Abstract
In gene targeting experiments, the importance of genetic background is now widely appreciated, and knockout alleles are routinely backcrossed onto a standard inbred background. This produces a congenic strain with a substantial segment of embryonic stem (ES)-cell-derived chromosome still flanking the knockout allele, a phenomenon often neglected in knockout studies. In cholecystokynin 2 (Cckbr) knockout mice backcrossed with C57BL/6, we have found a clear ‘congenic footprint’ of expression differences in at least 10 genes across 40 Mb sequence flanking the Cckbr locus, each of which is potentially responsible for aspects of the ‘knockout’ phenotype. The expression differences are overwhelmingly in the knockout-low direction, which may point to a general phenomenon of background dependence. This finding emphasizes the need for caution in using gene knockouts to attribute phenotypic effects to genes. This is especially the case when the gene is of unknown function or the phenotype is unexpected, and is a particular concern for large-scale knockout and phenotypic screening programmes. However, the impact of genetic background should not be simply viewed as a potential confound, but as a unique opportunity to study the broader responses of a system to a specific (genetic) perturbation.
Keywords: Cckbr, congenic footprint, gene expression, knockout, microarray, mouse mutant
A gene expression profile is the most comprehensive snapshot of the state of a sample of cells or tissue currently available. It allows analysis of the cell as a system at an entirely new level. In the simplest case, if a gene is knocked out and the resulting strain is viable, adjustments in the expression of other genes of related function would be required to compensate. If such differences in gene expression profile are detected between knockout and control, these are probable parts of the same biological subsystem. There are few studies in the literature to date employing this systems biology approach and our original aim was to make use of a powerful dataset to investigate an example with a well characterized Cckbr (cholecystokynin receptor 2) knockout strain (Nagata et al. 1996). We did detect functional neighbours of Cckbr, but saw many more strong differences because of the ES-cell-derived chromosomal context of the knockout. We describe this very striking finding involving the physical neighbourhood of the Cckbr locus and its consequences.
Effects of genetic background on the phenotype of targeted mutations in mice (knockouts) are widely appreciated, and most are backcrossed into C57BL/6 or another suitable strain. Inevitably, a substantial fragment of the flanking embryonic stem (ES)-cell-derived chromosome is retained (discussed further below). The genetic problem (Gerlai 1996; Wolfer et al. 2002) or opportunity (Bolivar et al. 2001) that this represents has been well discussed among geneticists, but the possible consequences are rarely considered in studies of knockouts (Crusio 2004). One reason for this may be that it is hard to estimate the likelihood that phenotypic differences are because of flanking sequences rather than the targeted allele, especially as the phenotype(s) being studied are typically selected with a bias towards identifying or confirming a known function of the gene. In contrast, gene expression profiling does not make any a priori assumption about the effect of the targeted allele. In the example we present here, there are numerous gene expression differences in what we call the congenic footprint. Each of these could be responsible for the ‘knockout’ phenotype, and this has implications for the interpretation of the phenotypes of the thousands of knockout strains that have been made and for large-scale plans to generate knockouts and screen them for phenotypic effects (Austin et al. 2004; Auwerx et al. 2004).
We have been studying a cholecystokinin receptor 2 (Cckbr) knockout in connection with a model of neuropathic pain (Kurrikoff et al. 2004). Using Affymetrix MOE430av2 arrays, which have 22 690 sets of 11 oligonucleotides (probesets) representing about 14 000 different genes, we have compared knockout and wildtype gene expression profiles in midbrain and medulla. To maximize the statistical power to see gene expression differences related to the knockout, we pooled all of our microarray data to compare knockout (extensively backcrossed to C57BL/6) animals (16 arrays) with C57BL/6 (16 arrays) other variables such as surgical treatment are balanced across these groups. This number of samples is much greater than is used in most microarray experiments, and this gives us good statistical power to detect expression differences.
Materials and methods
Cckbr was originally knocked out in J1 ES cells, derived from 129S4/Jae mice (Nagata et al. 1996), subsequently backcrossed N = 10 to C57BL/6Bkl (Scanbur-BK) in Tartu. Homozygous knockout and wildtype animals were obtained by intercrossing heterozygous stock. Housing was at 20 ± 2°C under a 12-h, lights on at 0700 cycle, water and food ad libitum. The University of Tartu Animal Care Committee approved all animal procedures (EC Directive 86/609/EEC).
Sixteen, 3-month-old males from 12 litters were used. Nine days after surgery (neuropathic pain model or sham; Kurrikoff et al. 2004), mice were killed by cervical dislocation and tissues frozen in liquid nitrogen. RNA was extracted with TRIzol (Invitrogen, UK). From 4 μg of total RNA, complementary DNA and labelled complementary RNA (cRNA) were made using the One-Cycle Kit (Affymetrix, Santa Clara, CA, USA). Fragmented cRNA was hybridized to the Mouse 430A 2.0 array according to Affymetrix protocols.
The .cel files were analysed using dChip (PM-MM model) (Li & Wong 2001) and the statistical package R (Ihaka & Gentleman 1996). We used a nested analysis of variance (anova) model to test for differences between knockout and control across all 32 samples, while recognizing that the two tissue samples from each mouse may not be independent. Hybridization date and surgical treatment were nuisance factors, and pairs of tissues (midbrain and medulla) from each animal were treated as replicates. The R command for anova applied to each probeset was aov(signal∼date+surgery+genotype+Error(mouse)).
The Shapiro–Wilk test does not reject normality for most probesets, and spot checks using permutation of the genotype factor gave similar P-values. False discovery rate (FDR) control (Benjamini & Hochberg 1995) using Q = 0.05 was used on the whole-genome and chromosome 7 levels.
The National Institute of Environmental Health Sciences (NIEHS)/Perlegen resequencing data for 129S1/SvImJ, a reasonably close relative of 129S4/Jae, shows that the observed differences are not because of poor hybridization resulting from sequence differences between strains except for one possible case. A total of 22 187 polymorphisms were discovered between 129S1/SvImJ and C57BL/6 in the footprint region but only 12 are in the probe sequences of the 11 differing probesets. Six are in one probeset: 1417961_a_at (Trim30; tripartite motif protein 30).
Genotyping: Single nucleotide polymorphisms (SNPs) (rs13479420, rs13479422, rs13479512, rs13479517, rs3657451 and rs3677347) from dbSNP were genotyped using primer extension (MegaBACE SNuPe, GE Healthcare, UK).
Results and discussion
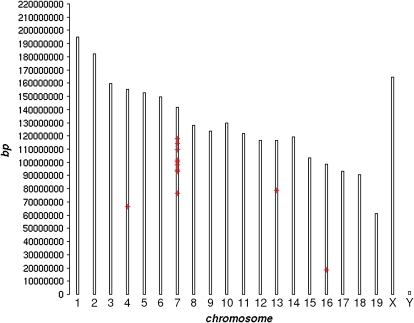
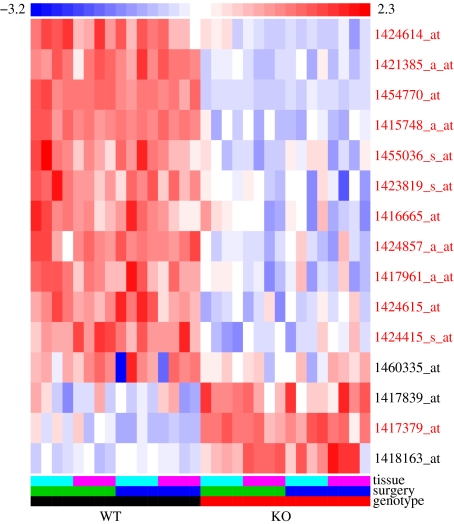
We did a conservative analysis of the 32 samples, treating the two tissues as replicate samples and every probeset as a separate test. We found 15 probesets with very strong evidence for an expression difference between knockout and control mice, one of which represents Cckbr itself. Across the entire array, the false discovery rate for these 15 is estimated at <0.05 (FDR, Benjamini & Hochberg 1995; Storey & Tibshirani 2003; Storey et al. 2004), these methods give the same result in this case). To our surprise, 12 of them, representing 10 different genes in addition to Cckbr, map in a 40-Mbp region around the Cckbr locus on chromosome 7 (Fig. 1). All but one differed in the same direction, knockout-low (Fig. 2).
Figure 1. Genomic locations of the 15 probesets most significantly differing between wildtype and Cckbr knockout mice.
This figure was generated using Webgestalt (Zhang et al. 2005).
Figure 2. Heatmap of the probesets most significantly differing between wildtype (left) and Cckbr knockout (right) mice.
Each column represents one of the 32 Affymetrix MOE430A 2.0 arrays, and each row represents a probeset (transcript). The probeset names on the right are colour coded red for chromosome 7 and black for others. Each row is centred and scaled, and the colour scale at the top is calibrated in standard deviations, red for high values, white near the mean and blue for low values. The rows have been hierarchically clustered using |1 − R| (where R is the Pearson correlation coefficient) and a complete linkage criterion. The colour bar at the bottom shows the identities of the samples applied to each array. Tissue: pale blue, medulla; pink, midbrain. Surgery: blue, sham; green, neuropathic pain model. Strain: black, C57BL/6; red, Cckbr knockout. KO, knockout; WT, wildtype.
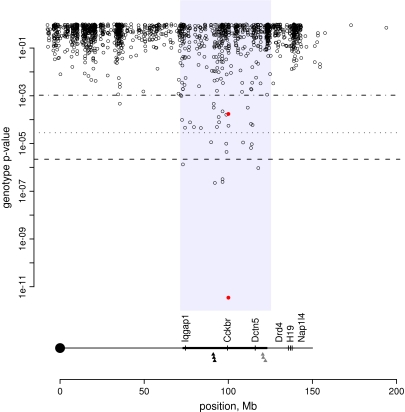
Inspection of the data indicates that there are many more differences in the same region. Plotting the P-values for difference of chromosome 7 probesets against their position on the chromosome (Fig. 3) shows a cloud of values above 0.1 with a distinctive rain shower of differing expression between 73 and 123 Mb (indicated in pale blue). Clearly, it is worth considering a slightly less conservative criterion here. Again using FDR Q = 0.05, but this time just considering chromosome 7, there are 33 probesets in the 40-Mb region that differ significantly and only one in the remainder of the chromosome. This is a very striking and specific pattern.
Figure 3. The congenic footprint: mouse chromosome 7.
Upper: uncorrected P-values for genotype effect plotted by location (Mbp) on chromosome 7. The two red filled points are Cckbr probesets. The three lines are significance thresholds with differing multiple-testing criteria (α = 0.05). Dashed: Bonferroni corrected (whole genome); dotted: FDR (whole genome); dot–dashed (FDR, chromosome 7). The shaded area indicates the footprint region. Lower: map of chromosome 7 on the same scale. The heavier line represents the congenic footprint, and the filled triangles are the locations of the six SNP markers. The grey triangles indicate that there were a few knockout animals with C57BL/6 alleles at these loci, indicating the presence of more than one breakpoint. All positions are from National Center for Biotechnology Information sequence build 34.
Hypothesizing that these expression differences were in ES-cell-derived sequences linked to the knockout allele (i.e. a differential chromosomal segment), we genotyped six informative SNPs outside of genes flanking Cckbr (Fig. 3, bottom). These confirmed that the region is indeed of non-C57BL/6 origin in the knockout strain. The three SNPs on the proximal side (black triangles in Fig. 3) are all homozygous 129 genotype in knockout animals. For the distal three SNPs (grey in Fig. 3), this is also true, except that three of the 16 knockout DNAs show a C57BL/6 genotype, indicating that there are different congenic breakpoints represented in our set of mice.
The pattern of expression differences that we see must be a consequence of the differences between sequences with an alternate strain of origin. We call the pattern a congenic footprint.
The ten genes that most strongly define the congenic footprint are diverse in function and do not have an obvious common functional theme. They are Dctn5 (dynactin 5, actin-associated, 1415748_a_at), Coq7 (demethyl-Q 7, ubiquinone biosynthesis, 1416665_at), Trim30 (tripartite motif, T-lymphocyte regulatory, 1417961_a_at), Trim34(1424857_a_at), Myo7a (myosin VIIa, 1421385_a_at), Arl6ip1 (adenosine diphosphate-ribosylation factor-like 6 interacting protein 1, 1423819_s_at), Spon1 (spondin 1, extracellular matrix, 1424415_s_at), Ndufc2 (nicotinamide adenine dinucleotide (reduced) dehydrogenase (ubiquinone) 1, subcomplex unknown, 2, 1455036_s_at), Iqgap1 (IQ motif containing guanosine triphosphatase activating protein 1, 1417379_at) and Frag1 (fibroblast growth factor receptor activating 1, 1424615_at, 1424614_at).
The affected genes that are not on chromosome 7, on the other hand, do have a probable common theme related to inflammation. They are Cldn5 (claudin 5), Tlr4 (toll-like receptor 4) and Lysmd3 (LysM, putative peptidoglycan-binding, domain containing 3). Tlr4 is a candidate for inflammatory response and neuropathic pain (Poltorak et al. 1998; Tanga et al. 2005) and very plausible as a gene whose regulation is linked to Cckbr. Cldn5 is probably also involved in inflammation, through its role in endothelial cell junctions, which are involved in the process of leukocyte migration across the epithelium (Cook-Mills & Deem 2005). Lysmd3 is a poorly characterized gene with recently annotated sequence similarity to lysozyme.
One possibility is that the expression differences observed in certain transcripts could be because of poor hybridization due to sequence differences rather than genuine expression differences. Although a single nucleotide change can make a large difference in the hybridization of a 25-mer probe as used here, multiple differences in a gene would be required to cause an apparent expression difference because each transcript is represented by a set of 11 probes. J1 ES cells, used to prepare the Cckbr knockout, were derived from the strain 129S4/Jae, which shares a relatively recent common ancestor with 129S1/SvImJ with no known outcrossings (Simpson et al. 1997). 129S1/SvImJ has been extensively resequenced by hybridization in the NIEHS/Perlegen project (http://mouse.perlegen.com/mouse/download.html, downloaded 2 February 2006). They detect 22 187 polymorphisms between 129S1/SvImJ and C57BL/6 in the footprint region. Among these, however, only 12 are found in the probe sequences of the 11 significantly different probesets, and six of these are in a single probeset: 1417961_a_at, ‘tripartite motif protein 30’. This probeset is thus the only one where there is a likelihood that there is a sequence difference rather than a true expression difference.
Standard practise currently expects confirmation or replication (by a second method such as quantitative PCR) of expression differences detected by microarray. We find the congenic footprint presented here compelling in the absence of such a follow-up, chiefly because of its specificity, evident from Fig. 1. The cluster of expression differences on chromosome 7 cannot be a coincidence (P = 6.7 × 10−20, Fisher’s exact test). From the statistical point of view we have been able to take a stringent approach, and as such, less than one of these findings is expected to be a false positive (examination of Fig. 2 indicates that there are numerous false negatives). This contrasts with many microarray studies in the literature which have poor statistical power to detect true positives and thus yield a high proportion of false positives because of multiple testing. The most interesting further experiments would involve similar tests on other knockout strains and indeed other congenic strain pairs, to confirm that congenic footprints in gene expression are a general phenomenon resulting from genetic background effects.
The size of the congenic region in knockouts that have been backcrossed, even extensively, is unlikely to be small. Without selection, the fraction of loci heterozygous at the Nth generation of backcrossing is (1/2)N−1 (Silver 1995), but loci linked to the knockout are not so readily lost. The probability of a recombination within 10 cM on one side is by definition 10% per generation. Even at N = 10 generations as in this case, the probability that there has been no recombination and thus a flank of ES-cell-derived chromosome of at least 10 cM on a given side would be 63% [100 × (1 − 1/e1)], i.e. one-fold sampling assuming recombination is a Poisson process). Similarly, the probability of having >3.3 cM of flank on one side is 95% [100 × (1 − 1/e3)]. The minimum differential chromosome region at backcross 10 is thus 6.6 cM, which corresponds to roughly 13 Mbp of sequence, with on average 110 genes. The average size is approximately 200/N or 20 cM (Silver 1995), approximately 40 Mbp, 250 genes that could differ from the controls. The congenic footprint we identified in this study is 40 Mbp and contains 573 genes.
The footprint genes with showed expression differences must logically be considered candidates for causing phenotypic differences between the knockout strain and C57BL/6 in addition to Cckbr. The point here is not to show that these differences actually do cause the phenotype in this case, but rather to point out that there is every reason to believe that this situation, in which multiple expression differences must be considered, is typical. A recent paper reports a similar congenic footprint observation with a Rab3a knockout (chromosome 8) (Yang et al. 2006). Gene knockouts are extremely valuable, but are not as clean a system for testing function as is commonly assumed. In this case, the majority of the phenotypic effects attributed to the Cckbr knockout are in accordance with what is known of the biochemistry of the receptor’s ligands (Kurrikoff et al. 2004). The complexity of the pain-related phenotype, and particularly the apparent dominance of the knockout effect on both pain and mechanical sensitivity (Kurrikoff et al. 2004), however, indicates that further investigation of candidates for this phenotype may be fruitful. For characterizing knockouts in general, we believe far more use should be made of genetic tests for congenic effects (Crusio 2004; Gerlai 1996; Wolfer et al. 2002). In future more targeted mutations may be made directly in strains of interest as methods are refined (e.g. Ware et al. 2003). Similar considerations apply of course to congenic strains constructed deliberately as a way of isolating quantitative trait loci.
The expression levels seen in the differential chromosomal segment of the knockout strain may or may not reflect the situation in the sequence’s strain of origin. A striking feature of the congenic footprint is that almost all of the expression differences are low in the knockout. We hypothesize that there is a mismatch between coevolved sets of interacting sequences in the two parental inbred strains, for example transcription factors and proteins that activate them, that leads to this bias. Sets of genes like this will have diverged in the different subspecies that are combined in the ancestry of the domestic mouse, and working combinations were probably selected during inbreeding (Petkov et al. 2005). If this proves to be true, the phenomenon could prove very useful for exploring the regulation of transcription of many genes, and it would be interesting to investigate strains created from various proportions of different inbred strains such as congenics, recombinant congenic and consomic panels. A recent analysis of recombinant congenic lines derived from strains A/J and C57BL/6J found over 1200 genes in the genome as a whole whose expression was affected by the predominant genetic background, although no directional bias in expression was reported (Lee et al. 2006). This is a similar proportion to our observations, because our congenic footprint is roughly 1% of the genome. Analysis of consomic strain panels would also be very interesting in this context. One such study using liver has appeared (Shockley & Churchill 2006).
Acknowledgments
This work was supported by the Estonian Science Foundation GARFS5688, by the Marie Curie TOK fellowship MTKD-CT-2004-517176 (S.K.) and the UK Medical research Council (G0000170). Comments from the editors and a reviewer were very helpful.
References
- Austin CP, et al. The knockout mouse project. Nat Genet. 2004;36:921–924. doi: 10.1038/ng0904-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auwerx J, et al. The European dimension for the mouse genome mutagenesis program. Nat Genet. 2004;36:925–927. doi: 10.1038/ng0904-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y. Controlling the false discovery rate – a practical and powerful approach to multiple testing. J Roy Stat Soc B Met. 1995;57:289–300. [Google Scholar]
- Bolivar VJ. Mapping of quantitative trait loci with knockout/congenic strains. Genome Res. 2001;11:1549–1552. doi: 10.1101/gr.194001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook-Mills JM. Active participation of endothelial cells in inflammation. J Leukoc Biol. 2005;77:487–495. doi: 10.1189/jlb.0904554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crusio WE. Flanking gene and genetic background problems in genetically manipulated mice. Biol Psychiatry. 2004;56:381–385. doi: 10.1016/j.biopsych.2003.12.026. [DOI] [PubMed] [Google Scholar]
- Gerlai R. Gene-targeting studies of mammalian behavior: is it the mutation or the background genotype? Trends Neurosci. 1996;19:177–181. doi: 10.1016/s0166-2236(96)20020-7. [DOI] [PubMed] [Google Scholar]
- Ihaka R. R: a language for data analysis and graphics. J Comput Graph Stat. 1996;5:299–314. [Google Scholar]
- Kurrikoff K. Deletion of the CCK2 receptor gene reduces mechanical sensitivity and abolishes the development of hyperalgesia in mononeuropathic mice. Eur J Neurosci. 2004;20:1577–1586. doi: 10.1111/j.1460-9568.2004.03619.x. [DOI] [PubMed] [Google Scholar]
- Lee PD. Mapping cis-acting regulatory variation in recombinant congenic strains. Physiol Genomics. 2006;25:294–302. doi: 10.1152/physiolgenomics.00168.2005. [DOI] [PubMed] [Google Scholar]
- Li C. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata A. G protein-coupled cholecystokinin-B/gastrin receptors are responsible for physiological cell growth of the stomach mucosa in vivo. Proc Natl Acad Sci USA. 1996;93:11825–11830. doi: 10.1073/pnas.93.21.11825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkov PM. Evidence of a Large-Scale Functional Organization of Mammalian Chromosomes. PLoS Genet. 2005;1:e33. doi: 10.1371/journal.pgen.0010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poltorak A. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- Shockley KR. Gene expression analysis of mouse chromosome substitution strains. Mamm Genome. 2006;17:598–614. doi: 10.1007/s00335-005-0176-y. [DOI] [PubMed] [Google Scholar]
- Silver L. Mouse Genetics. New York: Oxford University Press; 1995. [Google Scholar]
- Simpson EM. Genetic variation among 129 substrains and its importance for targeted mutagenesis in mice. Nat Genet. 1997;16:19–27. doi: 10.1038/ng0597-19. [DOI] [PubMed] [Google Scholar]
- Storey JD. Strong control, conservative point estimation, and simultaneous conservative consistency of false discovery rates: a unified approach. J R Stat Soc B. 2004;66:187–205. [Google Scholar]
- Storey JD. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanga FY. The CNS role of Toll-like receptor 4 in innate neuroimmunity and painful neuropathy. Proc Natl Acad Sci USA. 2005;102:5856–5861. doi: 10.1073/pnas.0501634102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware CB. Utility of a C57BL/6 ES line versus 129 ES lines for targeted mutations in mice. Transgenic Res. 2003;12:743–746. doi: 10.1023/b:trag.0000005246.35812.c8. [DOI] [PubMed] [Google Scholar]
- Wolfer DP. Knockout mice: simple solutions to the problems of genetic background and flanking genes. Trends Neurosci. 2002;25:336–340. doi: 10.1016/s0166-2236(02)02192-6. [DOI] [PubMed] [Google Scholar]
- Yang S. Biochemical, molecular and behavioral phenotypes of Rab3A mutations in the mouse. Genes Brain Behav. 2006;6:77–96. doi: 10.1111/j.1601-183X.2006.00235.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33:W741–748. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]