Abstract
Retinochrome is a member of the rhodopsin family having a chromophore retinal and functioning as a retinal photoisomerase in squid photoreceptor cells. Unlike vertebrate rhodopsins, but like many invertebrate rhodopsins, retinochrome does not have a glutamic acid at position 113 to serve as a counterion for the protonated retinylidene Schiff base. Here we investigated possible counterions in retinochrome by site-specific mutagenesis. Our results showed that the counterion is the glutamic acid at position 181, at which almost all the pigments in the rhodopsin family, including vertebrate and invertebrate rhodopsins, have a glutamic or aspartic acid. The remarkable exceptions are the long-wavelength visual pigments that have a histidine that, together with a nearby lysine, serves as a chloride-binding site. Replacement of Glu-181 of bovine rhodopsin with Gln caused a 10-nm red-shift of absorption maximum. Because the position at 181 is in the extracellular loop connecting the transmembrane helices VI and V, these results demonstrate the importance of this loop to function for spectral tuning in the rhodopsin family.
Retinochrome is a seven-transmembrane α-helical protein that acts as a retinal photoisomerase; it is found in cephalopod photoreceptor cells (1, 2). It contains an all-trans-retinal as a chromophore and produces 11-cis-retinal after absorption of light (3, 4). The 11-cis-retinal is then supplied to photoconverted rhodopsin via a retinal-shuttle protein, intracellular retinal-binding protein (5, 6), to complete the visual cycle of these photoreceptor cells. Squid retinochrome has an amino acid sequence ≈20% identical to those of vertebrate and invertebrate rhodopsins (7), and its absorption maximum (495 nm) is similar to the maxima of rhodopsins. Recently, a homologue of retinochrome, retinal G protein-coupled receptor (RGR), has been found in the mammalian pigment epithelium and Müller cells (8, 9): retinochrome and RGR are classified into one of the subgroups in the rhodopsin family (10–12). The physiological roles of retinochrome and rhodopsin are different, but they function with absorption of a visible light that isomerizes the retinal chromophore, the common light-driven mechanism of action in the rhodopsin family. Thus, it is of interest to investigate the molecular architecture of retinochrome.
It has been well known that the retinal chromophore free in solution exhibits absorption maximum in the ultra-violet region, and protonation of retinylidene Schiff base is a major mechanism to shift its maximum (2) to the visible region (13). Like in all the pigments including vertebrate rhodopsins, the retinal chromophore in retinochrome binds to the lysine residue at position 296§ in the helix VII (Fig. 1) through a protonated Schiff-base linkage (14, 15). The positive charge on the protonated Schiff base is energetically unstable in the protein interior, and therefore, a negatively charged amino acid residue called counterion should be present in the protein. In vertebrate visual pigments such as bovine rhodopsin, the glutamic acid at position 113 in the helix III serves as a counterion to stabilize the protonation of the retinylidene Schiff base (16–18). However, like the other pigments including invertebrate rhodopsins (19), retinochrome has an amino acid residue different from glutamic acid at this position (14), and therefore, these pigments may have a counterion at a different position. Thus, we have tried to identify the residue that acts as a counterion in retinochrome by site-directed mutagenesis.
Figure 1.
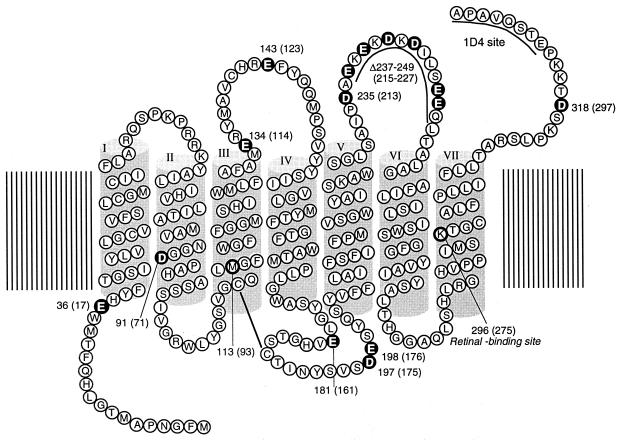
Secondary structural model of retinochrome showing the location of Glu and Asp residues (white letters). For affinity purification with an anti-rhodopsin antibody, the amino acid sequence of monoclonal antibody Rho1D4 epitope (ETSQVAPA) is introduced to the C terminus of retinochrome. On the basis of sequence alignment, the bovine rhodopsin amino acid residue numbering system is used. The retinochrome numbering system is shown in parentheses.
Our results clearly showed that the glutamic acid at position 181 in the extracellular IV–V loop acts as the counterion in retinochrome. Interestingly, we found that the glutamic acid at position 181 is highly conserved in all the rhodopsin groups including vertebrate and invertebrate rhodopsins. The remarkable exceptions are the long-wavelength visual pigments including iodopsin and human red that have the histidine, which, together with a nearby lysine, serves as a chloride-binding site (20) to shift the spectrum to the red further (21–23). From these results, the diversity of the spectral tuning mechanism among the rhodopsin family will be discussed.
Materials and Methods
Preparation of Retinochrome and Rhodopsin Mutants.
Retinochrome cDNA (7) was a generous gift from Ikuko Hara-Nishimura (Kyoto University) and Mikio Nishimura (National Institute for Basic Biology, Okazaki, Japan). The coding region of the cDNA was isolated by PCR in which it was tagged by the monoclonal antibody Rho 1D4 epitope-sequence (ETSQVAPA; 24). The tagged cDNA was inserted into a HindIII site of a plasmid vector SRα (25). A point mutation was made by using a commercial site-directed mutagenesis kit, Quikchange (Stratagene), according to the manufacturer's instructions. A deletion mutant was prepared by using the standard PCR method. The method for the construction of the bovine rhodopsin mutant genes E181Q and E113Q was performed as described (26).
Expression and Purification of Retinochrome and Rhodopsin.
cDNA was transfected into HEK 293S cells by using the calcium-phosphate method as described (26). The transfected cells were harvested for 2 days and collected by centrifugation. After the addition of excess all-trans-retinal, retinochrome was extracted with 1% dodecylmaltoside (DM) in Hepes buffer (pH 7.0) containing 140 mM NaCl (buffer A), bound to 1D4-agarose and eluted with buffer A containing 0.02% DM and 0.1 mg/ml C-terminal peptide of bovine rhodopsin, followed by washing with 0.02% DM in buffer A. The method used for the expression of the bovine rhodopsin mutant genes E181Q and E113Q in HEK 293S cells was as described (26).
Removal of chloride from retinochrome and rhodopsin samples was carried out by dialysis against a 500-fold volume of 50 mM Hepes buffer (pH 7.0; buffer B) with a 4× exchange of the buffer.
Spectrophotometer.
Absorption spectra were recorded at 0°C with a Shimadzu Model MPS-2000 spectrophotometer interfaced with an NEC PC-9801 computer (27). The temperature of the sample was regulated to within 0.1–0.5°C by a temperature controller (Neslab, Tokyo, Japan).
Results and Discussion
Because retinochrome contains nine Glu and six Asp in its sequence (Fig. 1), we speculated that one of these residues would act as a counterion in retinochrome. Thus, we have expressed single amino acid mutants of retinochrome where each Glu or Asp in the molecule, except in the V–VI cytoplasmic loop region, was replaced with Gln or Asn. To examine the residues in the V–VI loop as possible contribution candidates, we prepared a deletion mutant (Δ237–249) that lacks this region.
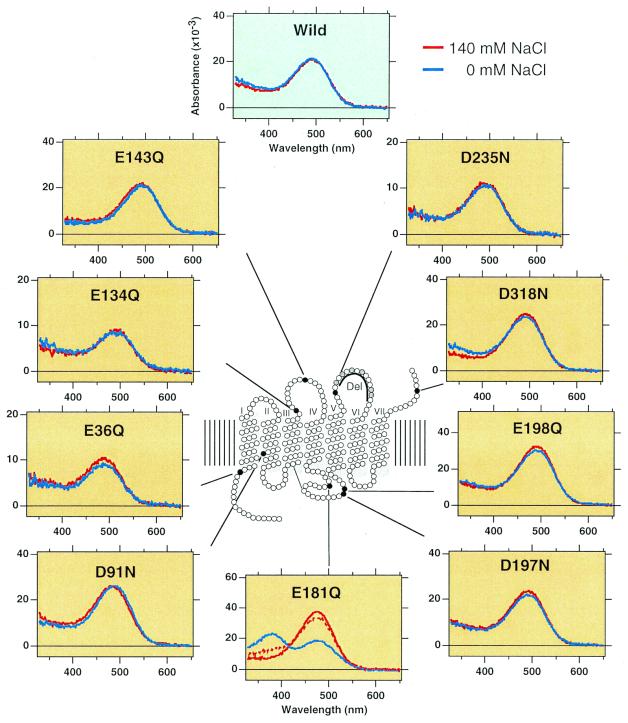
Identification of the counterion by site-directed mutagenesis has been performed successfully in bacteriorhodopsin (28) and vertebrate rhodopsins (16–18). In these studies, replacements of Asp-85 in bacteriorhodopsin and Glu-113 in vertebrate rhodopsins caused the formation of a deprotonated Schiff-base chromophore, thereby resulting in the shift of absorption maximum to ≈380 nm. On the other hand, the mutants show similar maxima as the original pigments in the presence of chloride, because chloride can act as a surrogate counterion. Fig. 2 shows absorption spectra of the expressed wild-type (WT) and mutant retinochromes in the presence or absence of chloride. The deletion mutant did not form an active protein, although the low-level expression was confirmed by immunoblotting analysis. Thus, the spectra of this mutant are not shown in the figure. All the active mutants except D91N and E181Q exhibited almost the same absorption maxima (493 nm) as WT, irrespective of the presence or absence of chloride. The absorption maximum (490 nm) of the D91N mutant was slightly different from that of the WT in the absence of chloride, and showed a blue-shifted absorption maximum (484 nm) in the presence of chloride. We first thought that this aspartic acid could be a counterion of retinochrome, because a mutagenesis study on bovine rhodopsin indicated that a glutamic acid artificially introduced into the 90 position (next to 91) could act as a counterion (29). However, our results clearly showed that the replacement of this residue did not match the criteria of counterion, although this residue may be located near the chromophore. The criteria of counterion are clearly applicable to the E181Q mutant; that is, the mutant showed absorption maximum at about 380 nm in the absence of chloride, and it shifted to the visible region (480 nm) in the presence of chloride (Fig. 2). These results strongly suggested that glutamic acid at position 181 acts as a counterion in retinochrome.
Figure 2.
Absorption spectra of mutant retinochromes. Red and blue lines show the spectra in the presence and absence of chloride, respectively. In the E181Q mutant, the addition of chloride to a chloride-free sample restored the visible absorption (broken line). The numbering is shown as bovine rhodopsin system.
To eliminate the possibility that E181Q mutant is denatured in the absence of chloride, we added chloride in the chloride-free sample and observed an increase in absorbance at the visible region (Fig. 2). Furthermore, the pKa of the Schiff base was estimated to be 7.0 by changing the pH of the sample in the absence of chloride (data not shown). The value is very similar to that observed in the counterion-depleted mutant of bovine rhodopsin (pKa = ≈6.0; ref. 16).
The above results indicated that the glutamic acid at position 181 in the extracellular loop connecting the transmembrane helices IV and V acts as the counterion in retinochrome, whereas in vertebrate rhodopsins, the counterion is the glutamic acid at position 113 in the transmembrane helix III. Interestingly, vertebrate rhodopsins also have the glutamic acid at position 181 (Fig. 3). Furthermore, almost all the pigments in the rhodopsin family whose amino acid sequences have been reported thus far have a glutamic or aspartic acid at this position, with the only exceptions being the long-wavelength visual pigments (iodopsin and human red) and VA opsins (Fig. 3). In this context, it should be noted that the histidine at position 181 and the lysine at position 184 of the long-wavelength visual pigments are the chloride-binding site (20), and binding of chloride causes about 40-nm red shift of the pigments (21–23). Fourier transform infrared spectroscopy indicated that the chloride binds to this site with water molecules to situate near the retinal chromophore (T. Hirano, H. Imai, H. Kandori, and Y.S., unpublished work). Recently, the bovine rhodopsin structure as determined by x-ray crystallography was published (30). In the structure, one of the oxygen atoms of the glutamic acid at position 181 is situated close (about 4.5 Å) to the C12 of the retinal chromophore, and it may also interact with Glu-113 via the peptide carbonyl and amide of Cys-187. Therefore, the amino acid residue at position 181 could play a crucial role in the rhodopsin family in shifting the absorption maximum.
Figure 3.
Comparison of amino acids at position 113 and 181. Phylogenetic relationships among the members of the rhodopsin family are schematically represented based on previous reports (9–11). All the members of vertebrate-rhodopsin group possess a glutamic or aspartic acid at position 113 in helix III, whereas retinochrome or invertebrate-rhodopsin groups do not. On the other hand, all have a glutamic or aspartic acid at position 181 in the extracellular IV–V loop, except for the long-wavelength visual pigments (iodopsin and human red) and VA opsins. Note that Tyr-113 is neutral in the case of invertebrate octopus rhodopsin (19). GenBank accession numbers for the respective sequences are; for squid retinochrome, X57143; for bovine retinal G protein-coupled receptor, S67535; for human retinal G protein-coupled receptor, U15790; for mouse peropsin, AF012271; for human peropsin, AF012270; for scallop Go-rhodopsin, AB006455; for squid rhodopsin, X70498; for Drosophila Rh1, K02315; for Drosophila Rh3, Y00043; for Xenopus melanopsin, AF014797; for bovine rhodopsin, K00506; for human rhodopsin, U49742; for chicken green, M92038; for chicken blue, M92037; for iodopsin (chicken red), X57490; for human red, M13305; for chicken pinopsin, U15762; for salmon VA opsin, AF001499; and for human encephalopsin, AF140242.
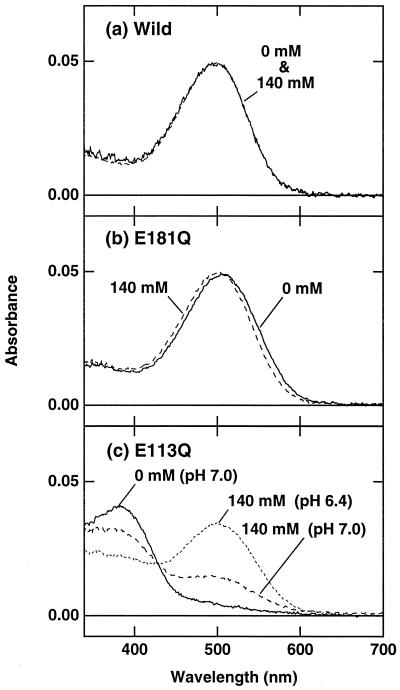
To examine whether the glutamic acid at position 181 affects the absorption characteristics of vertebrate rhodopsins, we replaced the glutamic acid with glutamine in bovine rhodopsin (Fig. 4). The replacement caused an ≈10-nm red shift of absorption maximum, but the addition of chloride shifted the maximum back to that of the WT. This absorption maximum of E181Q in the presence of chloride is consistent with a previous report on this mutant (18). In contrast, the E113Q mutant had its absorption maximum at ≈380 nm, and the addition of chloride caused an increase in absorbance at ≈500 nm (Fig. 4). Taken together with the recent rhodopsin structure (30), these results suggested that Glu-181 in bovine rhodopsin is not the counterion but is located near the region including Schiff base to affect absorption characteristics. The electrostatic character of Glu-181 is compensated by the addition of chloride when Glu-181 was replaced with glutamine. Interestingly, the effect of replacement of Glu-181 in bovine rhodopsin is phenomenologically similar to that of Asp-91 in retinochrome (Fig. 4), although the apparent dissociation constants of chloride are somewhat different from each other. Asp-91 is next to Gly-90 in bovine rhodopsin, and the glutamic acid introduced at this position acts as a counterion in bovine rhodopsin (29). In this region, two glycines (Gly-89 and Gly-90) are situated tandemly, such that some distortion of the peptide backbone around these residues is induced (30). Taken together, in the rhodopsin family including retinochrome, the retinal chromophore is close to the amino acid residues at positions 91 and 181, and Glu-181 acts as the counterion in retinochrome, whereas the specifically acquired Glu-113 acts as the counterion in vertebrate rhodopsins. It is of interest to note that some variation in amino acid residue is seen at position 181 in some vertebrate visual pigments, possibly because the role of Glu-181 as a counterion may be weakened by the newly acquired counterion at position 113. We emphasize here that this is, to our knowledge, the first invertebrate retinal pigment to be expressed, but the expression of invertebrate “rhodopsins” in cell culture has been unsuccessful thus far. Therefore, it is unclear yet whether the glutamic or the aspartic acid at position 181 acts as the counterion in invertebrate rhodopsins. Further experiments will shed a light on the mechanism of visible-light absorption and its relation to the diversity of function in the rhodopsin family.
Figure 4.
Absorption spectra of WT bovine rhodopsin (a) and its mutants E181Q (b) and E113Q (c). Solid and broken lines show the spectra in the presence (140 mM NaCl) and absence of chloride at pH 7.0, respectively. The dotted line in c shows the spectrum in 140 mM NaCl at pH 6.4.
Acknowledgments
We thank Prof. J. Nathans for his gift of the 293S cell line, Prof. F. Tokunaga for providing a pUSRα expression vector, and Prof. R. S. Molday for his gift of the rho 1D4-producing hybridoma. This work was supported in part by Grant-in-Aid for Scientific Research from the Japanese Ministry of Education, Science, Sports, and Culture (to Y.S. and A.T.). T.Y. is supported by the Japanese Society for the Promotion of Science Research Fellowship for Young Scientists. This paper is dedicated to Prof. Tomiyuki Hara and Dr. Reiko Hara.
Abbreviation
- WT
wild type
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
In the retinochrome numbering system, the lysine residue is at position 275. Based on the sequence alignment, Lys-275 in retinochrome corresponds to Lys-296 in bovine rhodopsin. Hereafter, we describe the residue number of retinochrome by using the bovine rhodopsin numbering system.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.260349597.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.260349597
References
- 1.Hara T, Hara R. Nature (London) 1965;206:1331–1334. doi: 10.1038/2061331a0. [DOI] [PubMed] [Google Scholar]
- 2.Hara T, Hara R, Takeuchi J. Nature (London) 1967;214:572–573. doi: 10.1038/214572a0. [DOI] [PubMed] [Google Scholar]
- 3.Hara T, Hara R. Nature (London) 1968;219:450–454. doi: 10.1038/219450a0. [DOI] [PubMed] [Google Scholar]
- 4.Hara T, Hara R. Nature (London) 1973;242:39–43. doi: 10.1038/newbio242039a0. [DOI] [PubMed] [Google Scholar]
- 5.Ozaki K, Terakita A, Hara R, Hara T. Vision Res. 1987;27:1057–1070. doi: 10.1016/0042-6989(87)90020-4. [DOI] [PubMed] [Google Scholar]
- 6.Terakita A, Hara R, Hara T. Vision Res. 1989;29:639–652. doi: 10.1016/0042-6989(89)90026-6. [DOI] [PubMed] [Google Scholar]
- 7.Hara-Nishimura I, Matsumoto T, Mori H, Nishimura M, Hara R, Hara T. FEBS Lett. 1990;271:106–110. doi: 10.1016/0014-5793(90)80383-t. [DOI] [PubMed] [Google Scholar]
- 8.Jiang M, Pandey S, Fong H K W. Invest Opthalmol Visual Sci. 1993;34:3669–3678. [PubMed] [Google Scholar]
- 9.Hao W, Fong H K W. J Biol Chem. 1999;274:6085–6090. doi: 10.1074/jbc.274.10.6085. [DOI] [PubMed] [Google Scholar]
- 10.Kojima D, Terakita A, Ishikawa T, Tsukahara Y, Maeda A, Shichida Y. J Biol Chem. 1997;272:22979–22982. doi: 10.1074/jbc.272.37.22979. [DOI] [PubMed] [Google Scholar]
- 11.Miyata T. In: Explosive Evolution of Bioorganism Viewed from DNA (in Japanese) Miyata T, editor. Tokyo: Iwanami; 1998. pp. 31–57. [Google Scholar]
- 12.Blackshaw S, Snyder S H. J Neurosci. 1999;19:3681–3690. doi: 10.1523/JNEUROSCI.19-10-03681.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pitt G A J, Collins F D, Morton R A, Stok P. Biochem J. 1955;59:122–128. doi: 10.1042/bj0590122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hara-Nishimura I, Kondo M, Nishimura M, Hara R, Hara T. FEBS Lett. 1993;335:94–98. doi: 10.1016/0014-5793(93)80447-3. [DOI] [PubMed] [Google Scholar]
- 15.Sekiya N, Kishigami A, Naoki H, Chang C-W, Yoshihara K, Hara R, Hara T, Tokunaga F. FEBS Lett. 1991;280:107–111. doi: 10.1016/0014-5793(91)80215-o. [DOI] [PubMed] [Google Scholar]
- 16.Zhukovsky E A, Oprian D D. Science. 1989;246:928–930. doi: 10.1126/science.2573154. [DOI] [PubMed] [Google Scholar]
- 17.Sakmar T P, Franke R R, Khorana H G. Proc Natl Acad Sci USA. 1989;86:8309–8313. doi: 10.1073/pnas.86.21.8309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nathans J. Biochemistry. 1990;29:9746–9752. doi: 10.1021/bi00493a034. [DOI] [PubMed] [Google Scholar]
- 19.Nakagawa M, Iwasa T, Kikkawa S, Tsuda M, Ebrey T G. Proc Natl Acad Sci USA. 1999;96:6189–6192. doi: 10.1073/pnas.96.11.6189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Z, Asenjo A B, Oprian D D. Biochemistry. 1993;32:2125–2130. doi: 10.1021/bi00060a001. [DOI] [PubMed] [Google Scholar]
- 21.Knowles A. Vision Res. 1980;20:475–483. doi: 10.1016/0042-6989(80)90122-4. [DOI] [PubMed] [Google Scholar]
- 22.Fager L Y, Fager R S. Exp Eye Res. 1979;29:401–408. doi: 10.1016/0014-4835(79)90056-3. [DOI] [PubMed] [Google Scholar]
- 23.Shichida Y, Kato T, Sasayama S, Fukada Y, Yoshizawa T. Biochemistry. 1990;29:5843–5848. doi: 10.1021/bi00476a028. [DOI] [PubMed] [Google Scholar]
- 24.Molday R S, MacKenzie D. Biochemistry. 1983;22:653–660. doi: 10.1021/bi00272a020. [DOI] [PubMed] [Google Scholar]
- 25.Kayada S, Hisatomi O, Tokunaga F. Comp Biochem Physiol B Biochem Mol Biol. 1995;110:599–604. doi: 10.1016/0305-0491(94)00179-x. [DOI] [PubMed] [Google Scholar]
- 26.Nagata T, Terakita A, Kandori H, Kojima D, Shichida Y, Maeda A. Biochemistry. 1997;36:6164–6170. doi: 10.1021/bi962920t. [DOI] [PubMed] [Google Scholar]
- 27.Shichida Y, Tachibanaki S, Mizukami T, Imai H, Terakita A. Methods Enzymol. 2000;315:347–363. doi: 10.1016/s0076-6879(00)15853-7. [DOI] [PubMed] [Google Scholar]
- 28.Marti T, Rosselet S J, Otto H, Heyn M, Khorana H G. J Biol Chem. 1991;267:16922–16927. [PubMed] [Google Scholar]
- 29.Rao V R, Cohen G B, Oprian D D. Nature (London) 1994;367:639–642. doi: 10.1038/367639a0. [DOI] [PubMed] [Google Scholar]
- 30.Palczewski K, Kumasaka T, Hori T, Behnke C A, Motoshima H, Fox B A, Le Trong I, Teller D C, Okada T, Stenkamp R E, et al. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]