Abstract
Activation of β-catenin has been causatively linked to the etiology of colon cancer. Conditional stabilization of this molecule in pro-T cells promotes thymocyte development without the requirement for pre-TCR signaling. We show here that activated β-catenin stalls the developmental transition from the double-positive (DP) to the single-positive (SP) thymocyte stage and predisposes DP thymocytes to transformation. β-Catenin–induced thymic lymphomas have a leukemic arrest at the early DP stage. Lymphomagenesis requires Rag activity, which peaks at this developmental stage, as well as additional secondary genetic events. A consistent secondary event is the transcriptional up-regulation of c-Myc, whose activity is required for transformation because its conditional ablation abrogates lymphomagenesis. In contrast, the expression of Notch receptors as well as targets is reduced in DP thymocytes with stabilized β-catenin and remains low in the lymphomas, indicating that Notch activation is not required or selected for in β-catenin–induced lymphomas. Thus, β-catenin activation may provide a mechanism for the induction of T-cell–acute lymphoblastic leukemia (T-ALL) that does not depend on Notch activation.
Introduction
The canonical β-catenin/TCF-LEF signaling is stimulated by wnts, a family of secreted cysteine-rich glycoproteins that bind to cell surface Frizzled receptors. In unstimulated cells, newly synthesized β-catenin is captured by a large cytoplasmic complex consisting of the tumor suppressor adenomatous polyposis coli (APC), the constitutively active kinase glycogen synthase kinase 3β (GSK-3β), and Axin. In this complex, β-catenin is phosphorylated by GSK-3β at 4 N-terminal serine and threonine residues and targeted for degradation.1–4 Activation of the Wnt/β-catenin cascade results in inhibition of the constitutive activity of GSK-3β5 by the cytoplasmic protein Dishevelled (Dvl).6–9 Consequently, β-catenin is no longer phosphorylated and can accumulate in the cytoplasm and nucleus. Once in the nucleus, β-catenin binds to members of the TCF/LEF family of transcription factors, the most downstream components of the Wnt-signaling pathway. For reviews, see van de Wetering10 and Staal and Clevers.11
The Wnt/β-catenin signaling cascade has been implicated in multiple stages of hematopoietic development. It was proposed that Wnt signaling controls the self-renewal of hematopoietic stem cells (HSCs).12 More recently it was shown that deregulated activation of this pathway enforced cell cycle entry in HSCs leading to the exhaustion of the long-term stem cell pool and a multilineage developmental block.13,14 In the thymus, loss- and gain-of-function studies have indicated that at least 2 stages of thymopoiesis require Wnt/β-catenin signaling. Thymocytes express the TCF-1 and LEF-1 effectors of the canonical Wnt/β-catenin signaling pathway. Ablation of TCF-1 activity affects all proliferating stages of thymocyte development including the CD44+CD25+ DN2 and the pre-TCR–dependent CD44−CD25− DN4 and CD8+TCRβ− immature single-positive (ISP) stage.15 Enforced expression of inhibitors of Wnt signaling such as soluble Frizzled or Dickkopf proteins in T-cell progenitors leads to a specific block in the DN1 to DN2 developmental transition.16,17 The concomitant ablation of LEF-1 and a TCF-1 hypomorph results in a complete block of embryonic thymocyte development at the ISP stage.18 The TCF-1−/− developmental block at the DN4 stage is β-catenin dependent since it can be relieved only by transgenic reconstitution with versions of TCF-1 that contain an intact β-catenin–binding domain.19 Conditional ablation of β-catenin20 or down-regulation of its activity by the expression of the inhibitor ICAT21 impacts negatively the DN to DP thymocyte transition. On the other hand, we have shown that conditional stabilization of β-catenin, at the DN3 stage of thymocyte development, promotes aberrant development of DP and SP thymocytes that have reduced TCRβ gene VDJ type rearrangements and Notch activity, and are devoid of pre-TCR and αβTCR.22,23
β-Catenin has only recently been implicated in the etiology of hematopoietic malignancies by studies showing that up-regulation of this signaling pathway marks the cancer stem cell for chronic myelogenous leukemia.24 Stabilization of β-catenin was also linked to B-cell chronic lymphocytic leukemia.25 Earlier studies linked genes targeted by β-catenin signaling in the etiology of leukemogenesis. Thus, up-regulation of the Wnt/β-catenin target c-Myc is probably the most uniform feature of hematopoietic malignancies and in particular Burkitt lymphomas,26 as well as T-cell acute lymphoblastic leukemia (T-ALL) of the HOX11 subtype.27 Transgenic mice overexpressing c-Myc develop both B- and T-cell lymphomas.28–31 c-Myb, another Wnt/β-catenin target was shown to cooperate with c-Myc to cause T-cell lymphoma in transgenic mice.32 FRAT1 activity, which leads to the stabilization of β-catenin, synergizes with c-Myc to promote T-cell leukemogenesis.33
Perhaps the most striking recent finding in T-ALL was the detection of Notch activating mutations in more than 50% of the examined cases.34 Notch activating mutations were found to superimpose with the activation of other T-ALL–related oncogenic pathways and to span the entire spectrum of the identified T-ALL subtypes.34 Notch appears to be a key player in mouse models of T-cell leukemia as well. Transgenic expression of activated Notch135 or Notch336 mediates thymocyte transformation that is linked with the modulation of pre-TCR signaling, inhibition of the E2A pathway, and up-regulation of c-Myc, as well as E2A-PBX.37 Conversely, lymphomas induced in E2A- or P53-deficient mice or in mice expressing activated Tal1/SCL appear to induce activation of Notch or select for secondary Notch-activating mutations.38–42 The monoclonal nature of these leukemias points to the requirement for additional genetic events. Although activation of Notch1 is a frequent event in human T-ALL as well as mouse lymphoma models, it does not constitute the common denominator for this disease. This is emphasized by the substantial fraction of T-ALL samples that do not show activation of Notch, and the requirement for secondary events in the Notch-dependent animal models of leukemia. It is therefore essential to evaluate pathways of lymphomagenesis that do not depend on Notch.
In this report, we provide evidence for the implication of β-catenin activation in lymphomagenesis. Activation of β-catenin stalled the developmental transition of DP thymocytes to the SP stage and predisposed these cells to transformation, which required Rag activity, expression of c-Myc, as well as additional genetic events. Significantly, β-catenin–mediated transformation did not require Notch activation. Our observations indicate that deregulated activation of β-catenin may be etiologically linked to T-ALL and suggest that β-catenin may be a suitable target for therapy of a subset of leukemias in which Notch is not activated.
Materials and methods
Mice
The CtnnbΔex3 mice43 were crossed with LckCre or CD4Cre transgenic mice44 to produce LckCre-CtnnbΔex3 and CD4Cre-CtnnbΔex3 compound mutant mice. LckCre-CtnnbΔex3-Rag2−/− mice have been described before.22 LckCre-CtnnbΔex3-TCR(CL4)-Rag2−/− and LckCre-CtnnbΔex3-TCR(6.5)-Rag2−/− were generated by crossing LckCre-CtnnbΔex3-Rag2−/− mice transgenic TCR-6.545 or TCR-CL4 mice.46 CD4Cre-CtnnbΔex3-pTα−/− and CD4Cre-CtnnbΔex3-Mycfl/fl mice were generated by crossing CD4Cre-CtnnbΔex3 with pTα−/−47 or c-Mycfl/fl mice, respectively.48
Flow cytometry and antibodies
FITC-, PE-, CyChrome-, or APC-conjugated antibodies were purchased from BD PharMingen (San Diego, CA). Antibodies used were as follows: anti–CD4-FITC, -CyChrome, -PE, or -APC (RM4-5); anti–CD8-FITC, -CyChrome, -PE, or -APC (53.6.7); anti–TCRβ-PE or -CyChrome (H57); anti–B220-CyChrome (RA3-6B2) or -CD19-PE (1D3); anti–CD44-FITC (IM7) or –PE (IM7); anti–CD25-APC (PC61); anti–pan NK-PE or biotinylated (DX5); anti–Gr1-PE or biotinylated (RB6.782); anti–γδ-PE or biotinylated; and anti–Ter-PE or biotinylated (Ly-76). Anti–β-catenin-FITC was from BD Transduction Laboratories (Lexington, KY). Biotinylated antibodies were revealed with streptavidin-PE or streptavidin-CyChrome. Staining was in fluorescence-activated cell sorter (FACS) buffer (PBS with 2% fetal bovine serum) for 30 minutes on ice. Samples were washed with FACS buffer. Intracellular staining for β-catenin or 7AAD was performed after surface staining according to Ioannidis.19 For intracellular β-catenin staining, thymocytes were fixed in 2% paraformaldehyde (PFA) for 15 minutes and stained for 45 minutes on ice with anti–β-catenin-FITC in FACS/0.1% saponin (FACS buffer supplemented with 0.1% saponin), before washing, and resuspending in FACS/0.1% saponin. A Cyan-flow cytometer (DakoCytomation, Fort Collins, CO) was used to analyze cells. Data were analyzed using the FlowJo software (Tree Star, Ashland, OR).
BrdU labeling
Mice were injected intraperitoneally with 1.8 μg freshly resuspended BrdU in water every day. BrdU was also supplied in their drinking water at 0.8 μg/mL, and fresh solutions were provided daily. Mice were processed to estimate BrdU incorporation 2 hours after injection. Thymocytes from BrdU-treated mice were surface stained, permeabilized, and stained intracellularly using the BrdU labeling kit from BD PharMingen.
In vivo tumor induction
Lymphoma or thymocyte suspensions were prepared aseptically. Cells (2 × 105) were injected in the tail vein of sublethally irradiated (4 Gy) Rag2−/−γc−/− mice (Taconic Farms, Germantown, NY). The mice were bled weekly starting at 2 weeks after injection, and the presence of CD4+CD8+ DP cells in their blood was investigated by flow cytometry. The health of the mice was monitored daily after the first bleeding with respect to body weight.
Northern blot analysis
Total RNA was isolated from LckCre, LckCre-CtnnbΔex3 and CD4Cre-CtnnbΔex3 thymocytes or from tumors using TRIZOL reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Total RNA (7 μg) was separated on a 1% formaldehyde gel and blotted to Hybond-N+ Nylon membrane (Amersham Biosciences, Arlington Heights, IL). Probes were generated by reverse-transcription–polymerase chain reaction (RT-PCR) using the following primers: c-Myc-F, TGCGATCCTGACGACGAGACCT; c-Myc-R, GGTGGGCGGTGTCTCCTCATGC; ID2-F, TCTGAGCTTATGTCGAATGATAGC; ID2-R, CACAGCATTCAGTAGGCTCGTGTC; ID3-F, CGCACTGTTTGCTGCTTTAGG; ID3-R, GTAGCAGTGGTTCATGTCGTC; P21-F, CAGATCCACAGCGATATCCA; P21-R, GCAGGCAGCGTATATACAGGA; P27-F, AGCCTGGAGCGGATGGACGC; P27-R, CACCTTGCAGGCGCTCTTGG; Rag2-F, CACATCCACAAGCAGGAAGTACAC; Rag2-R, GGTTCAGGGACATCTCCTACTAA; HPRT-F, GTTCTTTGCTGACCTGCTGG; HPRT-R, TGGGGCTGTACTGCTTAACC; Tal1-F, CTCCGGGTACCTTGATATTTG; Tal1-R, AACTTTTATTTAGAAATCTTCCCCA. Probes were labeled with α-[32P]−dCTP using Klenow polymerase, purified with Micro P-30 Chromatography Columns (BioRad, Hercules, CA).
Southern blot analysis
DNA (10 μg) was digested with EcoRI overnight at 37°C, separated on a 0.8% agarose gel, and blotted onto nitrocellulose. The blots were hybridized with a P32-labeled probe from the Jβ2 region of the TCRβ gene.
Microarray analysis
Thymocytes or tumor masses were homogenized for RNA isolation using the TRIZOL reagent (Invitrogen) and purified using Qiagen RNeasy minicolumns. RNA samples were processed for hybridization on the Mouse Expression-Array-430 Genechips (Affymetrix, Santa Clara) by the microarray core facility of the Dana-Farber Cancer Institute. Data from all 18 microarrays were loaded onto the DNA-Chip Analyzer (dChip, http://www.dchip.org) program for normalization and quantification. Normalization was performed using the default settings of the dChip software, which is based on perfect match (PM)/mismatch (MM) difference for each gene in each sample.49 The expression values were quantified using the PM-only model and genes were filtered according to 0.30 less than standard deviation/mean less than 10.00 and P call in the array used more than 50%. After filtering, genes were selected for expression differences higher than 3-fold by t test (P < .05) or for significant changes (P < .05).
Semiquantitative RT-PCR
cDNA was prepared by oligo-dT priming using the Superscript II RT kit (Invitrogen). cDNAs were equilibrated by real-time β-actin PCR using SYBR-green PCR master mix (Applied Biosystems, Foster City, CA) and an OpticonII (BioRad) PCR machine. Serial 1:5 dilutions were used. PCR products were separated on 1.2% agarose gels. Primers were as follows: β-actin-F, TGGAATCCTGTGGCATCCATGAAAC; β-actin-R, TAAAACGCAGCTCAGTAACAGTCCG; Notch1-F, CGGTGTGAGGGTGATGTCAATG; Notch1-R, GAATGTCCGGGCCAGCGCCACC; Notch3-F, GAGGCTACCTTGGCTCTGCT; Notch3-R, GGCAGCCTGTCCAAGTGATCT; Deltex1-F, CCCTCGCCACTGCTACCTA; Deltex1-R, AAAGGGAAGGCGGGCAACTC; Hes1-F, CAGCCAGTGTCAACACGACAC; Hes1-R, TCGTTCATGCACTCGCTGAG; Lunatic-Fringe-F, TTCATGAGCACGGCAGAGCGCATCC; Lunatic-Fringe-R, TCCTGTCCAGAATGGCAGCCTGTGG.
Western blot
Thymocytes were lysed in radioimmunoprecipitation assay (RIPA) buffer supplemented with Protease Inhibitor Cocktail (Roche Applied Science, Indianapolis, IN). Lysates (80 μg) were separated on 10% SDS-PAGE and transferred onto nitrocellulose membranes (GE-Healthcare Bio-Sciences, Piscataway, NJ). Nonspecific binding was blocked by incubation in blocking buffer (5% nonfat milk in PBST), followed by incubation with the primary antibodies and the appropriate horseradish peroxidase (HRP)–conjugated secondary antibodies diluted in blocking buffer. Antimouse anti–β-catenin was from BD-Biosciences (Franklin Lakes, NJ), and anti-GAPDH (1:3000) was from Abcam (Cambridge, United Kingdom). The signal was detected using the enhanced chemiluminescence Plus (ECL-Plus kit; GE-Healthcare Bio-Sciences).
Results
Stabilization of β-catenin induces a developmental block in the DP to SP transition
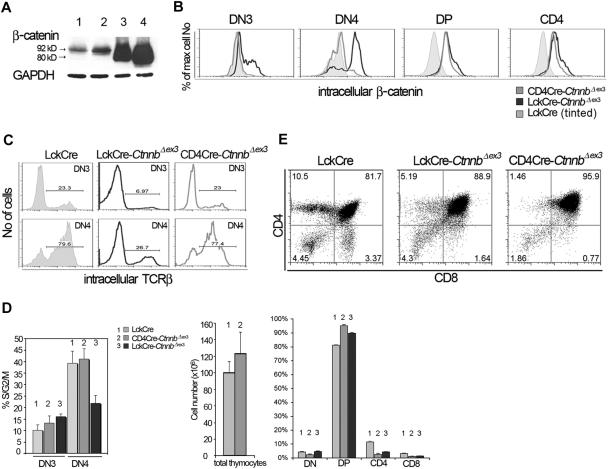
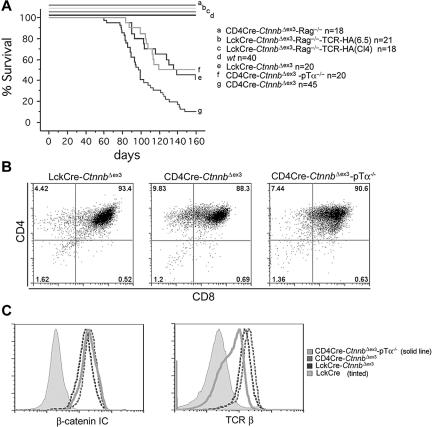
We have addressed the role of β-catenin in thymocyte development using a mouse model (CtnnbΔex3) that allows conditional Cre-mediated deletion of the third exon of the β-catenin gene, which includes critical phosphorylation sites involved in the degradation of the protein.43 Crossing to LckCre mice resulted in β-catenin activation starting at the DN3 stage of thymocyte development and led to the maturation of thymocytes without pre-TCR or αβ-TCR as well as the reduction in the fraction of SP thymocytes.22,23 This reduction could either result from the selective elimination of αβ-TCR–deficient DP thymocytes in these mice or it could be directly related to the activation of β-catenin signaling.
To distinguish between these 2 possibilities, we crossed the CtnnbΔex3 allele onto transgenic mice expressing Cre under the control of the CD4 gene promoter. Cre-mediated β-catenin stabilization in CD4Cre-CtnnbΔex3 thymi was detectable at the DP stage (Figure 1A-B). As a result, the fraction of CD4Cre-CtnnbΔex3 DN4-stage thymocytes expressing intracellular TCRβ chains and able to assemble the pre-TCR (77.4%) was comparable to that of the respective LckCre control cells (79.6%) (Figure 1C). Moreover, the proliferation rate of CD4Cre-CtnnbΔex3 DN4 stage cells analyzed by intracellular staining with the DNA dye 7AAD was comparable to that of the respective LckCre control cells (41.16% [n = 4] and 39.32% [n = 5], respectively, Figure 1D), suggesting that they had an active pre-TCR and were receiving pre-TCR–dependent proliferation signals. This was in contrast to LckCre-CtnnbΔex3 thymocytes that had a reduced fraction of DN4 cells with intracellular TCR-β chains and showed decreased cycling at this stage due to the lack of pre-TCR signaling.22
Figure 1.
Thymocyte development in CD4Cre-CtnnbΔex3 versus LckCre-CtnnbΔex3 mice. (A) β-Catenin protein levels in total thymocyte extracts from LckCre control (lanes 1-2), LckCre-CtnnbΔex3 (lane 3), and CD4Cre-CtnnbΔex3 (lane 4) mice revealed by Western blot analysis. Arrows depict wild-type and mutated β-catenin. Numbers indicate the predicted molecular weight in kilodaltons. (B) Course of β-catenin stabilization during thymocyte development. Thymocytes from the indicated mice were surface stained for expression of CD4 and CD8, or with a cocktail of lineage-specific antibodies (“Materials and methods”) combined with staining for surface expression of CD44 and CD25. Cells were then permeabilized and stained with anti–β-catenin FITC. Histogram overlays of electronically gated cells in the indicated subsets are representative of 4 independent experiments. (C) Intracellular TCRβ expression of DN3- and DN4-stage thymocytes. Thymocytes from the indicated mouse strains were surface stained as in panel B prior to permeabilization and staining for intracellular TCRβ expression (TCRβ-PE). Histograms depict intracellular TCRβ expression in the indicated electronically gated subsets. Numbers in the histograms indicate the fraction of TCRβ-positive cells and are representative of 5 independent experiments. (D) Fraction of DN3- and DN4-stage thymocytes in S/G2/M phases of the cell cycle. Thymocytes were surface stained as in panel B prior to permeabilization and intracellular staining with the DNA binding dye 7AAD to estimate their DNA content. Histogram bars represent the average values of 5 independent experiments; error bars represent standard deviation. (E, upper panels) CD4/CD8 profiles of thymocytes from the indicated mouse strains; the numbers in the quadrants indicate the percentage of cells in each. Profiles are representative of 6 independent experiments. (E, lower panels) Thymic cellularity was estimated in 4- to 8-week-old mice. Percent distribution of the different thymocyte subsets was determined after surface staining for CD4 and CD8. Data represent average values from 6 LckCre, 6 CD4Cre-CtnnbΔex3, and 5 LckCre-CtnnbΔex3. Standard errors are included. While LckCre-CtnnbΔex3 thymi have reduced cellularity,22,23 the cellularity of CD4Cre-CtnnbΔex3 thymi (P = .455) and the number of DPs (P = .2348) were comparable to LckCre controls in contrast to the absolute number of SPs, which was reduced in CD4Cre-CtnnbΔex3 mice (P = .013 for CD4 and P = .007 for CD8). The fraction of DPs significantly increased in CD4Cre-CtnnbΔex3 (P = .001) and LckCre-CtnnbΔex3 (P = .001) compared to LckCre mice. The fraction of SPs was significantly reduced in CD4Cre-CtnnbΔex3 (P = .001 for CD4+ and CD8+) and LckCre-CtnnbΔex3 (P = .001 for CD4+ and CD8+). Reduction in the fraction of SPs was comparable between CD4Cre-CtnnbΔex3 and LckCre-CtnnbΔex3 mice (P = .14 for CD4+ and P = .34 for CD8+).
Although the overall thymic cellularity of CD4Cre-CtnnbΔex3 mice was comparable to that of LckCre control mice, like LckCre-CtnnbΔex3 mice they had a reduced number of SP thymocytes (P = .013 for CD4+ and P = .007 for CD8+). The average fraction of CD4+ cells was 2.45% and 4.2% in CD4Cre-CtnnbΔex3 and LckCre-CtnnbΔex3 mice, respectively, and of CD8+ cells was 1% and 1.4% in CD4Cre-CtnnbΔex3 and LckCre-CtnnbΔex3 mice, respectively, indicating that CD4Cre-CtnnbΔex3 and LckCre-CtnnbΔex3 mice had similarly reduced fraction of SP thymocytes (P = .14 for CD4+ and P = .34 for CD8+). This correlated with an increased fraction of CD4+CD8+ DP thymocytes in CD4Cre-CtnnbΔex3 (P = .001) and LckCre-CtnnbΔex3 (P = .001) mice compared to LckCre mice. Thus, the thymic profile of CD4Cre-CtnnbΔex3 and LckCre-CtnnbΔex3 mice suggested that in both instances the stabilization of β-catenin induced a developmental block in the transition from the DP to the SP stage.
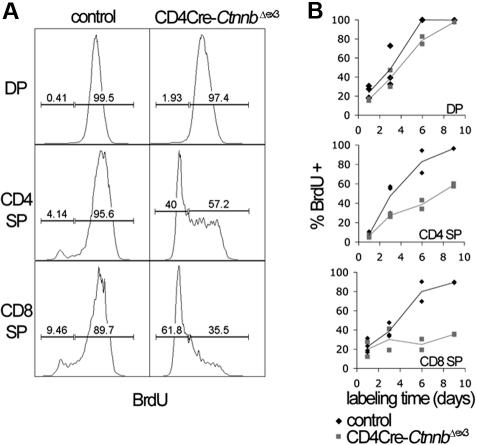
To determine the nature of the developmental block, we compared the rate of generation and loss of DP and SP thymocytes in control and CD4Cre-CtnnbΔex3 thymocytes. To this aim, we carried out incorporation studies with the thymidine analog bromodeoxyuridine (BrdU). Control and CD4Cre-CtnnbΔex3 littermates were injected intraperitoneally with BrdU every 24 hours and were exposed to BrdU in their drinking water. Thymocytes from the treated mice were isolated and analyzed for CD4, CD8, TCRαβ, and BrdU fluorescence after 1, 3, 6, and 9 days (Figure 2). The rapid labeling of DP thymocytes, which was complete in fewer than 6 days, indicates that they had a fast turnover. Most DP cells are eliminated through death by neglect, and only a small fraction of them develop into the nondividing mature (TCRαβ−) CD4 or CD8 SP cells. Therefore, the percentage of BrdU+ mature thymocytes (TCRβ high, CD4+CD8−, or CD4−CD8+) at a given time point represents the fraction of DP cells that have developed into mature SP cells. By labeling with BrdU for 9 days, we detected that although DPs were generated at similar rates in CD4Cre-CtnnbΔex3 compared to control mice (P = .25) the production rate of CD4+ SP (P = .01) and CD8+ SP (P = .05) thymocytes was significantly reduced in CD4Cre-CtnnbΔex3mice (Figure 2B). This indicates that the stabilization of β-catenin suppresses the production rate of mature SP thymocytes.
Figure 2.
Stabilization of β-catenin induces a DP to SP developmental block. (A) The kinetics of developing CD4Cre-CtnnbΔex3 thymocytes were investigated by BrdU incorporation studies. Analysis of the BrdU content in the indicated subsets was done after electronic gating on subsets defined by expression of CD4, CD8, and TCRβ. Histograms are representative of BrdU incorporation at day 9 of treatment. Bars indicate gates for BrdU+ or BrdU− cells and the numbers above the bars indicate percentages of labeled or unlabeled cells in each subset. (B) Turnover of CD4+CD8+ (DP) and production rate of mature single-positive (CD4 SP and CD8 SP) thymocytes. Mature thymocytes are defined as TCRβ hi and either CD4+CD8− or CD4−CD8+. The percentages of labeled cells of control and CD4Cre-CtnnbΔex3 mice are shown. Two to 4 control and CD4Cre-CtnnbΔex3 mice were in each time point.
Stabilization of β-catenin induces T-cell lymphomas
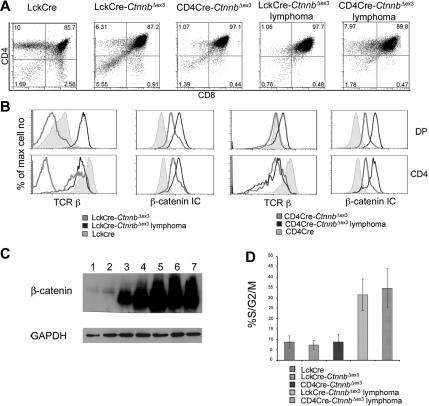
Both LckCre-CtnnbΔex3 and CD4Cre-CtnnbΔex3 mice developed massive highly mitotic thymic lymphomas that occupied the entire thoracic cavity, and overwhelmed the lungs and the heart (Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). Transformed cells were localized in the thymus and were not detectable in peripheral lymphoid organs. Despite the different onset of β-catenin stabilization in LckCre-CtnnbΔex3 and CD4Cre-CtnnbΔex3 mice, lymphomas from both strains were remarkably similar with a CD4+CD8+ (DP) surface phenotype and a few CD4+CD8lo (SP) cells (Figure 3A). Intracellular β-catenin staining and Western blot analyses revealed that the lymphoma cells had higher levels of β-catenin than did LckCre control, or LckCre-CtnnbΔex3 and CD4Cre-CtnnbΔex3 pretransformed thymocytes (Figure 3B-C). Although most Lck-Cre-CtnnbΔex3 DP and SP thymocytes are devoid of αβ-TCR, all the examined lymphomas (n > 10) showed surface expression of αβTCR, indicating that transformation targeted cells that had undergone beta selection and expressed TCRβ and TCRα chains (Figure 3C).
Figure 3.
β-catenin induces T-cell lymphomas. (A) CD4/CD8 profile of β-catenin–dependent lymphomas. Thymocytes from the indicated mice or thymic lymphomas were stained with antibodies against CD4, CD8 and analyzed by FACS. (B) Surface expression of TCRβ and intracellular expression of β-catenin in the indicated subsets analyzed by FACS. Thymocytes were surface stained with antibodies against CD4, CD8 as well as TCRβ. To evaluate intracellular β-catenin expression, surface-stained thymocytes were permeabilized and stained with anti–β-catenin FITC. Profiles presented in panels B-C are representative of 4 independent experiments. (C) Western blot analysis of β-catenin protein expression in total thymocytes isolated from LckCre (lanes 1-2), LckCre-CtnnbΔex3 (lane 3), and CD4Cre-CtnnbΔex3 (lane 4) mice, as well as LckCre-CtnnbΔex3 (lane 5) and CD4Cre-CtnnbΔex3 lymphomas (lanes 6-7). GAPDH was used as loading control. (D) Fraction of DP thymocytes and transformed cells in S/G2/M phase of the cell cycle. Thymocytes or lymphomas were stained with antibodies to CD4, CD8 followed by intracellular staining with 7AAD. DP cells were gated and the percentage of cells in the S/G2/M phase from 4 to 6 independent experiments was averaged and plotted in the bar histograms. Error bars represent standard error values.
Lymphomas in both mouse strains consisted of highly proliferative cells, as measured ex vivo by surface staining for CD4 and CD8 followed by intracellular staining with 7AAD and FACS analysis. More than 30% of the cells in LckCre-CtnnbΔex3 (n = 4) and CD4Cre-CtnnbΔex3 (n = 5) lymphomas were actively dividing compared to less than 10% in LckCre (n = 6) control thymocytes. Proliferation increased sharply upon transformation since pretransformed LckCre-CtnnbΔex3 (n = 7) and CD4Cre-CtnnbΔex3 (n = 6) DP thymocytes (Figure 3D) showed similar proliferation rates as control thymocytes.
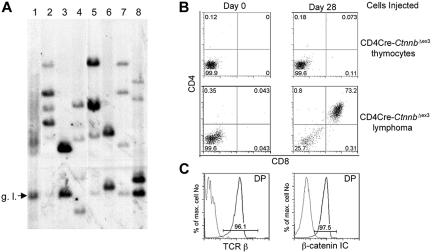
The increased proliferation only after transformation indicated that the process of lymphomagenesis was likely selective for secondary events. To confirm this hypothesis we determined the clonality of β-catenin–induced lymphomas by examining the diversity of TCRβ gene rearrangements. DNA isolated from 3 LckCre-CtnnbΔex3 and 4 CD4Cre-CtnnbΔex3 tumors was used in Southern blot analyses and hybridized with a probe derived from the Jb2 region of the TCRβ gene to determine the status of Db-to-Jb2 and Vb-to-DJb2 rearrangements (“Materials and methods”). In contrast to DNA from an LckCre control thymus showing a heterogeneous mix of bands characteristic of a polyclonal cell population (Figure 4A lane 1), all lymphoma samples had between 2 and 6 distinct bands, indicating that they consisted of 1 to 3 independent clones. This finding suggested that secondary mutations were selected for during lymphomagenesis (Figure 4A).
Figure 4.
β-Catenin lymphomas are malignant as well as oligoclonal. (A) Genomic DNA was prepared from LckCre thymocytes (lane 1), LckCre-CtnnbΔex3 lymphomas (lanes 2-4), and CD4Cre-CtnnbΔex3 lymphomas (lanes 5-8) and digested with EcoRI. Digested DNAs were electrophoresed through agarose gels and blotted onto nitrocellulose. Southern blots were probed with a P32-labeled 1.2-kb EcoRI-ClaI genomic fragment recognizing the Jβ2 region of the TCRβ gene locus (“Materials and methods”). g.l. indicates the fragment expected for the germ-line TCRβ gene configuration. (B) CD4Cre-CtnnbΔex3 thymocytes or CD4Cre-CtnnbΔex3– and LckCre-CtnnbΔex3–derived lymphomas (2 × 105 cells) were injected into sublethally irradiated Rag2−/−γc−/− double knock-out mice by tail vein injection. Injected mice were bled on the day of the injection and then weekly starting at 2 weeks after injection. CD4/CD8 profiles of white blood cells from injected Rag2−/−γc−/− mice. Profiles are representative of 2 independent transfer experiments involving 9 recipients and 3 independent tumors (2 CD4Cre-CtnnbΔex3 and 1 LckCre-CtnnbΔex3). (C) Expression of TCRβ (black) compared to isotype control (gray) or β-catenin (black) compared to isotype control (gray) in DP splenocytes at 5 weeks after adoptive transfer.
The malignant nature of the β-catenin–induced lymphomas, and their ability to grow autonomously and invade the organs of secondary recipients, was examined by transferring 2 × 105 tumor cells or a similar number of pretransformed thymocytes from CD4Cre-CtnnbΔex3 mice to sublethally irradiated Rag2−/−γc−/− recipients (4 Gy). Three independent lymphomas (2 CD4Cre-CtnnbΔex3 and 1 LckCre-CtnnbΔex3) were injected into 9 independent recipients. Tumor-derived CD4+CD8+ DP cells were detectable in the blood of all mice injected with lymphoma cells but not those receiving pretransformed CD4Cre-CtnnbΔex3 thymocytes 3 weeks after transfer (Figure 4B). These DP cells were of donor origin showing surface expression of αβTCR and high levels of β-catenin (Figure 4C). Recipient mice showed signs of poor health 3 to 5 weeks after transfer and were killed. Histologic analyses indicated that lymphoma cells had invaded most of the recipient's tissues including spleen, bone marrow, liver, lungs, and brain (data not shown). Thus, β-catenin–dependent lymphomas were malignant and could cause leukemia when transferred to healthy recipients.
β-Catenin–mediated lymphomagenesis depends on Rag activity
The incidence and latency of leukemogenesis varied significantly among the different mouse strains with stabilized β-catenin. The median latency of leukemia in LckCre-CtnnbΔex3 mice was 114 days and the incidence 66%, whereas in CD4Cre-CtnnbΔex3 mice it was 99 days and 88% (Figure 5A). Statistical analyses using the log-rank test indicated that the tumor-free survival curves between CD4Cre-CtnnbΔex3 and LckCre-CtnnbΔex3 were significantly different (P = .007). One major difference between CD4Cre-CtnnbΔex3 and LckCre-CtnnbΔex3 thymocytes is that LckCre-CtnnbΔex3 thymi contain fewer αβ-TCR+ DP cells, which appear to be selectively targeted for β-catenin–dependent thymocyte transformation. Leukemias did not develop in LckCre-CtnnbΔex3-Rag2−/− mice in which stabilization of β-catenin induced progression to the DP stage in the complete absence of Rag activity as well as pre-TCR or αβTCR.22 These 2 findings indicated that pre-TCR/αβTCR signaling and/or Rag activity may be needed for β-catenin–dependent thymocyte transformation.
Figure 5.
β-Catenin–mediated lymphomagenesis requires Rag activity. (A) Kaplan-Meyer tumor-free survival curves of LckCre-CtnnbΔex3, CD4Cre-CtnnbΔex3, LckCre-CtnnbΔex3-TCR(CL4)-Rag2−/−, LckCre-CtnnbΔex3-TCR(6.5)-Rag2−/−, and CD4Cre-CtnnbΔex3-pTα−/− mice. Mice were observed for 160 days after birth. The median latency of lymphoma development in LckCre-CtnnbΔex3, CD4Cre-CtnnbΔex3, and CD4Cre-CtnnbΔex3-pTα−/− mice was 99, 114, and 104 days, respectively, with a penetrance of 88%, 66%, and 53%, respectively. Log-rank tests indicated that CD4Cre-CtnnbΔex3 and LckCre-CtnnbΔex3 mice had significantly different disease-free survival curves (P = .007), while LckCre-CtnnbΔex3 and CD4Cre-CtnnbΔex3-pTa−/− mice have similar disease-free survival curves (P = .88). No lymphomas developed in LckCre-CtnnbΔex3-Rag2−/−, LckCre-CtnnbΔex3-TCR(CL4)-Rag2−/−, and LckCre-CtnnbΔex3-TCR(6.5)-Rag2−/− mice during the same period. (B) CD4/CD8profiles of lymphomas from the indicated mice. (C) (Histogram overlay left) Intracellular levels of β-catenin. (Histogram overlay right) Surface expression of TCRβ. The profiles presented are representative of 3 independent experiments.
To distinguish between the requirement for pre-TCR/αβTCR or Rag activity, we generated Rag-deficient mice that expressed LckCre-CtnnbΔex3 as well as the MHC class II–selected transgenic TCR-6.545 or the MHC class I–selected transgenic TCR-CL446 (LckCre-CtnnbΔex3-TCR(CL4)-Rag2−/− and LckCre-CtnnbΔex3-TCR(6.5)-Rag2−/−). These animals express transgenic αβTCRs that recognize epitopes of the influenza hemagglutinin (HA) protein. Thymocytes from these compound mutant mice progressed to the DP and SP stages through signaling from the pre-TCR and the transgenic αβTCRs (data not shown). We also generated mice lacking the pre-TCR (pTα−/−) but expressing CD4Cre and CtnnbΔex3(CD4Cre-CtnnbΔex3-pTα−/−), and therefore stabilized β-catenin in the few thymocytes that bypass the pre-TCR–dependent DN3 block and reach the DP stage. While LckCre-CtnnbΔex3-TCR(CL4)-Rag2−/− and LckCre-CtnnbΔex3-TCR(6.5)-Rag2−/− mice did not develop leukemia (Figure 5A), CD4Cre-CtnnbΔex3-pTα−/− mice developed thymic lymphomas. The latency (average latency: 101 days) and incidence of lymphomagenesis were similar to that of LckCre-CtnnbΔex3 mice (P = .88 by log-rank test), while CD4Cre-CtnnbΔex3 mice showed a reduced incidence and latency. Both LckCre-CtnnbΔex3 and CD4Cre-CtnnbΔex3-pTα−/− strains have a reduced subset of αβTCR+ DP cells compared to CD4Cre-CtnnbΔex3 mice, supporting the notion that although αβTCR may not be sufficient (ie, in the absence of Rag there is no transformation) it may be necessary for transformation.
Of interest, the leukemic profile of the CD4Cre-CtnnbΔex3-pTα−/− mice was similar to that of CD4Cre-CtnnbΔex3 and LckCre-CtnnbΔex3 mice, including a DP surface phenotype, αβTCR expression, and high levels of β-catenin (Figure 5B-C). These findings indicated that Rag activity was needed for transformation, while αβTCR signaling was not sufficient, and pre-TCR signaling was neither necessary nor sufficient to assist β-catenin in transformation.
β-catenin–dependent lymphomagenesis does not require Notch activation
To determine expression changes associated with stabilization of β-catenin and the resulting transformation, we compared the expression profiles of thymocytes from LckCre and CD4Cre-CtnnbΔex3 mice as well as lymphomas. RNA from 5 independent control thymi and 5 independent CD4Cre-CtnnbΔex3 thymi prior to transformation as well as from 8 independent CD4Cre-CtnnbΔex3–derived lymphomas was labeled and hybridized to the Mouse Expression Array 430 2.0 microarray chip from Affymetrix. The predominant population of cells in all the samples was the DP subset, which represented 82% to 85% of the control samples, 90% to 94% of the CD4Cre-CtnnbΔex3 samples, as well as 92% to 96% of the CD4Cre-CtnnbΔex3 lymphomas. Both the DP and CD4 subsets in the lymphomas consisted of highly proliferative transformed cells (Figure 3D and data not shown), indicating that the tumor load exceeded 89%. Thus, expression profiles obtained from these analyses are expected to represent the DP stage (ie, the phenotypic stage of development of the lymphomas).
We compared the obtained microarray data using the DNA-Chip Analyzer (dChip) software. Samples were normalized and filtered according to the default parameters of the program and genes with more than 3-fold expression differences were determined. Comparison of LckCre controls to CD4Cre-CtnnbΔex3 pretransformed thymocytes yielded 100 such genes, while comparisons of CD4Cre-CtnnbΔex3 pretransformed thymocytes to lymphomas yielded 244 genes (Figure S2; Tables S1–S2). Differential expression of genes detected by microarray analyses of unpurified samples needs careful interpretation and validation. Thus, significant differences in expression levels identified by these analyses in gene targets of the Wnt/β-catenin pathway or genes involved in proliferation, TCR signaling, or implicated in human T-ALL were confirmed by Northern blots, semiquantitative RT-PCR, or FACS analyses. Any further discussion of the microarray data focuses on gene expression changes that have been validated by additional methods.
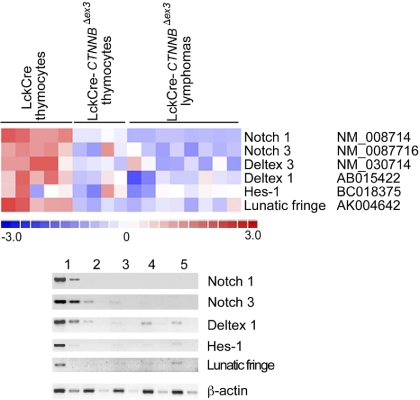
An intriguing observation from the microarray analyses was the more than 3-fold reduction in the levels of Notch1 expression in thymocytes with stabilized β-catenin and the resulting lymphomas. The Notch-positive regulator Lunatic-Fringe was also in the list of genes with more than 3-fold reduced expression in thymocytes with stabilized β-catenin (Tables S1–S2). This was in line with our previous observation that DN4-stage thymocytes with activated β-catenin have down-regulated Notch signaling.23 However, Notch1-activating mutations have been detected in more than 50% of all examined cases of human T-ALL.34 To determine Notch-dependent expression in β-catenin–induced lymphomas, we interrogated the microarray analyses for significant expression changes (P < .05) in members of the Notch family as well as target genes.50–52 These analyses indicated that the expression of Notch1 and Notch3, as well as the targets Deltex-1, Hes-1, and the positive regulator Lunatic-Fringe, was reduced in CD4Cre-CtnnbΔex3pretransformed thymocytes as well as lymphomas when compared to LckCre controls (Figure 6A). This information was confirmed by semiquantitative RT-PCR analyses using RNA extracted from sorted control and CD4Cre-CtnnbΔex3 DP thymocytes as well as 3 independent CD4Cre-CtnnbΔex3 lymphomas (Figure 6B). To further establish the integrity of the Notch1 gene in β-catenin–mediated lymphomas, we sequenced exons 26 and 27 encoding the heterodimerization domain and exon 34 encoding the PEST domain of the Notch1 protein. Both regions are frequently found mutated in T-ALL samples leading to oncogenic Notch activation. Sequencing of DNA from 10 independent β-catenin–induced lymphomas confirmed that Notch1 was not mutated (data not shown). Our findings indicated that β-catenin–induced transformation did not depend or benefit from Notch activation, and segregated the β-catenin–induced lymphomas from those that are etiologically linked to Notch activation.
Figure 6.
β-Catenin lymphomas do not show Notch activation. (A) Illustration of the expression of Notch family members and target genes that show significant (P < .05) expression changes when comparing microarray data from control LckCre to CD4Cre-CtnnbΔex3 pretransformed thymocytes or the resulting lymphomas. Expression changes are color coded; red indicates up-regulation and blue indicates down-regulation. Columns represent independent RNA preparations as indicated. Rows are independent genes; the identity and accession number of the genes is indicated. (B) Semiquantitative RT-PCR of Notch1, Notch3, Deltex1, Hes1, and Lunatic-Fringe transcription (“Materials and methods”). RNA was isolated from sorted DP thymocytes of LckCre control (lane 1), and CD4Cre-CtnnbΔex3 thymocytes (lane 2), as well as 3 independent CD4Cre-CtnnbΔex3 lymphomas (lanes 3-5). RT-PCR for β-actin was used to equilibrate the samples. Two 5-fold serial dilutions are shown.
c-Myc up-regulation marks β-catenin lymphomas and is required for transformation
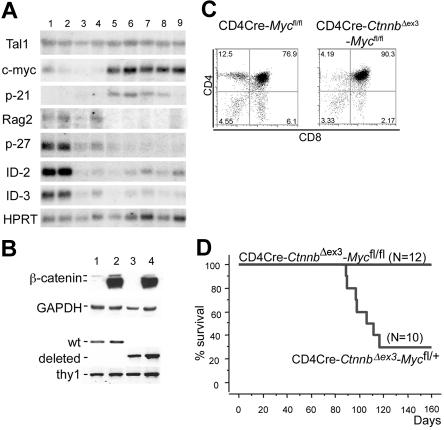
Additional expression changes were detected by Northern blot analyses using RNAs extracted from LckCre, LckCre-CtnnbΔex3, CD4Cre-CtnnbΔex3 thymocytes as well as from 2 LckCre-CtnnbΔex3 and 3 CD4Cre-CtnnbΔex3 (Figure 7A) lymphomas. Deregulation of β-catenin resulted in reduced expression of p27KIP, ID2, and ID3, both before and after transformation. The most consistent lymphoma-specific expression change detected in all examined samples including the microarray analyses as well as in the Northern blots was the up-regulation of c-Myc (arrays [n = 8]; Northern blot [n = 5]). p-21CIP/WAF was found up-regulated in most lymphomas examined by Northern blot analysis but appeared highly variable in the microarray assays (Figure 7A; Table S2).
Figure 7.
c-Myc is induced during β-catenin lymphomagenesis and it is required for transformation. (A) Northern blot analyses using RNAs extracted from LckCre (lanes 1-2), LckCre-CtnnbΔex3 (lane 3), CD4Cre-CtnnbΔex3 (lane 4) thymocytes, as well as from 2 independent LckCre-CtnnbΔex3 (lanes 5-6) and 1 CD4Cre-CtnnbΔex3 (lanes 7-9) lymphomas. Blots were hybridized with P32-labeled probes for the indicated genes (“Materials and methods”). (B) Western blot analysis showing stabilization of β-catenin and DNA-PCR showing deletion of c-Myc floxed allele in sorted DP thymocytes from CD4Cre (lane 1), CD4Cre-CtnnbΔex3 (lane 2), CD4Cre-Mycfl/fl (lane 3), and CD4Cre-CtnnbΔex3-Mycfl/fl (lane 4) mice. (C) CD4/CD8 profiles of CD4Cre-Mycfl/fl and CD4Cre-CtnnbΔex3-Mycfl/fl thymi. (D) Kaplan-Meyer tumor-free survival curves of CD4Cre-CtnnbΔex3-Mycfl/+ and CD4Cre-CtnnbΔex3-Mycfl/fl mice. The median latency of lymphoma was 101 days and the incidence of lymphoma was 70% in CD4Cre-CtnnbΔex3-Mycfl/+ mice.
Since c-Myc has been found up-regulated in a variety of leukemias and its transgenic overexpression was shown to lead to T- and B-cell lymphomas in mice, we sought to determine the requirement for c-Myc activity in β-catenin lymphomagenesis. To this aim, we crossed mice that allow Cre-mediated ablation of c-Myc onto the CD4Cre-CtnnbΔex3 background.48 The resulting compound mutant mice showed efficient and simultaneous deletion of the floxed Myc allele and stabilization of β-catenin (Figure 7B). CD4Cre-Mycfl/fl mice showed thymocyte development and cellularity comparable to wild-type mice (Figure 7C). The thymic profile of the CD4Cre-CtnnbΔex3-Mycfl/fl mice (Figure 7C) closely resembled that of CD4Cre-CtnnbΔex3 mice (Figure 2A). While heterozygous loss of c-Myc (CD4Cre-CtnnbΔex3-Mycfl/+) resulted in frequent lymphomas, mice with homozygous ablation of this protein (CD4Cre-CtnnbΔex3-Mycfl/fl) did not develop lymphomas. This finding demonstrated that c-Myc activity was required for β-catenin–mediated lymphomagenesis.
Discussion
We showed here that thymocyte specific activation of β-catenin induced a developmental block in the DP to SP transition and led to the development of malignant T-cell thymic lymphomas. These consisted of highly proliferative CD4+CD8+ DP cells with intermediate αβTCR surface expression. The DP stage of leukemic arrest coincided with the developmental stalling of preleukemic thymocytes expressing stabilized β-catenin. Transformation targeted cells with surface expression of αβTCR and required Rag activity as well as further genetic changes, including the overexpression of c-Myc. Ablation of c-Myc abolished the ability of β-catenin to transform thymocytes, establishing its requirement in this process. Notch-dependent transcription was reduced in DP thymocytes with activated β-catenin and remained low in the lymphomas, arguing against the involvement of the Notch pathway in this transformation process.
The conclusion that β-catenin transformation specifically targets early DP thymocytes with surface expression of αβTCR was supported by several observations in addition to the surface phenotype of the lymphomas. First, the highest incidence of leukemia was observed in CD4Cre-CtnnbΔex3 mice, which stabilized β-catenin at the DP stage (ie, after β selection) and contained the largest number of αβTCR+ DP cells. LckCre-CtnnbΔex3 mice in which β-catenin stabilization started before the pre-TCR checkpoint showed significantly lower incidence of transformation. Second, in CD4Cre-CtnnbΔex3 and CD4Cre-CtnnbΔex3-pTα−/− mice, Rag activity, which was required for lymphomagenesis, overlapped with β-catenin stabilization only at the DP stage. Third, LckCre-CtnnbΔex3 mice provided an overlap of Rag activity with stabilization of β-catenin at an earlier DN3 stage. However, even these tumors were DP with αβTCR expression and shared both expression and surface profile characteristics with the CD4Cre-CtnnbΔex3 and CD4Cre-CtnnbΔex3-pTα−/− that could be initiated only at the DP stage. The requirement of αβTCR for the transformation event is further supported by the altered expression of several genes recording TCR activation in pretransformed thymocytes with activated β-catenin and/or in the resulting lymphomas. These changes, which were confirmed by FACS analysis, included the up-regulation of CD44 and CD5 as well as the down-regulation of L-selectin and were consistent with activated TCR signaling (data not shown).
Additional secondary events were required and selected for during transformation, as indicated by the oligoclonality of the lymphomas. This, combined with the need for Rag activity, supports the notion that the developmental stalling at the DP stage, which corresponds to peaked Rag activity, may be deleterious. It is possible that Rag activity and/or the related double-strand repair processes are impacted in the developmentally blocked DP thymocytes, leading to genetic damage and instability. A consistent secondary event was the up-regulation of c-Myc expression, which may, at least in part, account for the enhanced proliferation of the lymphomas. The requirement for c-Myc in β-catenin transformation was established by the failure of stabilized β-catenin to transform c-Myc–deficient thymocytes in compound mutant mice. This did not result from developmental deficiencies related to c-Myc ablation since CD4Cre-Mycfl/fl thymocytes showed a normal developmental profile (Figure 7C) and CD4Cre-CtnnbΔex3-Mycfl/fl thymi had a profile similar to CD4Cre-CtnnbΔex3. c-Myc is found up-regulated in a variety of malignancies and in particular those of hematopoietic origin, and may be the common denominator in the etiology of leukemia.26,27 Several reports have recently pointed to Myc as an important downstream target of Notch in T-cell transformation.50,51 It is possible that the up-regulation of c-Myc we observed in β-catenin–induced lymphomas overrides the requirement for Notch in this transformation event.
More than 50% of the examined human leukemias harbor activating mutations of Notch1, which may be promoting and/or maintaining transformation by providing survival and proliferation signals.34 Activation of β-catenin in immature thymocytes resulted in the suppression of Notch-dependent transcription,23 and transformation did not require activation of Notch signaling. The β-catenin–induced lymphoma represents an animal model of leukemia that does not require or select for Notch activation. It is possible that uncontrolled activation of β-catenin provides sufficient proliferation and/or survival signals not to require or benefit from further activation of the Notch signaling pathway. Of interest, both β-catenin stabilization and constitutive Notch activation induce a developmental block at the DP stage.53 It has recently been proposed that this developmental block may be at the basis of Notch's ability to transform thymocytes.37 By analogy, it is possible that transformation induced by β-catenin is assisted by the developmental block at the DP stage.
In light of activated β-catenin inducing thymocyte transformation and the general genetic correspondence of human T-ALL and mouse T-cell leukemia, we predict that activation of β-catenin will have a significant role in the etiology of human T-ALL, specifically in the cases that do not show Notch activation. This is supported by recent evidence that Wnt signaling inhibitors may be epigenetically inactivated in a fraction of ALL samples (including B-cell ALL [B-ALL] and T-ALL).54 The prospects of linking our findings to human T-ALL and providing therapeutic solutions are reasonable, in part due to the availability of specific first-generation inhibitors of β-catenin such as CGP049090 and PKF 115-584 that inhibit the interaction of β-catenin with TCF55 as well as ICG-001 that specifically inhibits the interaction of β-catenin with CBP.56 Our observations indicate that the Wnt/β-catenin signaling pathway may provide a promising molecular therapeutic target for the Notch-independent subset of T-ALL.
Supplementary Material
Acknowledgments
This work was supported by NIH:R01 AI059676-01; the Smith Family New Investigator Award from the Medical Foundation; the GRASP Center P30 DK-34928 Award (F.G.); NIH:R01 CA104547-01A1; and the DOD AMD17-03-1-0210 (K.K.).
The authors wish to thank A. Parmelee and S. Kwok at the Tufts Laser Cytometry facility for providing invaluable theoretical and practical help with cell sorting.
Footnotes
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisment” in accordance with 18 U.S.C. section 1734.
Authorship
Contribution: Z.G. designed and performed experiments and analyzed data; M.D. designed and performed experiments and analyzed data; D.K. designed and performed experiments; R.C. provided technical assistance; J.O. performed experiments; A.T.L. contributed to the interpretation of the data; H.B. supported the initiation of the research project; K.K. contributed to the design of the experiments and participated in the preparation of the paper; F.G. planned, coordinated, and supported the work, analyzed the data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Fotini Gounari, Tufts-New England Medical Center, 750 Washington St, Tufts-NEMC #5602, Boston, MA 02111; e-mail: fgounari@tufts-nemc.org.
References
- 1.Behrens J, Jerchow BA, Wurtele M, et al. Functional interaction of an axin homolog, conductin, with beta-catenin, APC, and GSK3beta. Science. 1998;280:596–599. doi: 10.1126/science.280.5363.596. [DOI] [PubMed] [Google Scholar]
- 2.Kishida S, Yamamoto H, Ikeda S, et al. Axin, a negative regulator of the wnt signaling pathway, directly interacts with adenomatous polyposis coli and regulates the stabilization of beta-catenin. J Biol Chem. 1998;273:10823–10826. doi: 10.1074/jbc.273.18.10823. [DOI] [PubMed] [Google Scholar]
- 3.Ikeda S, Kishida S, Yamamoto H, Murai H, Koyama S, Kikuchi A. Axin, a negative regulator of the Wnt signaling pathway, forms a complex with GSK-3beta and beta-catenin and promotes GSK-3beta-dependent phosphorylation of beta-catenin. Embo J. 1998;17:1371–1384. doi: 10.1093/emboj/17.5.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. beta-catenin is a target for the ubiquitin-proteasome pathway. Embo J. 1997;16:3797–3804. doi: 10.1093/emboj/16.13.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cook D, Fry MJ, Hughes K, Sumathipala R, Woodgett JR, Dale TC. Wingless inactivates glycogen synthase kinase-3 via an intracellular signalling pathway which involves a protein kinase C. Embo J. 1996;15:4526–4536. [PMC free article] [PubMed] [Google Scholar]
- 6.Noordermeer J, Klingensmith J, Perrimon N, Nusse R. dishevelled and armadillo act in the wingless signalling pathway in Drosophila. Nature. 1994;367:80–83. doi: 10.1038/367080a0. [DOI] [PubMed] [Google Scholar]
- 7.Kishida S, Yamamoto H, Hino S, Ikeda S, Kishida M, Kikuchi A. DIX domains of Dvl and axin are necessary for protein interactions and their ability to regulate beta-catenin stability. Mol Cell Biol. 1999;19:4414–4422. doi: 10.1128/mcb.19.6.4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smalley MJ, Sara E, Paterson H, et al. Interaction of axin and Dvl-2 proteins regulates Dvl-2-stimulated TCF-dependent transcription. Embo J. 1999;18:2823–2835. doi: 10.1093/emboj/18.10.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Itoh K, Antipova A, Ratcliffe MJ, Sokol S. Interaction of dishevelled and Xenopus axin-related protein is required for wnt signal transduction. Mol Cell Biol. 2000;20:2228–2238. doi: 10.1128/mcb.20.6.2228-2238.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van de Wetering M, de Lau W, Clevers H. WNT signaling and lymphocyte development. Cell. 2002;109(Suppl):S13–19. doi: 10.1016/s0092-8674(02)00709-2. [DOI] [PubMed] [Google Scholar]
- 11.Staal FJ, Clevers HC. Wnt signaling in the thymus. Curr Opin Immunol. 2003;15:204–208. doi: 10.1016/s0952-7915(03)00003-7. [DOI] [PubMed] [Google Scholar]
- 12.Reya T, Duncan AW, Ailles L, et al. A role for Wnt signalling in self-renewal of haematopoietic stem cells. Nature. 2003;423:409–414. doi: 10.1038/nature01593. [DOI] [PubMed] [Google Scholar]
- 13.Scheller M, Huelsken J, Rosenbauer F, et al. Hematopoietic stem cell and multilineage defects generated by constitutive beta-catenin activation. Nat Immunol. 2006;7:1037–1047. doi: 10.1038/ni1387. [DOI] [PubMed] [Google Scholar]
- 14.Kirstetter P, Anderson K, Porse BT, Jacobsen SE, Nerlov C. Activation of the canonical Wnt pathway leads to loss of hematopoietic stem cell repopulation and multilineage differentiation block. Nat Immunol. 2006;7:1048–1056. doi: 10.1038/ni1381. [DOI] [PubMed] [Google Scholar]
- 15.Schilham MW, Wilson A, Moerer P, Benaissa-Trouw BJ, Cumano A, Clevers HC. Critical involvement of Tcf-1 in expansion of thymocytes. J Immunol. 1998;161:3984–3991. [PubMed] [Google Scholar]
- 16.Staal FJ, Meeldijk J, Moerer P, et al. Wnt signaling is required for thymocyte development and activates Tcf-1 mediated transcription. Eur J Immunol. 2001;31:285–293. doi: 10.1002/1521-4141(200101)31:1<285::AID-IMMU285>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 17.Weerkamp F, Baert MR, Brugman MH, et al. Human thymus contains multipotent progenitors with T/B lymphoid, myeloid, and erythroid lineage potential. Blood. 2006;107:3131–3137. doi: 10.1182/blood-2005-08-3412. [DOI] [PubMed] [Google Scholar]
- 18.Okamura RM, Sigvardsson M, Galceran J, Verbeek S, Clevers H, Grosschedl R. Redundant regulation of T cell differentiation and TCRalpha gene expression by the transcription factors LEF-1 and TCF-1. Immunity. 1998;8:11–20. doi: 10.1016/s1074-7613(00)80454-9. [DOI] [PubMed] [Google Scholar]
- 19.Ioannidis V, Beermann F, Clevers H, Held W. The beta-catenin—TCF-1 pathway ensures CD4(+)CD8(+) thymocyte survival. Nat Immunol. 2001;2:691–697. doi: 10.1038/90623. [DOI] [PubMed] [Google Scholar]
- 20.Xu Y, Banerjee D, Huelsken J, Birchmeier W, Sen JM. Deletion of beta-catenin impairs T cell development. Nat Immunol. 2003;4:1177–1182. doi: 10.1038/ni1008. [DOI] [PubMed] [Google Scholar]
- 21.Pongracz JE, Parnell SM, Jones T, Anderson G, Jenkinson EJ. Overexpression of ICAT highlights a role for catenin-mediated canonical Wnt signalling in early T cell development. Eur J Immunol. 2006;36:2376–2383. doi: 10.1002/eji.200535721. [DOI] [PubMed] [Google Scholar]
- 22.Gounari F, Aifantis I, Khazaie K, et al. Somatic activation of beta-catenin bypasses pre-TCR signaling and TCR selection in thymocyte development. Nat Immunol. 2001;2:863–869. doi: 10.1038/ni0901-863. [DOI] [PubMed] [Google Scholar]
- 23.Gounari F, Chang R, Cowan J, et al. Loss of adenomatous polyposis coli gene function disrupts thymic development. Nat Immunol. 2005;6:800–809. doi: 10.1038/ni1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jamieson CH, Ailles LE, Dylla SJ, et al. Granulocyte-macrophage progenitors as candidate leukemic stem cells in blast-crisis CML. N Engl J Med. 2004;351:657–667. doi: 10.1056/NEJMoa040258. [DOI] [PubMed] [Google Scholar]
- 25.Lu D, Zhao Y, Tawatao R, et al. Activation of the Wnt signaling pathway in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2004;101:3118–3123. doi: 10.1073/pnas.0308648100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoffman B, Amanullah A, Shafarenko M, Liebermann DA. The proto-oncogene c-myc in hematopoietic development and leukemogenesis. Oncogene. 2002;21:3414–3421. doi: 10.1038/sj.onc.1205400. [DOI] [PubMed] [Google Scholar]
- 27.Ferrando AA, Neuberg DS, Staunton J, et al. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell. 2002;1:75–87. doi: 10.1016/s1535-6108(02)00018-1. [DOI] [PubMed] [Google Scholar]
- 28.Spanopoulou E, Early A, Elliott J, et al. Complex lymphoid and epithelial thymic tumours in Thy1-myc transgenic mice. Nature. 1989;342:185–189. doi: 10.1038/342185a0. [DOI] [PubMed] [Google Scholar]
- 29.Marinkovic D, Marinkovic T, Mahr B, Hess J, Wirth T. Reversible lymphomagenesis in conditionally c-MYC expressing mice. Int J Cancer. 2004;110:336–342. doi: 10.1002/ijc.20099. [DOI] [PubMed] [Google Scholar]
- 30.Felsher DW, Bishop JM. Reversible tumorigenesis by MYC in hematopoietic lineages. Mol Cell. 1999;4:199–207. doi: 10.1016/s1097-2765(00)80367-6. [DOI] [PubMed] [Google Scholar]
- 31.van Lohuizen M, Verbeek S, Scheijen B, Wientjens E, van der Gulden H, Berns A. Identification of cooperating oncogenes in E mu-myc transgenic mice by provirus tagging. Cell. 1991;65:737–752. doi: 10.1016/0092-8674(91)90382-9. [DOI] [PubMed] [Google Scholar]
- 32.Badiani PA, Kioussis D, Swirsky DM, Lampert IA, Weston K. T-cell lymphomas in v-Myb transgenic mice. Oncogene. 1996;13:2205–2212. [PubMed] [Google Scholar]
- 33.Jonkers J, Korswagen HC, Acton D, Breuer M, Berns A. Activation of a novel proto-oncogene, Frat1, contributes to progression of mouse T-cell lymphomas. Embo J. 1997;16:441–450. doi: 10.1093/emboj/16.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weng AP, Ferrando AA, Lee W, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 35.Pear WS, Aster JC, Scott ML, et al. Exclusive development of T cell neoplasms in mice transplanted with bone marrow expressing activated Notch alleles. J Exp Med. 1996;183:2283–2291. doi: 10.1084/jem.183.5.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bellavia D, Campese AF, Alesse E, et al. Constitutive activation of NF-kappaB and T-cell leukemia/lymphoma in Notch3 transgenic mice. Embo J. 2000;19:3337–3348. doi: 10.1093/emboj/19.13.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zweidler-McKay PA, Pear WS. Notch and T cell malignancy. Semin Cancer Biol. 2004;14:329–340. doi: 10.1016/j.semcancer.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 38.Deftos ML, Bevan MJ. Notch signaling in T cell development. Curr Opin Immunol. 2000;12:166–172. doi: 10.1016/s0952-7915(99)00067-9. [DOI] [PubMed] [Google Scholar]
- 39.Reschly EJ, Spaulding C, Vilimas T, Graham WV, Brumbaugh RL, Aifantis I, Pear WS, Kee BL. Notch 1 promotes survival of E2A-deficient T-cell lymphomas through pre-T cell receptor-dependent and -independent mechanisms. Blood. 2006;107:4115–4121. doi: 10.1182/blood-2005-09-3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O'Neil J, Shank J, Cusson N, Murre C, Kelliher M. TAL1/SCL induces leukemia by inhibiting the transcriptional activity of E47/HEB. Cancer Cell. 2004;5:587–596. doi: 10.1016/j.ccr.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 41.Nacht M, Jacks T. V(D)J recombination is not required for the development of lymphoma in p53-deficient mice. Cell Growth Differ. 1998;9:131–138. [PubMed] [Google Scholar]
- 42.O'Neil J, Calvo J, McKenna K, et al. Activating Notch1 mutations in mouse models of T-ALL. Blood. 2005 doi: 10.1182/blood-2005-06-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harada N, Tamai Y, Ishikawa T, et al. Intestinal polyposis in mice with a dominant stable mutation of the beta-catenin gene. Embo J. 1999;18:5931–5942. doi: 10.1093/emboj/18.21.5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wolfer A, Bakker T, Wilson A, et al. Inactivation of Notch 1 in immature thymocytes does not perturb CD4 or CD8T cell development. Nat Immunol. 2001;2:235–241. doi: 10.1038/85294. [DOI] [PubMed] [Google Scholar]
- 45.Kirberg J, Baron A, Jakob S, Rolink A, Karjalainen K, von Boehmer H. Thymic selection of CD8+ single positive cells with a class II major histocompatibility complex-restricted receptor. J Exp Med. 1994;180:25–34. doi: 10.1084/jem.180.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morgan DJ, Liblau R, Scott B, et al. CD8(+) T cell-mediated spontaneous diabetes in neonatal mice. J Immunol. 1996;157:978–983. [PubMed] [Google Scholar]
- 47.Fehling HJ, Krotkova A, Saint-Ruf C, von Boehmer H. Crucial role of the pre-T-cell receptor alpha gene in development of alpha beta but not gamma delta T cells. Nature. 1995;375:795–798. doi: 10.1038/375795a0. [DOI] [PubMed] [Google Scholar]
- 48.de Alboran IM, O'Hagan RC, Gartner F, et al. Analysis of C-MYC function in normal cells via conditional gene-targeted mutation. Immunity. 2001;14:45–55. doi: 10.1016/s1074-7613(01)00088-7. [DOI] [PubMed] [Google Scholar]
- 49.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci U S A. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Palomero T, Lim WK, Odom DT, et al. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc Natl Acad Sci U S A. 2006;103:18261–18266. doi: 10.1073/pnas.0606108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weng AP, Millholland JM, Yashiro-Ohtani Y, et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 2006;20:2096–2109. doi: 10.1101/gad.1450406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weerkamp F, Luis TC, Naber BA, et al. Identification of Notch target genes in uncommitted T-cell progenitors: No direct induction of a T-cell specific gene program. Leukemia. 2006;20:1967–1977. doi: 10.1038/sj.leu.2404396. [DOI] [PubMed] [Google Scholar]
- 53.Izon DJ, Punt JA, Xu L, et al. Notch1 regulates maturation of CD4+ and CD8+ thymocytes by modulating TCR signal strength. Immunity. 2001;14:253–264. doi: 10.1016/s1074-7613(01)00107-8. [DOI] [PubMed] [Google Scholar]
- 54.Roman-Gomez J, Cordeu L, Agirre X, et al. Epigenetic regulation of WNT signaling pathway in acute lymphoblastic leukemia. Blood. 2006 doi: 10.1182/blood-2006-09-047043. [DOI] [PubMed] [Google Scholar]
- 55.Lepourcelet M, Chen YN, France DS, et al. Small-molecule antagonists of the oncogenic Tcf/beta-catenin protein complex. Cancer Cell. 2004;5:91–102. doi: 10.1016/s1535-6108(03)00334-9. [DOI] [PubMed] [Google Scholar]
- 56.Emami KH, Nguyen C, Ma H, et al. A small molecule inhibitor of beta-catenin/CREB-binding protein transcription [corrected]. Proc Natl Acad Sci U S A. 2004;101:12682–12687. doi: 10.1073/pnas.0404875101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.