n × 117 kb. A similar periodicity is observed in the distances between the SC genes located on different arcs of the chromosome (SI Fig. 7b).
Fig. 2.
Distributions of distances between SC paired genes in the E. coli genome. The distribution displays periodic peaks, indicating a preferred chromosomal distance (≈117 kb) between genes belonging to an SC pair. A 117-kb spaced grid is overlapped with the histogram. The first three peaks, cut for visualization purposes, reach the values of 628, 649, and 645, respectively. (Inset) Discrete Fourier transform analysis of the distribution, displaying a major 117-kb peak (dashed vertical line). Statistical significance lines are plotted at 3σ (dashed line) and 5σ (dotted line) from the mean of 10,000 randomizations.
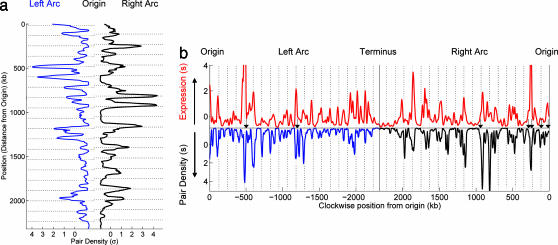
Many different distributions of locations for the SC genes in E. coli could generate the above distance distribution (Fig. 2). For example, the SC pairs could be distributed uniformly along the genome. Alternatively, they could be located in a small number of clusters separated by intervals of n × 117 kb. To assess the distribution of locations, we calculated a position-dependent pair density, the number of SC pairs involving genes located in a particular chromosomal window (Fig. 3a). We found that the SC genes are located in a series of sharp peaks that are spread across the entire chromosome. As shown in Fig. 3a, the major peaks fit closely to grid lines of n × 117 kb (P = 0.012; see SI Fig. 8) and maintain phase along the length of an entire chromosome arc (for example, in Fig. 3a, the last major peak on the right arc is n = 15 periods from the first peak). Thus the SC paired genes tend to be localized in a specific set of regularly spaced islands along each half of the genome.
Fig. 3.
Distributions of pair density along chromosomal arcs, and correlation with expression. (a) Density of the SC paired genes along the chromosome. The distance from the central axis reflects the number of times a gene at this position is in an SC pair with another gene, i.e., the total number of pairs involving this position. The distributions for the right arc (black) and for the left arc (blue) are placed facing each other to emphasize the symmetry. Prominent peaks occur preferentially in phase with the 117-kb spaced grid (horizontal lines, P = 0.012; see SI Fig. 8). (b) Comparison of transcription level with pair density along the chromosome. Absolute expression level (red, on top) during log phase growth is plotted as a function of chromosomal position. Expression is compared with pair density (bottom distribution) along the left (blue) and right (black) arcs of the chromosome. Distance above the horizontal indicates increasing expression and below the horizontal indicates increasing pair density (in units of σ). Many of the highest peaks in the transcription profile fall near the 117-kb grid lines defined by the SC pairs. In addition, the locations of six of the seven rRNA operons (black arrows) fit this same grid (P = 0.054, calculated as for the SC pair density grid fit, by using 1,000 randomization of the rRNA operon positions). Note that the rRNA positions were not included in the pair selection because of their multiplicity in many genomes and were not included in the transcription profile because of their high transcript levels.
To explore the functional basis of these distributions, we examined the relationship between SC genes and transcription in E. coli. Transcription has long been hypothesized to play a role in condensing the chromosome (3). In addition, many of the genes that belong to a large number of SC pairs are known to be highly transcribed. We found that the pair density mirrors the log-phase transcript level (16) along the chromosome (Pearson correlation coefficient R = 0.67, P < 10−44) (Fig. 3b), and this correlation decreases steadily from log phase into stationary phase (SI Fig. 9), suggesting that the pair density reflects a component of transcription specific to log-phase growth. We also found that many of the major peaks in the log-phase transcription profile fall along the 117-kb grid defined by the SC pairs. Furthermore, we found that six of the seven rRNA operons (the most highly transcribed regions of the genome), although not included in either the pair density or transcription profiles (see Fig. 3b legend), fit the same 117-kb grid (P = 0.054; black arrows in Fig. 3b). Together, these observations indicate that the SC pair distributions are intimately linked with transcription.
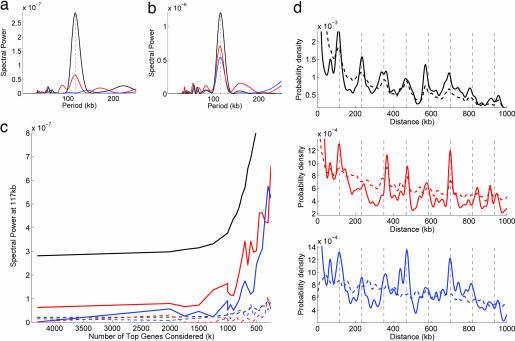
We next sought to understand this link in more detail, in particular the relationship between the periodicities in the SC pairs and the positioning of highly transcribed genes in E. coli; in addition, because the SC pairs were chosen by using orthologs conserved over many genomes, we simultaneously examined the connection between SC pairs and the positioning of highly conserved genes. We therefore constructed two new pair sets in which the gene pairs were selected randomly from E. coli by using probabilities proportional to their level of transcription (transcription pairs) or conservation (conservation pairs) (see Methods). The distance distributions of these new pair sets are therefore enriched in distances that separate highly transcribed or conserved genes along the chromosome, allowing us to examine preferences in the chromosomal spacing of these genes in a manner similar to the SC pairs. We first compared the distance distributions of these two new pair sets with the SC pairs. In contrast to the SC pairs, we found no periodicity in conservation pairs (Fig. 4 a and d) and a weak 117-kb periodicity in transcription pairs [Fig. 4 a and d, consistent with previous observations of 115 kb in transcription (18, 19)].
Fig. 4.
Comparison of periodicity in the distance distribution of SC paired genes with periodicities in the distance distributions of transcription and conservation pairs in E. coli (see Methods). (a) Fourier spectra of the distance distribution of the three pair sets when all genes in E. coli are considered. The SC pairs (black) display a strong periodicity peak at 117 kb, whereas the transcription pairs (red) show a weak peak at 117 kb and the conservation pairs (blue) do not show a peak. (b) The Fourier spectra for pair sets constructed from the top 250 SC, conserved, or transcribed genes. These sets each show a strong peak at 117 kb with SC pairs (black), the strongest followed by transcription (red) and conservation (blue). (c) Strength of the 117-kb periodicity peak in the Fourier transform (y axis, solid lines) as each of the pair sets is restricted to the top k genes (x axis) in that set. Note that as k decreases, the periodicity at 117 kb grows stronger in all three sets (with SC pairs always strongest), indicating that the distances between the top genes in each set tend to be spaced at n × 117 kb along the chromosome. The dotted lines are a control indicating the strength of the Fourier spectrum at 100 kb; this does not show the same sharp rise as k decreases. (d) Distance distributions for each of the three pair sets with all genes (dotted line with corresponding Fourier spectrum shown in a) and k = 250 top genes (solid line with Fourier spectrum shown in b). Grid lines at n × 117 kb are shown for reference. At k = 250, the 117-kb periodicity can be clearly seen in each set (peaks along grid lines), whereas for all genes, only the SC pairs show a clear periodicity.
However, as we gradually restrict the pair sets to the top genes in each set (i.e., the genes with the highest levels of transcription, conservation, or number of SC pairings), a 117-kb periodicity steadily emerges in both transcription and conservation and grows stronger in all three pair sets (Fig. 4c). In addition, the locations of the top transcribed and top conserved genes fit the 117-kb grid defined by the SC pairs (SI Fig. 10). Thus, all three metrics converge to similar distance and position distributions but only for the most highly conserved and transcribed genes, and the periodicity in the SC pairs is two to five times as strong (Fig. 4). Finally, we examined the interdependence of these three sets by gradually excluding from each set the top genes from other sets. We found that the periodicities in all sets depend strongly on the presence of the top SC paired genes, whereas the periodicity in the SC pairs is largely independent of the top transcribed or conserved genes (SI Fig. 11).
Discussion
The SC gene pairs, which were chosen based only on their evolutionary patterns of chromosomal proximity and co-occurrence across many species, therefore display a strong 117-kb periodicity in genomic distances and locations in E. coli. In addition, the density of SC pairs is highly correlated with transcription levels. Could simple constraints on the sizes or sites of genome rearrangements generate these patterns? General models based on repulsion of gene clusters or preferred sizes for recombination or horizontal gene transfer could account for local spacing along the chromosome; for example, a strong preferred recombination distance of ≈117 kb could generate several gene clusters spaced at 117 kb by splitting a single initial gene cluster by 117-kb recombination events. However, the clusters generated from splitting two different initial clusters would not naturally be in phase with each other. In general, such local constraints cannot easily explain a global periodicity of positions that extends in almost perfect phase along each half of the genome. Even an extreme case of recombination hotspots spaced at n × 117 kb along the chromosome could maintain the 117-kb periodicity only if the SC paired genes were constrained to the very center or edges of each 117-kb stretch. Otherwise, a single inversion would destroy the periodicity. In addition, any horizontal gene transfer would disrupt the periodicity unless the fragment were small (≪117 kb) or ≈117 kb long with SC genes at the center or edges. We cannot rule out the possibility that such rearrangement processes contribute to the observed patterns, e.g., symmetric inversions about the origin of replication (24–26) could explain the observed symmetry of SC paired genes. However, the localization of SC genes in an in-phase set of periodically spaced islands suggests that some selective pressure beyond these processes maintains these genes at these specific locations.
Structural constraints due to the spatial organization of the chromosome offer a simple explanation. In-phase positional periodicities in amino acid sequences are a canonical structural motif seen in proteins, where they indicate the presence of a specific face on a periodic structure (27), for example, the face of hydrophobic residues in contact with the membrane in a transmembrane domain (27). Because of the regular period, these spatially contiguous structural faces are composed of residues that are separated by periodic intervals along the sequence. In the E. coli chromosome, the periodic distributions suggest an analogous structural organization, a regular 117-kb looping, and a single structural face of each chromosome half, along which SC pairs are predominantly localized.
In Fig. 5, we depict three conformations consistent with these constraints. Note that these conformations are simplified backbones rather than exact structures; for example, the coiled backbone in each panel represents 21-fold compacted DNA, which may consist of more complicated substructure (SI Fig. 12b), e.g., of 10–12 (possibly irregular and stochastic) topological domains (7, 8). The basic feature of each configuration is a regular 117 kb coiling along the backbone of the circular chromosome, which creates regions of high pair density along a face or faces of the structure. This 117- kb looped circular chromosome could be arranged in multiple ways within the cell. It could be flattened with the origin and terminus positioned at opposite cell poles (Fig. 5 a and b), compatible with previous observations in C. crescentus (12) and E. coli (11). Alternatively, as suggested by recent experimental data in E. coli (13, 14), the origin could be positioned at midcell, placing the right and left arcs in different cell halves (Fig. 5c). Other arrangements are also possible, including longer-range periodicities (SI Figs. 12a and 13). In addition, the structure could undergo dynamical transitions (Fig. 5d), e.g., between a longitudinally symmetric configuration (Fig. 5 a or b) and a transverse symmetric one (Fig. 5c), while constantly maintaining the structural features suggested by our analysis. In fact, the origin has been observed to move from cell pole to midcell before replication (11, 14). Additional experimental data, however, will be required to discriminate between these different possibilities.
Fig. 5.
Three chromosome conformations consistent with the SC pair distributions. Each displays the topological features of regular 117-kb loops and preferred structural faces of SC pair localization. The pair density (above 1.1 SD) is indicated by increasing dot size (464 total dots, or 10% of the genome, for density >1.1 σ). Note that the curves represent genomic DNA that is compacted 21-fold and may consist of substructures including topological domains. The dimensions of the helices are 2.5 μm high with a radius of 0.3 μm, compatible with an average E. coli cell. Consecutive dots along the helices represent ≈1 kb of compacted genomic DNA. The first two conformations are symmetric along the origin-terminus axis and involve two structural faces in which the dark SC pair dense regions are either rotated inward to meet in a single interface (a) or point outward (b). In an alternative configuration (c), supported by recent data (13), most of the chromosome would be coiled into a single stack of 117-kb loops, with the origin and terminus at midcell. Configurations a, b, and c may be viewed as different arrangements of a toroidal coil, such that dynamical changes could continuously occur (d). In each configuration, the origin and terminus regions may be structured differently than the remaining chromosome (11, 13). Helical moments (arrows in a and b) for transcriptional activity are found to be oriented toward the pair dense faces, indicating that the faces of each arc with maximal transcriptional activity are close to the SC pair dense faces. If i is the index of the ith kb along the genome, located at coordinates (xi, yi) in the structure, with expression level wi, the helical moment is defined as a vectors starting at (0, 0), and ending at xmoment = Σ(wixi)/Σ(wi), and ymoment = Σ(wiyi)/Σ(wi).
We evaluated the agreement of the SC gene pairs with the structures above by using the 3D distances between the pairs as a metric, while varying the size of the loops. We found a helical period of 117 kb to be the optimum for both arcs (SI Fig. 14). Two important properties emerge from these structural representations. First, the 117-kb looping causes a dramatic concentration of the pair dense regions in space, in a few patches along each face, consistent with spatial colocalization of subsets of SC paired genes. Second, many pairs, for example those separated by >1 Mb, are not physically close on the structure but share the feature of localization in the pair-dense faces. This suggests that the correlations in the SC pairs reflect confinement to these faces, rather than mere physical distance. This would be analogous to the correlations observed in protein residues, which often reflect important substructures regardless of spatial vicinity (20).
If the distributions of the SC pairs are the product of a structural periodicity and the localization of SC pairs along specific structural faces of the E. coli chromosome, what could be responsible for these features? Given the correlation between SC pairs and transcription, transcription is an attractive possibility for causing localization: the localization of certain highly transcribed genes along the structural faces (see helical moments in Fig. 5) would have the advantage of creating spatial subregions in which highly transcribed genes could be accessed by limited diffusion of RNA polymerase or RNA polymerase fixed in factories. Such subregions are consistent with experimental observations of foci of RNA polymerase in E. coli (28, 29) [which, like the transcription correlation, we observe are specific to log-phase growth (28)] and with transcription factories in the eukaryotic nucleus (3). If the pair dense faces are oriented inward (Fig. 5a), the density of transcription could help mechanically hold the chromosome arms together (explaining the symmetry in SI Figs. 6 and 7). Alternatively, if oriented on the surface (Fig. 5b), they could allow nascent transcripts to be accessed by ribosomes and the membrane (30).
Spatial localization of highly transcribed genes alone, however, would not generate periodicity. Rather, periodicity requires a second constraint, a regular loop size analogous to the 3.5-residue turn of an α-helix. This suggests an intrinsic property of the chromosome or of its binding proteins (e.g., H-NS MukBEF) (6); for example, the association of chromosomal DNA with proteins that induce a regular curvature would create periodic loops. Helices are also known to be the energetically optimal way of confining a string to certain geometrical spaces (31); thus, a 117-kb looping may be the spontaneous outcome of physicochemical properties including macromolecular crowding, supercoiling, DNA persistence length, and cell dimension.
The bacterial chromosome, however, is known to be a highly dynamic structure (2, 3, 12, 13) and the proposed models (Fig. 5) are based on patterns in gene positioning that are inherently static. Two points should be emphasized. First, the proposed features may relate to a specific portion of the cell cycle, perhaps log-phase growth, as indicated in SI Fig. 9. However, such features could be maintained despite dynamical changes (e.g., a reorientation from Fig. 5 a to c). In addition, it is likely that replication plays a dominant role in determining the structure of the chromosome (13, 14). In particular, the proposed looping would be advantageous for replication, allowing the chromosome to rapidly unwind and rewind with minimal local entanglement, like two coaxial [Fig. 5c (13)] or parallel [Fig. 5 a and b (12)] stacks of ropes.
The relationship of the proposed features to existing experimental data also bears discussion. These features are consistent with confocal microscopy (10, 12, 13), recombination (32), transposon insertion (33), and atomic force microscopy (34). In addition, our findings may yield insight into previously observed periodicities of 96 kb in transcription factor-binding sites (17), 90–120 kb in wavelet analysis (15, 16), and 115 kb in transcription levels (18, 19) in E. coli. In particular, our analysis suggests that these periodicities reflect an in-phase 117-kb grid, occupied by top-transcribed, top-conserved, and SC gene pairs. The significance of the pattern in the SC pairs may be due to the special vantage point of comparative genomics, where loci are identified based on the combined results of evolution acting on multiple genomes.
Independent of structure, our approach reveals significant gene organization on the chromosome. More general comparative analyses of how genes, gene pairs, or higher multiplets of genes are positioned in the genome should yield further insight into chromosome architecture. Similar methods should also be applicable to eukaryotes. The particular structural faces we propose and the chromosome-wide structural periodicity make specific predictions, which must be tested experimentally. To this effect, we are examining other bacteria for similar patterns (see SI Fig. 15 for C. crescentus, which displays a similar strong periodicity at 113 kb) and have developed a multiplex method of chromosome conformation capture (3C) (35) to measure the distances between thousands of chromosomal loci simultaneously at the resolution of our model. Ultimately, genome sequences and their structures may be highly interdependent aspects of a single finely tuned system. Evolutionary conservation should provide a powerful means of unraveling this interdependence.
Methods
Selection of SC Gene Pairs.
We selected the SC gene pairs based on evolutionary preference for chromosomal proximity and phylogenetic co-occurrence across many genomes, as explained in Fig. 1 and in detail below. For all gene pairs in E. coli, we calculated a score for chromosomal proximity and for phylogenetic co-occurrence. We then selected all gene pairs with phylogenetic co-occurrence scores of P < 10−10 and chromosomal proximity scores of P < 10−4. The results do not vary significantly with P cutoff.
Genomic Data.
The genomes were obtained from GenBank and consisted of 105 bacterial and three eukaryotic genomes (Saccharomyces cerevisiae, Schizosaccharomyces pombe, and Caenorhabditis elegans, which were included to represent particularly distant species).
Chromosomal Proximity.
For a pair of genes x and y, we calculated the tendency toward chromosomal proximity by using the difference in the order in which genes appear along the chromosome (gene-order difference) (23). We evaluated the probability
that the pair and its orthologs have gene order difference D, less than or equal to the gene-order differences, dg(x, y), observed across a set G of genomes g. Pg was calculated numerically under the null hypothesis that orthologous genes are randomly ordered on the chromosome. In genomes with multiple chromosomes, the gene-order distance between genes on different chromosomes was assigned to be greater than the maximum number of genes on any chromosome.
To correct for the variable phylogenetic divergence of query genomes, we constructed a UPGMA (36) phylogenetic tree based on a phylogenetic distance ϕ(g1, g2) between genomes g1 and g2. Note that the use of an alternative phylogenetic reconstruction method (neighbor-joining) does not affect our conclusions. We used a phylogenetic distance based on gene content (37), specifically, the mutual information between E. coli ortholog occurrence vectors in two genomes. The probabilities Pg from each genome were weighted based on the phylogenetic tree, by using an approach similar to the method of phylogenetic contrasts (36) (see SI Methods for details). The orthology mapping was established by using best bidirectional orthologs from Kyoto Encyclopedic of Genes and Genomes (KEGG) Sequence Similarity Database (www.genome.ad.jp/kegg/ssdb).
Phylogenetic Co-Occurrence.
Phylogenetic profile cooccurrence probability was calculated by using the extended hypergeometric distribution method described in Kharchenko et al. (38), which also includes a correction for the phylogenetic divergence. The orthologs were determined by using best bidirectional BLASTP hits against National Center for Biotechnology Information NR protein data set. Organisms containing orthologs for <1% of E. coli genes were excluded from calculations.
Distributions of Distances and Positions and Fourier Transform.
We constructed a histogram of the distances between genes for all SC pairs in E. coli. The histogram was transformed into a continuous probability density by using a Gaussian smoothing window (σ = 4 kb) and normalizing the total density over the entire genome to 1. A discrete Fourier transform of the data were computed from 0 to 1,000 kb by using a Tukey window to taper the ends (ratio of 0.5 for tapered to untapered length). The periodicity is independent of the maximum distance value. We calculated the statistical significance by repeating the smoothing and Fourier analysis on 10,000 randomizations in which the positions of the operons involving SC paired genes [determined from Price et al. (39)] were randomized within their chromosomal arc. The P value was determined by counting the number of randomizations with a Fourier peak as strong as or stronger than the 117-kb SC pair peak.
The density of SC pairs was computed by counting the number of SC pairs involving genes at each position along the chromosome, smoothing with the Gaussian window (σ = 8 kb), and normalizing by the overall gene density. The 1D grid is defined as a set of positions nτ + p along the chromosome, where τ is the spacing between grid points (the period), p is the offset (or phase) (set separately for each arc), and n is an integer. We evaluate the fit of the distributions to the grid using the sum of the distances of each peak to the nearest grid point (over all choices of p for each τ) as the error measure (see SI Fig. 8).
Expression Correlation.
We calculated an average of the absolute transcript level for wild-type standard growth conditions (4-morpholinepropanesulfonic acid minimal glucose, doubling time 2–8 h) using 5 Affymetrix (Santa Clara, CA) microarrays data sets extracted from the ASAP database [www.genome.wisc.edu/tools/asap.htm, Allen et al. (16)]. These data were smoothed by using a Gaussian window σ = 6 kb and normalized by the overall gene density as above. We calculated the Pearson correlation coefficient of the smoothed data with the pair position density, sampling once every 12 kb to avoid smoothing artifacts (and averaging over all choices of the sampling phase). P was computed by using Student's t test with n−2 degrees of freedom (where n is the number of data points).
Transcription and Conservation Pair Sets.
We constructed pair sets based on the levels of transcription (Ti) and conservation (Ci) of genes in E. coli (GE.coli), with i ∈ GE.coli by using log-phase transcript level from Allen et al. (16) for transcription and the number of orthologs of a gene (using best bidirectional orthologs from KEGG Sequence Similarity Database) for conservation. Each pair in the transcription pair set was chosen by randomly selecting two genes from GE.coli, where the probability of selecting gene i is pi = Ti/Ttot, with Ttot = ΣTi. Similarly, for selecting pairs in the conservation pair set we used probabilities pi = Ci/Ctot. Distance distributions and Fourier spectra were calculated as for the SC pairs.
Pair sets limited to the top k transcribed genes were created by choosing i ∈ GE.coli(k, T), where GE.coli(k, T) is the set of top k transcribed genes. Similarly, we defined pairs for the top k conserved genes by sampling from a subset GE.coli(k, C) and for the top k SC genes by taking the subset of the initial SC pairs in which both genes are elements of GE.coli(k, SC), the set of k genes most represented in the initial SC pair set.
Supplementary Material
Acknowledgments
We thank F. J. Isaacs, D. Peer, N. Reppas, J. Shendure, M. Umbarger, and I. Yanai for advice and critical reading of the manuscript. Part of this work was funded by the Department of Energy and National Institutes of Health Grant P50 GM068763. D.S. is also a faculty scholar at Lawrence Livermore National Laboratory.
Abbreviation
- SC
statistically correlated.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0610776104/DC1.
References
- 1.Chakalova L, Debrand E, Mitchell JA, Osborne CS, Fraser P. Nat Rev Genet. 2005;6:669–677. doi: 10.1038/nrg1673. [DOI] [PubMed] [Google Scholar]
- 2.Travers A, Muskhelishvili G. Curr Opin Genet Dev. 2005;15:507–514. doi: 10.1016/j.gde.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 3.Cook PR. Nat Genet. 2002;32:347–352. doi: 10.1038/ng1102-347. [DOI] [PubMed] [Google Scholar]
- 4.Branco MR, Pombo A. PLoS Biol. 2006;4:e138. doi: 10.1371/journal.pbio.0040138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bates D, Kleckner N. Cell. 2005;121:899–911. doi: 10.1016/j.cell.2005.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dame RT. Mol Microbiol. 2005;56:858–870. doi: 10.1111/j.1365-2958.2005.04598.x. [DOI] [PubMed] [Google Scholar]
- 7.Postow L, Hardy CD, Arsuaga J, Cozzarelli NR. Genes Dev. 2004;18:1766–1779. doi: 10.1101/gad.1207504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Higgins NP, Yang X, Fu Q, Roth JR. J Bacteriol. 1996;178:2825–2835. doi: 10.1128/jb.178.10.2825-2835.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Segall A, Mahan MJ, Roth JR. Science. 1988;241:1314–1318. doi: 10.1126/science.3045970. [DOI] [PubMed] [Google Scholar]
- 10.Teleman AA, Graumann PL, Lin DC, Grossman AD, Losick R. Curr Biol. 1998;8:1102–1109. doi: 10.1016/s0960-9822(98)70464-6. [DOI] [PubMed] [Google Scholar]
- 11.Niki H, Yamaichi Y, Hiraga S. Genes Dev. 2000;14:212–223. [PMC free article] [PubMed] [Google Scholar]
- 12.Viollier PH, Thanbichler M, McGrath PT, West L, Meewan M, McAdams HH, Shapiro L. Proc Natl Acad Sci USA. 2004;101:9257–9262. doi: 10.1073/pnas.0402606101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X, Liu X, Possoz C, Sherratt DJ. Genes Dev. 2006;20:1727–1731. doi: 10.1101/gad.388406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nielsen HJ, Ottesen JR, Youngren B, Austin SJ, Hansen FG. Mol Microbiol. 2006;62:331–338. doi: 10.1111/j.1365-2958.2006.05346.x. [DOI] [PubMed] [Google Scholar]
- 15.Allen TE, Price ND, Joyce AR, Palsson BO. PLoS Comput Biol. 2006;2:e2. doi: 10.1371/journal.pcbi.0020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Allen TE, Herrgard MJ, Liu M, Qiu Y, Glasner JD, Blattner FR, Palsson BO. J Bacteriol. 2003;185:6392–6399. doi: 10.1128/JB.185.21.6392-6399.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kepes F. J Mol Biol. 2004;340:957–964. doi: 10.1016/j.jmb.2004.05.039. [DOI] [PubMed] [Google Scholar]
- 18.Jeong KS, Ahn J, Khodursky AB. Genome Biol. 2004;5:R86. doi: 10.1186/gb-2004-5-11-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carpentier AS, Torresani B, Grossmann A, Henaut A. BMC Genomics. 2005;6:84. doi: 10.1186/1471-2164-6-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Socolich M, Lockless SW, Russ WP, Lee H, Gardner KH, Ranganathan R. Nature. 2005;437:512–518. doi: 10.1038/nature03991. [DOI] [PubMed] [Google Scholar]
- 21.Vakoc CR, Letting DL, Gheldof N, Sawado T, Bender MA, Groudine M, Weiss MJ, Dekker J, Blobel GA. Mol Cell. 2005;17:453–462. doi: 10.1016/j.molcel.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 22.Pellegrini M, Marcotte EM, Thompson MJ, Eisenberg D, Yeates TO. Proc Natl Acad Sci USA. 1999;96:4285–4288. doi: 10.1073/pnas.96.8.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huynen MA, Bork P. Proc Natl Acad Sci USA. 1998;95:5849–5856. doi: 10.1073/pnas.95.11.5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eisen JA, Heidelberg JF, White O, Salzberg SL. Genome Biol. 2000;1:RESEARCH0011. doi: 10.1186/gb-2000-1-6-research0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Makino S, Suzuki M. Science. 2001;292:803. doi: 10.1126/science.292.5518.803a. [DOI] [PubMed] [Google Scholar]
- 26.Tillier ER, Collins RA. Nat Genet. 2000;26:195–197. doi: 10.1038/79918. [DOI] [PubMed] [Google Scholar]
- 27.Eisenberg D, Weiss RM, Terwilliger TC. Proc Natl Acad Sci USA. 1984;81:140–144. doi: 10.1073/pnas.81.1.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cabrera JE, Jin DJ. Mol Microbiol. 2003;50:1493–1505. doi: 10.1046/j.1365-2958.2003.03805.x. [DOI] [PubMed] [Google Scholar]
- 29.Liu M, Durfee T, Cabrera JE, Zhao K, Jin DJ, Blattner FR. J Biol Chem. 2005;280:15921–15927. doi: 10.1074/jbc.M414050200. [DOI] [PubMed] [Google Scholar]
- 30.Woldringh CL. Mol Microbiol. 2002;45:17–29. doi: 10.1046/j.1365-2958.2002.02993.x. [DOI] [PubMed] [Google Scholar]
- 31.Maritan A, Micheletti C, Trovato A, Banavar JR. Nature. 2000;406:287–290. doi: 10.1038/35018538. [DOI] [PubMed] [Google Scholar]
- 32.Valens M, Penaud S, Rossignol M, Cornet F, Boccard F. EMBO J. 2004;23:4330–4341. doi: 10.1038/sj.emboj.7600434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manna D, Breier AM, Higgins NP. Proc Natl Acad Sci USA. 2004;101:9780–9785. doi: 10.1073/pnas.0400745101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim J, Yoshimura SH, Hizume K, Ohniwa RL, Ishihama A, Takeyasu K. Nucleic Acids Res. 2004;32:1982–1992. doi: 10.1093/nar/gkh512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dostie J, Richmond TA, Arnaout RA, Selzer RR, Lee WL, Honan TA, Rubio ED, Krumm A, Lamb J, Nusbaum C, Green RD, Dekker J. Genome Res. 2006;16:1299–1309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Felsenstein J. Am Nat. 1985;125:1–15. [Google Scholar]
- 37.Snel B, Bork P, Huynen MA. Nat Genet. 1999;21:108–110. doi: 10.1038/5052. [DOI] [PubMed] [Google Scholar]
- 38.Kharchenko P, Chen L, Freund Y, Vitkup D, Church GM. BMC Bioinformatics. 2006;7:177. doi: 10.1186/1471-2105-7-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Price MN, Huang KH, Alm EJ, Arkin AP. Nucleic Acids Res. 2005;33:880–892. doi: 10.1093/nar/gki232. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.