Abstract
Anion transporting proteins of the CLC type are involved in anion homeostasis in a variety of organisms. CLCs from Arabidopsis have been shown to participate in nitrate accumulation and storage. In this study, the physiological role of the functional chloride transporter AtCLC-d from Arabidopsis was investigated. AtCLC-d is weakly expressed in various tissues, including the root. When transiently expressed as a GFP fusion in protoplasts, it co-localized with the VHA-a1 subunit of the proton-transporting V-type ATPase in the trans-Golgi network (TGN). Stable expression in plants showed that it co-localized with the endocytic tracer dye FM4-64 in a brefeldin A-sensitive compartment. Immunogold electron microscopy confirmed the localization of AtCLC-d to the TGN. Disruption of the AtCLC-d gene by a T-DNA insertion did not affect the nitrate and chloride contents. The overall morphology of these clcd-1 plants was similar to that of the wild-type, but root growth on synthetic medium was impaired. Moreover, the sensitivity of hypocotyl elongation to treatment with concanamycin A, a blocker of the V-ATPase, was stronger in the clcd-1 mutant. These phenotypes could be complemented by overexpression of AtCLC-d in the mutant background. The results suggest that the luminal pH in the trans-Golgi network is adjusted by AtCLC-d-mediated transport of a counter anion such as Cl− or NO3−.
Keywords: chloride, nitrogen, vesicular traffic, channel, immunoelectron microscopy, V-ATPase
Introduction
The protein family of voltage-regulated chloride channels and anion transporters (CLCs) is widely distributed in prokaryotes and eukaryotes. Many eukaryotic CLC proteins localize to the cell surface and function as chloride channels (Jentsch et al., 2002). A key feature of CLCs is their double pore structure, which results from the fact that CLCs are dimeric proteins and each monomer forms an anion pore (Jentsch et al., 2002). A common characteristic of the CLC chloride channels is that they are also permeable to nitrate (Pusch et al., 1995), but this is of little physiological importance in animals.
In contrast to the eukaryotic plasma membrane CLC channels, a prokaryotic CLC has been shown to function as an electrogenic chloride/proton exchanger (Accardi and Miller, 2004). Similarly, several mammalian CLCs have recently been recognized to function as chloride/proton exchangers (Picollo and Pusch, 2005; Scheel et al., 2005). The mammalian CLCs that function as anion/proton exchangers appear to be mainly localized to intracellular vesicles, but not to the plasma membrane. The vesicular anion transporters ClC-3, ClC-5 and ClC-7 from mammals co-localize with vacuolar V-ATPase and appear to be functionally linked to this proton pump. Several mammalian ClCs may thus provide an electric shunt for the efficient acidification of intracellular compartments by V-type H+-ATPase (Jentsch et al., 2002).
Seven CLC genes have been identified in the Arabidopsis genome. Mutant plants have given some insight into their physiological function. Recently, the plant homolog AtCLC-a was shown to function as a nitrate/proton exchanger on the tonoplast (De Angeli et al., 2006). A loss-of-function mutant plant for this transporter accumulates less nitrate than the wild-type (Geelen et al., 2000). A quantitative trait loci analysis of nitrate storage in Arabidopsis suggested that AtCLC-c might be involved in nitrate accumulation (Harada et al., 2004). Indeed, in a loss-of-function clc-c mutant, total anion homeostasis appeared to be affected (Harada et al., 2004). These results suggest that a physiological function of AtCLCs is in nitrate accumulation and anion homeostasis in Arabidopsis.
A clear drawback in assigning a role to AtCLCs was that the functional analysis of several AtCLCs, e.g. by heterologous expression in frog oocytes, was unsuccessful (Hechenberger et al., 1996). However, in contrast to some other CLCs, AtCLC-d restored the growth defect of the yeast gef1 mutant on iron-limiting medium (Hechenberger et al., 1996). GEF1 (ScCLC) is the only CLC identified in the genome of Saccharomyces cerevisiae. ScCLC is Golgi-localized and plays a role in vesicular cation homeostasis (Gaxiola et al., 1998). Whether AtCLC-d acts in a channel-like manner or as a proton-coupled chloride transporter is still unclear at this point.
In this study, the expression pattern of AtCLC-d and its cellular localization were determined and a loss-of-function mutant was isolated. AtCLC-d was weakly expressed, but was developmentally regulated in several tissues. GFP-tagged AtCLC-d co-localized with V-type ATPase in the trans-Golgi network (TGN). Loss of AtCLC-d in clcd-1 plants did not affect overall anion content, but impaired root growth. Furthermore, hypocotyl elongation was impaired by concanamycin A, a specific H+-V-ATPase inhibitor, and this effect was stronger in the clcd-1 mutant. These results suggest some cooperativity of the H+-V-ATPase and AtCLC-d. The data are in agreement with the hypothesis that the anion transporter AtCLC-d provides an electrical shunt for optimal acidification of the trans-Golgi network.
Results
Developmental regulation of AtCLC-d expression
To analyze the expression pattern of AtCLC-d, a 2 kb fragment 5′ of the ATG was cloned in front of the β-glucoronidase gene. The construct was transformed into Arabidopsis and the promoter activity was assayed in two independent homozygous lines from the T2 generation. Promoter activity was most prominent in the root of germinating seedlings and in cotyledons of 5-day-old seedlings (Figure 1). Although differential expression was observed, a weak staining, indicating weak promoter activity, was observed in most plant organs. In older plants, stronger staining was observed in the hydathodes and in flowers, especially in anthers and pollen (Figure 1).
Figure 1.
Expression pattern of AtCLC-d in Arabidopsis. Staining pattern of homozygous T2 plants expressing a 2 kb promoter fragment fused to GUS in (a, b) 2-day-old, (e) 5-day-old and (f) 10-day-old seedlings, plus (c) hydathodes and (d) flowers. (g) Overall expression pattern in 28-day-old plants. The numbers indicate days after germination.
Isolation of a homozygous knockout line
A homozygous T-DNA insertion line in the Col-0 ecotype was isolated from the Salk collection (SALK_042895). The T-DNA insertion was within the fourth intron of the AtCLC-d gene (At5g26240; Figure 2), and initial hybridization with an antisense probe covering the 3′ terminal fragment of AtCLC-d showed that this 3′ fragment was overexpressed in clcd-1 (data not shown). Such overexpression of transcripts in lines from the Salk collection is not unprecedented, as the T-DNA harbors a 35S promoter, which drives expression of genes located close to the left border. However, the partial transcript lacks the coding information for the first three transmembrane helices of AtCLC-d. Proteins that might be formed from this transcript are unlikely to be functional. Subsequent Northern blot hybridization with a 5′ probe confirmed that the full transcript was only detected in wild-type plants, but not in the homozygous clcd-1 mutant (Figure 2). The 5′ part of the transcript and the fragment covering the T-DNA insertion were not detectable using RT-PCR, which confirmed that the AtCLC-d transcript was at least strongly reduced, if not abolished, in clcd-1. RT-PCR with primers covering the 5′ fragment of the gene confirmed the strong expression of the 3′ fragment of the partial AtCLC-d transcript in the clcd-1 line. The resistance to kanamycin of progeny obtained from selfed heterozygous plants with a clcd-1 allele segregated roughly 3:1, indicating a single T-DNA insertion in clcd-1 mutants.
Figure 2.
Isolation of a T-DNA insertion line for AtCLC-d. (a) Schematic position of the insertion in the 4th intron and location of primers. Exons are indicated as black boxes. The indicated primers were used for genotypic screening. (b) RT-PCR showing absence of the full transcript but expression of the 3′ fragment of the transcript in the clcd-1/clcd-1 line. (c) Northern blot hybridization with total RNA from wt and homozygous clcd-1 plants confirming loss of the 5′ part of the transcript in the mutant line.
Loss of AtCLC-d did not affect anion homeostasis
Growth and development of homozygous clcd-1 plants was undistinguishable from that of wild-type plants on soil. Plants did not show developmental defects under long- or short-day conditions (data not shown), and germinated and grew normally on agar supplemented with various concentrations of nitrate or chloride (data not shown). Plants grown on soil or on agar media supplemented with 50 mm KNO3 or 60 mm NaCl, did not have altered chloride and nitrate concentrations compared with the wild-type (Figure 3). These results suggest that AtCLC-d is not involved in overall anion homeostasis or that AtCLC-d function is redundant in Arabidopsis.
Figure 3.
Anion profile of 19-day-old Col-0 and homozygous clcd-1 plants grown on various synthetic agar media. Nitrate (a) and chloride (b) concentrations per fresh weight for plants grown in the absence of NaCl on 1.5 mm NH4NO3 or 6 mm NO3 as the sole nitrogen source or on 1.5 mm NH4NO3 supplemented with 60 mm NaCl. The results are means and SE for triplicate determinations. Similar data were obtained in an independent experiment.
AtCLC-d was localized to the trans-Golgi network
Transient expression in Arabidopsis protoplasts resulted in localization of an AtCLC-d–GFP fusion in small dots that were distributed throughout the cytoplasm (Figure 4). AtCLC-d did not co-localize with ARA7–mRFP, which localizes to an endosomal compartment (Figure 4a–d; Ueda et al., 2004). ARA7 (also called AtRabF2b) belongs to the family of Rab-type GTPases and has been implicated in vacuolar trafficking (Kotzer et al., 2004). In contrast, AtCLC-d–GFP partially co-localized with the rat sialyl transferase (ST–mRFP; Figure 4e–h) that is localized to trans-Golgi cisternae and the TGN (Dettmer et al., 2006; Saint-Jore et al., 2002). The Qa SNARE SYP41 has been identified in the TGN (Bassham et al., 2000). In many protoplasts, a strict co-localization was observed with SYP41–mRFP (Figure 4i–l). However, the pattern of the SYP41–mRFP marker in transiently expressing protoplasts was not entirely consistent and varied somewhat between different protoplasts. An intense overall red fluorescence and less pronounced co-localization were also observed with this marker. Furthermore, co-localization was observed with co-expression of the mRFP-tagged VHA-a1 subunit of V-ATPase (Dettmer et al., 2006) (Figure 4m–p). The co-localization was not only visible in individual confocal sections, but was also identified in 3D reconstructions of cell sections. A maximum projection of part of a cell is shown in Figure 4(q). Punctate fluorescent structures were also observed in root cells of stably transformed AtCLC-d–GFP-expressing plants. Due to the weak activity of the endogenous promoter of AtCLC-d–GFP, the CaMV 35S promoter was used to drive expression. The fluorescence obtained from such plants was always rather weak, both in the T1 and T2 generations.
Figure 4.
Co-localization of AtCLC-d with TGN markers in protoplasts. (a, e, i, m) Confocal laser scanning microscopy of AtCLC-d–GFP showing punctate fluorescence. (b, f, j, n) Staining patterns of co-transformed (b) ARA7–mRFP, (f) ST–mRFP, (j) Syp41–mRFP and (n) the mRFP-labeled VHA-a1 subunit of V-ATPase. Overlays are shown in (c, g, k, o) and bright-field images in (d, h, l, p). Scale bars = 10 µm. (q) Maximum projection of serial sections of part of a protoplast co-expressing AtCLC-d–GFP and VHA-a1–mRFP.
Immunogold labeling of ultra-thin cryosections on root tissue was performed using an anti-GFP antibody to confirm the above results. Transmission electron microscopy revealed consistent staining of tubulovesicular structures close to the trans site of the Golgi stacks (Figure 5a,b). Gold particles labeled the same structures, which represent the TGN, in VHA-a1–GFP plants (Figure 5d), but not in controls not expressing AtCLC-d–GFP (Figure 5c). In both cases, no staining was observed on the Golgi itself. The results are consistent with a preferential localization of AtCLC-d in the trans-Golgi network.
Figure 5.
Immunogold labeling of ultra-thin root cryosections of plants stably expressing AtCLC-d–GFP. (a, b) Anti-GFP antibodies were detected with silver-enhanced Nanogold. Gold particles accumulated in the TGN region close to the trans-Golgi cisternae of cortex and epidermal cells of root tips. The trans-Golgi side is marked with (T) and is always shown on the lower side of the Golgi stack (G). (c) No labeling above background was seen in controls not expressing AtCLC-d–GFP. (d) Labeling in plants stably expressing VHA-a1–GFP. Scale bar = 200 nm.
Function of AtCLC-d in endosomal compartments
Endosomal vesicles aggregate upon treatment with the fungal toxin brefeldin A (BFA), which blocks certain ADP ribosylation factor guanidine nucleotide exchange factors (ARF-GEFs) involved in membrane trafficking (Geldner et al., 2001). Early endosomes may accumulate in the core (Geldner et al., 2001), while trans-Golgi markers may arrange at the periphery of such ‘BFA compartments’ (Grebe et al., 2003). The punctate fluorescence from AtCLC-d–GFP-expressing plants was lost after incubating with BFA (50 µm) for 2 h, but an aggregate of fluorescence in the center of each cell was observed (Figure 6a,c). Furthermore, the lipophilic styryl dye FM4-64, a widely used tracer for endocytic membrane trafficking (Bolte et al., 2004; Grebe et al., 2003; Meckel et al., 2004) co-localized with AtCLC-d–GFP. After the addition of FM4-64 (2 µm), the plasma membrane rapidly stained red. Punctate red fluorescence was additionally apparent after a few minutes (Figure 6f). These punctate structures partially overlapped with the AtCLC-d–GFP fluorescent dots in confocal microscopic sections (Figure 6g,j). This is consistent with AtCLC-d–GFP residing in an early endocytic compartment identical to the TGN (Dettmer et al., 2006).
Figure 6.
Brefeldin A sensitivity and co-localization with the endocytic marker FM4-64 in root cells. Fluorescence (a, c) and bright-field (b, d) images of stably AtCLC-d–GFP-expressing plant roots. (a, b) Untreated control, (c, d) treatment with BFA (50 µm). Scale bar = 50 µm. Green AtCLC-d–GFP fluorescence (e, h), red FM4-64 labeling (f, i) and overlay (g, j) taken after 6 min (e–g) and in a different experiment after about 10 min (h–j). Scale bars = 10 µm.
Root growth inhibition at neutral and acidic pH
As AtCLC-d co-localized with the VHA-a1 subunit of V-ATPase, we tested whether clcd-1 mutant plant growth was affected at different pH. Wild-type root growth was maximal at acidic pH, consistent with an important role for the acidic rhizosphere pH in plant growth. In contrast, plant development was entirely impaired on agar plates that were buffered with 20 mm MES at neutral pH (Figure 7). Compared with the wild-type, the mutant clcd-1/clcd-1 plants had a slightly reduced root length, which was fully restored in ectopic 35S::AtCLC-d–GFP overexpressors in the null background (Figure 7). Taking into account the rapid exchange of lipid dye and membrane between the plasma membrane and the AtCLC-d–GFP-positive early endocytic compartment (Figure 6), the external pH may have affected the pH homeostasis. Importantly, complementation of the clcd-1/clcd-1 plants with GFP-tagged AtCLC-d suggests that the tag did not affect its physiological function and localization.
Figure 7.
Root growth at acidic pH in the clcd-1/clcd-1 line and complementation by ectopic overexpression of AtCLC-d–GFP. Representative root length of seedlings after 8 days (upper panels). Lower panel: statistical analysis of primary root length. The root length was calculated from three independent experiments (means ± SE). More than 50 seedlings were analyzed in each experiment.
Reduced cell expansion by co-inhibition with V-ATPase
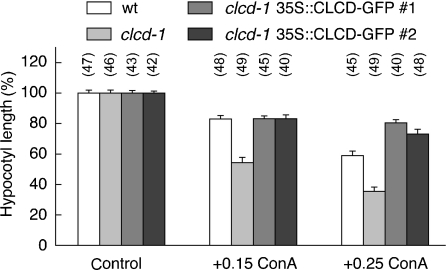
V-ATPase is required for cell elongation, a phenomenon that is apparent from the dwarf phenotype in the det3 mutant that shows a partial V-ATPase loss (Schumacher et al., 1999). Inhibition of V-ATPase by concanamycin A, a specific inhibitor, similarly impaired the hypocotyl elongation of etiolated seedlings in a dose-dependent manner (Figure 8). To confirm a more direct link between a functional V-ATPase and AtCLC-d, the effect of concanamycin A on hypocotyl elongation was assayed in clcd-1 and wild-type plants. In 5-day-old dark-germinated seedlings, the hypocotyls of clcd-1 plants were of similar length when germinated on pure agar in the dark (Figure 8) and when the pH of the medium was adjusted to pH 5 or 7 (data not shown). In contrast, clcd-1 hypocotyls were shorter than wild-type (P < 0.001) when the V-ATPase was partially blocked by concanamycin A (Figure 8), proving a close cooperativity of the proton pump and AtCLC-d. This phenotype was complemented by expression of the GFP-tagged AtCLC-d construct under the control of the 35S promoter.
Figure 8.
Impaired hypocotyl elongation by loss of AtCLC-d and by treatment with concanamycin A. Normalized hypocotyl length of wild-type 5-day-old seedlings (white bar), clcd-1 mutant (light gray bar) and two ectopic AtCLC-d–GFP overexpressers in the clcd-1 background (dark gray bars) without concanamycin A (left), and with 0.15 (middle) or 0.25 µm concanamycin A (right). The number of hypocotyls analyzed in this particular experiment is shown in parentheses on the top of each bar. Means and SE are given and were statistically different between clcd-1 and the other lines upon treatment with concanamycin A (P < 0.001, Student’s t-test). Similar differences between wild-type and clcd-1 were observed in four independent experiments; the results for one of these are shown. Although absolute values of hypocotyl length were almost identical for wild-type and clcd-1 in the absence on concanamycin A, normalized values are given here for better display. Absolute hypocotyl lengths (in mm) were: wild-type = 11.5; clcd-1 = 10.4; complemented clcd-1 = 11.5 and 10.8.
Discussion
Expression of the anion transporter AtCLC-d was developmentally regulated and relatively strong in roots of young seedlings. A weak, but relatively broad expression pattern was observed with the promoter–GUS construct. This matches the tissue distribution obtained for AtCLC-d by RT-PCR (Hechenberger et al., 1996) and the data from global expression profiling experiments using microarrays (available at http://www.genevestigator.org).
AtCLC-d had been shown previously to function as an anion transporter by complementation of a yeast strain with a deletion in the single, Golgi-localized endogenous ScCLC (Gaxiola et al., 1998; Hechenberger et al., 1996). The fluorescent AtCLC-d–GFP fusion protein was also functional in such assays, an observation that could be reproduced in our laboratory (Figure S1). When heterologously expressed in yeast, AtCLC-d–GFP localizes to punctate structures that represent the Golgi and possibly to vesicular structures (Hechenberger et al., 1996; Schwappach et al., 1998). In Arabidopsis, AtCLC-d–GFP localized preferentially to TGN membranes. Partial co-localization was observed with the trans-Golgi marker ST–mRFP (that can often be detected in the TGN), but not the endosomal marker ARA7–mRFP. Strict co-localization was observed with the TGN marker SYP41 (Bassham et al., 2000), and especially the VHA-a1 subunit of V-type ATPase (Dettmer et al., 2006). That AtCLC-d resides in the same membrane structures at the trans side of the Golgi that also contain a proton pump suggests a close functional cooperativity of the pump and the anion transporter. A close functional relationship between V-type ATPase and vesicular CLC chloride transporters has not previously been reported in plants, but has been observed in yeast and animals. Interestingly, loss of yeast ScCLC did lead to several secondary defects, including altered quality control mechanisms for membrane proteins (Li et al., 1999) and an iron-suppressible defect in respiration (Greene et al., 1993). Such a phenotype was also caused by defects in another gene, GEF2, that encodes a subunit of the vacuolar H+-ATPase (Greene et al., 1993). The link between vesicular CLC anion transporters and the H+-pump has been amply confirmed for the mammalian ClC-3, ClC-5 and ClC-7 transporters. For example, osteopetrosis, a severe bone disease, is associated with mutations in V-ATPase and CLC-7 (Jentsch et al., 2002). It has been proposed that endosomal CLCs provide an electric shunt for the efficient acidification of intracellular compartments (Jentsch et al., 2002). To build up the necessary proton gradients, the excess positive charge in these organelles has to be neutralized, which may be achieved by the import of anions. From studies on isolated animal endosomes, it is known that acidification is more efficient in the presence of extravesicular chloride (Fuchs et al., 1989). In situ studies with secretory and recycling endosomes of the trans-Golgi network indicated a dependence of the acidification rate on the level of chloride in the cytosol (Demaurex et al., 1998). This demonstrates the need for chloride conductance in the acidification of these intracellular eukaryotic organelles. Efficient endosomal acidification can be easily envisaged by the V-ATPase cooperating with an anion channel; however, endosomal and intracellular CLCs appear to be anion/proton exchangers. Only excess transport of anions, e.g. as 2 Cl−/1 H+ exchange, appears to efficiently electrically neutralize the vesicular lumen. Whether AtCLC-d acts as anion channel or as anion/proton exchanger cannot be answered at the moment. A molecular determinant that distinguishes CLC channels from CLC transporters appears to be position E259 (which corresponds to E203 in EcCLC) (Accardi et al., 2005). This residue is ‘transporter-like’ in AtCLC-d, and this may suggest that AtCLC-d acts as an anion/H+ exchanger, rather than as a channel. This also opens up the possibility that it is not acidification of the TGN per se, but the anion accumulation in these structures that is critical for their vesicular transport function, a hypothesis that may be clarified in future studies.
A defective AtCLC-d might thus disrupt normal cellular function by causing defective TGN acidification. This may subsequently impair vesicular traffic and affect optimal root growth. It remains unclear which cellular defect impairs root elongation at neutral or more alkaline external pH, but a defective pH homeostasis and impaired vesicular acidification may be involved.
The importance of TGN acidification for optimal root growth and cell elongation is compatible with the results of another study, in which the vacuolar subunit VHA-c1 was knocked-down by RNAi. In that study, the reduced activity of the V-ATPase resulted in shorter root length and impaired cell elongation in the hypocotyl of Arabidopsis (Padmanaban et al., 2004). The acidification of endosomal compartments appears to be essential for normal plant development, as mutants with defective V-ATPase are lethal (Dettmer et al., 2005).
Function of AtCLC-d in acidification of endosomal compartments
The luminal pH of endomembrane compartments in eukaryotic cells is generally acidic, although data on the exact luminal pH of plant endomembrane vesicles are scarce. An increasingly acidic luminal pH towards the vesicle destination appears to be required for correct membrane trafficking of endosomal vesicles in eukaryotes, as well as for import of cargo, such as ions, into these vesicles. A decrease in the luminal pH has been reported in a number of eukaryotic cells along the secretory pathway (Paroutis et al., 2004). The luminal pH in secretory granules and endosomes may be primarily regulated by the activity and number of V-ATPases, but the proton leak and counter-ion conductance provided by AtCLC-d may also be of fundamental importance.
Intracellular mammalian CLCs and the yeast ScCLC show close sequence similarity to the Arabidopsis CLC proteins, which is consistent with an intracellular localization. AtCLC-d is the first plant anion transporter localized to compartments involved in trafficking. As expected from this localization, the total anion accumulation in plants was not affected in a loss-of-function mutant.
The TGN has been recognized as a major sorting site in the secretory pathway, where plant proteins destined for the lysosome or vacuole are segregated from exocytic cargo (Jurgens, 2004). It has, however, been shown that the early endosome and the trans-Golgi network are at least partially identical (Dettmer et al., 2006). Such data are fully compatible with the data presented here and are supported by the finding that the AtCLC-d–GFP compartments rapidly stained with endocytosed FM4-64. The fluorescence pattern of AtCLC-d–GFP after BFA treatment is compatible with a function in the secretory pathway, as BFA at least partially blocks post-Golgi secretion and leads to rapid internalization of plasma membrane proteins into large aggregates containing Golgi compartments (Hawes, 2005). The data presented here indicate that AtCLC-d and V-ATPase support growth in expanding cells by tight cooperation within the TGN.
Experimental procedures
Plant material and growth conditions
Arabidopsis thaliana plants (ecotype Col-0) were grown in soil and transformed by spraying using the GV3101 Agrobacterium strain. Seeds were germinated in the dark for 4 days on agar plates (0.8%) containing 1.5 mm NH4NO3, 1 mm KH2PO4, 0.5 mm MgSO4, 100 µm NaSiO3, 1.25 mm CaSO4, 50 µm Na-Fe-EDTA, 50 µm H3BO3, 3 µm MnSO4, 1 µm ZnSO4, 1.3 µm CuSO4, 0.03 µm Mo7O24(NH4)6 and 20 mm MES, with the pH adjusted to 6.0 using Tris. Various nitrogen sources were added using appropriate NH4NO3 concentrations or exchanging NH4NO3 for KNO3 or (NH4)2SO4. Selection and segregation of transgenic plants for kanamycin resistance were performed on agar plates with 50 µg/ml kanamycin. Selection of BASTA resistance utilized plates supplemented with 4 µmN-methyl sulfoximine. Seeds germinated on agar nutrient medium were grown at 22°C with cycles of 16 h light/ 8 h dark. Soil-grown plants were maintained in greenhouse conditions.
Hypocotyl elongation was measured after germination for 5 days in the dark. A 4 h light stimulus was given after plating the seeds on agar plates (0.8% Phytagar, Duchefa (http://www.duchefa.com)) made with distilled water without nutrients. The means ± SE for 30–50 hypocotyls of each seed batch are given. The protocol for transient expression in protoplasts from suspension cells from Arabidopsis can be found at http://www.uni-tuebingen.de/ZMBP/centfac/transf/index.html.
Isolation of the T-DNA insertion allele
The T-DNA insertion disrupting AtCLC-d was identified in the Salk collection (SALK_042895). The genomic sequence was amplified by PCR with flanking primers (LB 5′-GCGTGGACCGCTTGCTGCAAC-3′, D1fw 5′-AGGAATTGATATTCCCGGCACC-3′, D1rev 5′-CTCCGATAACTCCTATTACAGC-3′, D2fw: 5′- CTGGAGTTGCTGCTGCCTTTAG-3′). Northern blot analysis revealed that this transgenic line overexpressed the C-terminal fragment (positions 653–2404 of the cDNA starting at the ATG). Sequence analysis of this PCR product showed that the T-DNA was inserted at position 926 of the genomic sequence starting at the ATG.
Expression analysis
RNA was isolated using an RNeasy plant mini kit (Qiagen; http://www.qiagen.com/). cDNA was transcribed from total RNA (2 µg) isolated from seedlings using reverse transcriptase MMLV-RTase (Fermentas, http://www.fermentas.com) and oligo(dT) primer. All PCR amplifications comprised 30 cycles. A 2 kb promoter fragment was isolated from genomic DNA using the following primers: 5′-GAGGTACCTTAGAGAACGAGGGGTACGG–3′ and 5′-GAGGGTACCGATCGAGAGTTCGACTTCTGG–3′. Both primers contained KpnI restriction sites (underlined) to facilitate cloning into pTkan-GUS (Su et al., 2004).
Molecular complementation
The AtCLC-d sequence was amplified from cDNA (Hechenberger et al., 1996). In this study, the d86 splice variant was investigated throughout. To obtain the rescue construct AtCLC-d–GFP, the stop codon of the AtCLC-d gene was deleted and ligated 5′ in-frame with the GFP(S65T) coding sequence. This sequence was inserted into pPTbar, a derivative of the pPZP212 vector (Hajdukiewicz et al., 1994), containing the 35S promoter and an rbcs terminator. The primers used were 5′-GAGTCGACATGTTATCGAATCATCTCCAGA-3′ and 5′-AGGTCGACTCTTATTTGTATAGTTCATCCA-3′. Plasmids were introduced into Agrobacterium strain GV3101::pMP90, and plants were transformed using standard procedures. Plants were assayed for GFP fluorescence in the T1 generation and were then transferred to soil. Homozygous T2 progeny were used.
Marker constructs and microscopy
mRFP-tagged constructs were derived from VHA-a1–GFP (Dettmer et al., 2006) or N-ST–YFP (Grebe et al., 2003) by replacing the XFP sequence by mRFP (Campbell et al., 2002). ARA7–mRFP has been described previously by Takano et al. (2005).
For bright-field and differential interference contrast microscopy, a Zeiss Axioplan microscope (Zeiss; http://www.zeiss.com/) equipped with a Diagnostics Spot RT slider camera were used, and for confocal laser scanning microscopy, a Leica TCS SP2 was used (Leica, http://www.leica.de). Images were processed in NIH image and Adobe Photoshop.
Immunogold labeling was performed on ultra-thin (80–100 nm) thawed Tokuyasu cryosections of formaldehyde-fixed (8%, 2 h) and sucrose-infiltrated (2.1 M) root tips using rabbit anti-GFP serum (1:250, 60 min; Abcam, http://www.abcam.com) and silver-enhanced (HQ Silver, 8 min; Nanoprobes, http://www.nanoprobes.com) goat (Fab′) anti-rabbit IgG coupled to Nanogold (1:50; no. 2004, Nanoprobes) (Völker et al., 2001). Staining with uranyl acetate and final embedding in uranyl acetate/methylcellulose (Sigma; http://www.sigmaaldrich.com/) was performed according to the method described by Griffiths (1993).
Analytical methods
The anion content was analyzed from liquid N2-homogenized material by HPLC using a Wescan ion analyzer equipped with an anion chromatography column (Gamma Analysen Technik GmbH, http://www.gatgmbh.com) using 2 mm phtalic acid, 10% methanol, pH 5, as eluent.
Acknowledgments
We thank P. Neumann and D Ripper for excellent technical help, B. Stadelhofer and H. Stransky for help with the HPLC analysis, G. Pilot (University of Bonn, Germany) for technical advice, T. Jentsch (University of Hamburg, Germany) for CLC constructs, and F. de Courcy (ZMBP, Tübingen, Germany) for critically reading the manuscript. This work was supported by the Deutsche Forschungsgemeinschaft (SFB446).
Supplementary material
The following supplementary material is available for this article online:
Functional complementation of the Dgef1 yeast strain by AtCLC-d.
This material is available as part of the online article from http://www.blackwell-synergy.com.
References
- Accardi A. Secondary active transport mediated by a prokaryotic homologue of ClC Cl− channels. Nature. 2004;427:803–807. doi: 10.1038/nature02314. [DOI] [PubMed] [Google Scholar]
- Accardi A. Separate ion pathways in a Cl−/H+ exchanger. J. Gen. Physiol. 2005;126:563–570. doi: 10.1085/jgp.200509417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassham D C. AtVPS45 complex formation at the trans-Golgi network. Mol. Biol. Cell. 2000;11:2251–2265. doi: 10.1091/mbc.11.7.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte S. FM-dyes as experimental probes for dissecting vesicle trafficking in living plant cells. J. Microsci. 2004;214:159–173. doi: 10.1111/j.0022-2720.2004.01348.x. [DOI] [PubMed] [Google Scholar]
- Campbell R E. A monomeric red fluorescent protein. Proc. Natl Acad. Sci. USA. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Angeli A. The nitrate/proton antiporter AtCLCa mediates nitrate accumulation in plant vacuoles. Nature. 2006;442:939–942. doi: 10.1038/nature05013. [DOI] [PubMed] [Google Scholar]
- Demaurex N. Mechanism of acidification of the trans-Golgi network (TGN). In situ measurements of pH using retrieval of TGN38 and furin from the cell surface. J. Biol. Chem. 1998;273:2044–2051. doi: 10.1074/jbc.273.4.2044. [DOI] [PubMed] [Google Scholar]
- Dettmer J. Essential role of the V-ATPase in male gametophyte development. Plant J. 2005;41:117–124. doi: 10.1111/j.1365-313X.2004.02282.x. [DOI] [PubMed] [Google Scholar]
- Dettmer J. Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell. 2006;18:715–730. doi: 10.1105/tpc.105.037978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs R. Acidification and ion permeabilities of highly purified rat liver endosomes. J. Biol. Chem. 1989;264:2212–2220. [PubMed] [Google Scholar]
- Gaxiola R A. The yeast CLC chloride channel functions in cation homeostasis. Proc. Natl Acad. Sci. USA. 1998;95:4046–4050. doi: 10.1073/pnas.95.7.4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geelen D. Disruption of putative anion channel gene AtCLC-a in Arabidopsis suggests a role in the regulation of nitrate content. Plant J. 2000;21:259–267. doi: 10.1046/j.1365-313x.2000.00680.x. [DOI] [PubMed] [Google Scholar]
- Geldner N. Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature. 2001;413:425–428. doi: 10.1038/35096571. [DOI] [PubMed] [Google Scholar]
- Grebe M. Arabidopsis sterol endocytosis involves actin-mediated trafficking via ARA6-positive early endosomes. Curr. Biol. 2003;13:1378–1387. doi: 10.1016/s0960-9822(03)00538-4. [DOI] [PubMed] [Google Scholar]
- Greene J R. The GEF1 gene of Saccharomyces cerevisiae encodes an integral membrane protein; mutations in which have effects on respiration and iron-limited growth. Mol. Gen. Genet. 1993;241:542–553. doi: 10.1007/BF00279896. [DOI] [PubMed] [Google Scholar]
- Griffiths G. Heidelberg, Germany: Springer Verlag; 1993. Fine Structure Immunocytochemistry; pp. 174–177. [Google Scholar]
- Hajdukiewicz P. The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 1994;25:989–994. doi: 10.1007/BF00014672. [DOI] [PubMed] [Google Scholar]
- Harada H. Quantitative trait loci analysis of nitrate storage in Arabidopsis leading to an investigation of the contribution of the anion channel gene, AtCLC-c, to variation in nitrate levels. J. Exp. Bot. 2004;55:2005–2014. doi: 10.1093/jxb/erh224. [DOI] [PubMed] [Google Scholar]
- Hawes C. Cell biology of the plant Golgi apparatus. New Phytol. 2005;165:29–44. doi: 10.1111/j.1469-8137.2004.01218.x. [DOI] [PubMed] [Google Scholar]
- Hechenberger M. A family of putative chloride channels from Arabidopsis and functional complementation of a yeast strain with a CLC gene disruption. J. Biol. Chem. 1996;271:33632–33638. doi: 10.1074/jbc.271.52.33632. [DOI] [PubMed] [Google Scholar]
- Jentsch T J. Molecular structure and physiological function of chloride channels. Physiol. Rev. 2002;82:503–568. doi: 10.1152/physrev.00029.2001. [DOI] [PubMed] [Google Scholar]
- Jurgens G. Membrane trafficking in plants. Annu. Rev. Cell Dev. Biol. 2004;20:481–504. doi: 10.1146/annurev.cellbio.20.082503.103057. [DOI] [PubMed] [Google Scholar]
- Kotzer A M. AtRabF2b (Ara7) acts on the vacuolar trafficking pathway in tobacco leaf epidermal cells. J. Cell Sci. 2004;117:6377–6389. doi: 10.1242/jcs.01564. [DOI] [PubMed] [Google Scholar]
- Li Y. Yeast mutants affecting possible quality control of plasma membrane proteins. Mol. Cell. Biol. 1999;19:3588–3599. doi: 10.1128/mcb.19.5.3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meckel T. Endocytosis against high turgor: intact guard cells of Vicia faba constitutively endocytose fluorescently labelled plasma membrane and GFP-tagged K-channel KAT1. Plant J. 2004;39:182–193. doi: 10.1111/j.1365-313X.2004.02119.x. [DOI] [PubMed] [Google Scholar]
- Padmanaban S. Differential expression of vacuolar H+-ATPase subunit c genes in tissues active in membrane trafficking and their roles in plant growth as revealed by RNAi. Plant Physiol. 2004;134:1514–1526. doi: 10.1104/pp.103.034025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paroutis P. The pH of the secretory pathway: measurement, determinants, and regulation. Physiology. 2004;19:207–215. doi: 10.1152/physiol.00005.2004. [DOI] [PubMed] [Google Scholar]
- Picollo A. Chloride/proton antiporter activity of mammalian CLC proteins ClC-4 and ClC-5. Nature. 2005;436:420–423. doi: 10.1038/nature03720. [DOI] [PubMed] [Google Scholar]
- Pusch M. Gating of the voltage-dependent chloride channel CIC-0 by the permeant anion. Nature. 1995;373:527–531. doi: 10.1038/373527a0. [DOI] [PubMed] [Google Scholar]
- Saint-Jore C M. Redistribution of membrane proteins between the Golgi apparatus and endoplasmic reticulum in plants is reversible and not dependent on cytoskeletal networks. Plant J. 2002;29:661–678. doi: 10.1046/j.0960-7412.2002.01252.x. [DOI] [PubMed] [Google Scholar]
- Scheel O. Voltage-dependent electrogenic chloride/proton exchange by endosomal CLC proteins. Nature. 2005;436:424–427. doi: 10.1038/nature03860. [DOI] [PubMed] [Google Scholar]
- Schumacher K. The Arabidopsis det3 mutant reveals a central role for the vacuolar H(+)-ATPase in plant growth and development. Genes Dev. 1999;13:3259–3270. doi: 10.1101/gad.13.24.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwappach B. Golgi localization and functionally important domains in the NH2 and COOH terminus of the yeast CLC putative chloride channel Gef1p. J. Biol. Chem. 1998;273:15110–15118. doi: 10.1074/jbc.273.24.15110. [DOI] [PubMed] [Google Scholar]
- Su Y H. Molecular and functional characterization of a family of amino acid transporters from Arabidopsis. Plant Physiol. 2004;136:3104–3113. doi: 10.1104/pp.104.045278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano J. Endocytosis and degradation of BOR1, a boron transporter of Arabidopsis thaliana, regulated by boron availability. Proc. Natl Acad. Sci. USA. 2005;102:12276–12281. doi: 10.1073/pnas.0502060102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda T. Functional differentiation of endosomes in Arabidopsis cells. Plant J. 2004;40:783–789. doi: 10.1111/j.1365-313X.2004.02249.x. [DOI] [PubMed] [Google Scholar]
- Völker A. Cell cycle-independent expression of the Arabidopsis cytokinesis-specific syntaxin KNOLLE results in mistargeting to the plasma membrane and is not sufficient for cytokinesis. J. Cell Sci. 2001;114:3001–3012. doi: 10.1242/jcs.114.16.3001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Functional complementation of the Dgef1 yeast strain by AtCLC-d.