Abstract
Only few small, regulatory RNAs encoded opposite another gene have been identified in bacteria. Here, we report the characterization of a locus where a small RNA (SymR) is encoded in cis to an SOS-induced gene whose product shows homology to the antitoxin MazE (SymE). Synthesis of the SymE protein is tightly repressed at multiple levels by the LexA repressor, the SymR RNA and the Lon protease. SymE co-purifies with ribosomes and overproduction of the protein leads to cell growth inhibition, decreased protein synthesis and increased RNA degradation. These properties are shared with several RNA endonuclease toxins of the toxin-antitoxin modules, and we show that the SymE protein represents evolution of a toxin from the AbrB fold, whose representatives are typically antitoxins. We suggest that SymE promotion of RNA cleavage may be important for the recycling of RNAs damaged under SOS-inducing conditions.
Introduction
In Escherichia coli, a combination of approaches based on sequence conservation, structural features and direct detection has led to the identification of approximately 80 small RNAs (sRNAs) that do not encode tRNAs, rRNAs or proteins but rather are thought to be regulators (Storz and Gottesman, 2006). While the functions of only a subset of these regulatory RNAs is known, most of the characterized sRNAs act to modulate mRNA stability and/or translation by base-pairing with mRNAs that are encoded at a different chromosomal position. In general, the region of complementarity between these trans-encoded sRNA and the mRNA target is limited, and all of these sRNAs have been found to bind to and require the RNA chaperone Hfq for function.
In contrast to the sRNAs encoded on the bacterial chromosome, most of the characterized plasmid sRNAs are encoded opposite the genes that they regulate and thus have perfect complementarity with their target mRNAs (Wagner et al., 2002). Many of these cis-encoded sRNAs modulate the synthesis of replication proteins and thus control the copy number of the plasmids. They act by blocking ribosome binding or by promoting the formation of an mRNA secondary structure that leads to transcription termination. Other cis-encoded plasmid sRNAs repress the synthesis of proteins that would otherwise be toxic to the cell (Gerdes et al., 1997). Because the protein toxin kills cells in which the plasmid is lost, these sense-antisense pairs have been termed addiction modules or post-segregational killing systems. Perhaps the best-characterized antitoxin sRNA is the Sok antisense RNA of plasmid R1 which represses translation of the hok mRNA. The Sok RNA is very unstable and is quickly degraded when the R1 plasmid is lost from the cell. Under these conditions, the more stable hok mRNA is translated, and the Hok protein kills the cells that no longer carry the plasmid.
In addition to the Hok-Sok systems for plasmid addiction, a number of plasmids also encode toxin-antitoxin modules where the antitoxin is a protein (Engelberg-Kulka and Glaser, 1999). There are multiple classes of these protein toxin-antitoxin modules which are encoded not only on plasmids but on bacterial chromosomes as well (Anantharaman and Aravind, 2003; Pandey and Gerdes, 2005). The majority of these systems share a common gene organization – usually operons of two closely linked genes where one gene encodes an antitoxin and the other a toxin. The antitoxins are typically transcription factors that repress the toxin-antitoxin operon and also bind to and block the activity of the toxins. The toxins encoded by these modules are a very diverse group of proteins that appear to kill cells by disrupting any of several different aspects of cell physiology. In the course of bacterial evolution, operons constituting these systems have undergone a wide range of ‘mixing and matching’ of toxin and antitoxin genes, while retaining the same organizational syntax (Anantharaman and Aravind, 2003).
Free-living organisms in particular encode an abundance of these protein toxin-antitoxin modules, but their physiological roles have been controversial. It has alternatively been proposed that toxins promote cell killing, cell stasis, long-term cell persistence or reuse of resources; although some of these roles may not be mutually exclusive (Buts et al., 2005; Gerdes et al., 2005; Condon, 2006). While the physiological roles of the toxin proteins remain under debate, the biochemical functions of some of the toxins are more clearly understood. Many toxins belong to the RelE, MazF/Kid, Doc and PIN domain superfamilies of predicted RNA endonucleases, and subsets of these toxins have been shown to promote mRNA cleavage either on their own or in association with ribosomes. As for the Hok-Sok example, the protein antitoxin generally is significantly less stable than the toxin such that toxin activity increases under conditions of stress when the antitoxin is degraded.
In our cloning-based screen for E. coli sRNAs, we identified a 77 nucleotide RNA, denoted RyjC based on its genomic position, which is encoded opposite the 5′ end of the yjiW mRNA (Kawano et al., 2005). The RyjC promoter is embedded in the yjiW coding sequence (Fig. 1A) and the sRNA and much of its sequence are conserved in Salmonella. Here we examine the function of the yjiW-RyjC sense-antisense pair which we have renamed SymE (SOS-induced yjiW gene with similarity to MazE) and SymR (symbiotic RNA). We present evidence that SymR serves an antitoxin function by repressing SymE synthesis and that the SymE protein has properties of the endonuclease toxins. We propose that SymE represents an instance of toxin evolution from a superfamily hitherto only known to contain antitoxin transcription factors. We also suggest that the SymE-SymR pair might represent the deployment of a chromosomal toxin-antitoxin module for the recycling of RNA damaged concomitantly with DNA.
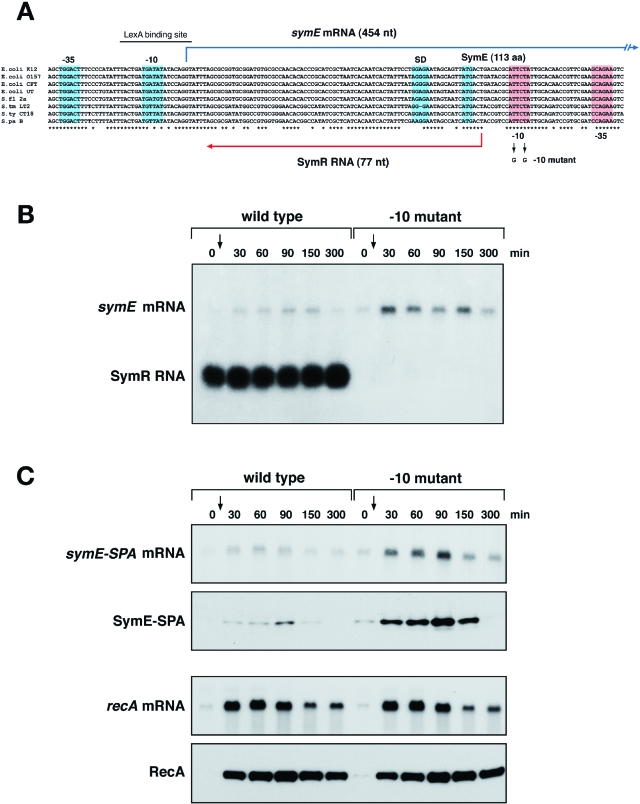
Fig. 1.
SymR RNA represses SymE translation. A. Genetic organization of the symER locus. Multiple sequence alignments of the different symER sequences were constructed using clustalw. The species correspond to E. coli strains K12, O157:H7, CFT073 and UT189, Shigella flexneri 2a, Salmonella typhimurium LT2, S. typhi CT18 and S. paratyphi B. The −10 and −35 promoter sequences, the Shine-Dalgarno sequence and initiation codon of symE are indicated by blue boxes. The −10 and −35 sequences of symR are indicated by red boxes. B. symE mRNA and SymR RNA levels in MG1655 and the −10 symR promoter mutant. Total RNA was isolated from wild-type MG1655 and −10 mutant strains grown in LB medium at 37°C at 0, 30, 60, 90, 150 and 300 min after treatment with 1 μg ml−1 mitomycin C. Samples (5 μg) were analysed by Northern hybridization using oligonucleotide probes specific to symE and SymR. C. symE-SPA mRNA and protein and recA mRNA and protein levels in MG1655 symE-SPA and the −10 symR promoter mutant. Total RNA and cell lysates were prepared from MG1655 symE-SPA and symE-SPA −10 mutant strains in grown in LB medium at 37°C at 0, 30, 60, 90, 150 and 300 min after treatment with 1 μg ml−1 mitomycin C. RNA samples (5 μg) were analysed by Northern hybridization using oligonucleotide probes specific to symE and recA, and cell lysates were analysed by immunoblot assays using monoclonal anti-FLAG M2-AP and polyclonal anti-RecA antibodies.
Results
SymR antisense RNA represses symE translation
To examine the effects of SymR RNA expression on the cis-encoded symE gene, we needed to construct a strain lacking the SymR RNA. Given that the two genes overlap and the promoter of SymR is embedded within the symE open reading frame, we had to eliminate SymR expression by mutating the symR promoter. This was achieved by introducing two point mutations onto the E. coli chromosome that disrupted the −10 sequence of the symR promoter but did not alter the SymE amino acid sequence (Fig. 1A). This −10 mutation effectively abolished expression of the SymR RNA (Fig. 1B). Previous studies had shown that the symE (yjiW) promoter has a LexA binding site and is strongly induced by DNA damaging agents (Fernández De Henestrosa et al., 2000). We therefore compared symE mRNA levels in wild-type MG1655 cells and the corresponding strain carrying the −10 promoter mutation at different times before and after cells were treated with the DNA damaging agent mitomycin C (Fig. 1B). For wild-type cells virtually no symE mRNA expression was observed in the absence of mitomycin C treatment. Low levels of the mRNA were detected 30 min after SOS induction with a peak between 60 and 90 min. Overall slightly higher symE mRNA levels were detected for the −10 mutant strain; more symE mRNA could be detected in the absence of DNA damage, and higher induction was observed at 30, 60, 90 and 150 min. Generally, the elimination of SymR expression resulted in approximately threefold higher symE mRNA levels.
We also examined the effects of SymR RNA on SymE protein levels by integrating a sequential peptide affinity (SPA) tag adjacent to the SymE stop codon on the chromosomes of the wild type and −10 mutant strains. When symE-SPA mRNA levels after mitomycin C treatment were compared in the wild type and −10 mutant backgrounds, we again observed an approximately threefold increase in symE-SPA mRNA levels in the −10 mutant compared with the wild-type strain (Fig. 1C). A greater than sevenfold difference in SymE-SPA protein levels was observed between the wild-type and −10 mutant strains. For the wild-type strain, no protein was detected in the absence of mitomycin C treatment and only low levels were detected after the treatment. As for the symE-SPA mRNA, the peak of SymE-SPA protein was at 90 min. For the −10 mutant, SymE-SPA was clearly present in uninduced cells and the levels were strongly increased upon mitomycin C treatment. In contrast, no difference in recA mRNA and protein levels was observed with the same wild type and −10 promoter extracts (Fig. 1C). Together these results indicate that while the presence of the SymR RNA has some effect on symE mRNA levels, there is a greater effect on protein levels. The presence of the antisense RNA however, does not impact the timing of SymE expression after SOS induction.
SymR RNA levels are 10-fold higher than symE mRNA levels
The relative levels of the symE mRNA and SymR RNA were determined by quantitative Northern analysis (Fig. 2A). The total RNA samples isolated for the 0 and 90 min time points in Fig. 1B and C were separated alongside known amounts of in vitro synthesized SymR and symE RNAs. Using this approach, the amount of symE mRNA in wild-type strain was determined to be 0.01 fmol μg−1 of total RNA at time 0 and 0.02 fmol μg−1 at 90 min, while the amount of SymR in wild-type strain was determined to be 0.2 fmol μg−1 of total RNA at both time points. Thus, the ratio of symE mRNA and SymR is nearly 1:10 suggesting that most of the SymR RNA in the cell is not base-paired with the symE mRNA. We also found the SymR RNA to be very stable; almost no decrease in SymR levels was observed even 60 min after inhibiting transcription with rifampicin treatment (data not shown).
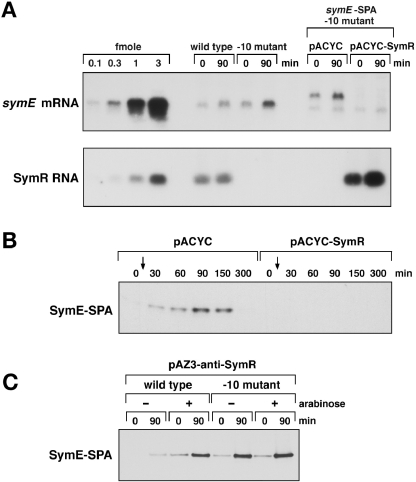
Fig. 2.
Effects of different SymR RNA levels. A. Quantitative Northern analysis of symE and SymR RNA levels. Total RNA (10 μg) isolated for the Northern analysis in Fig. 1B and total RNA (0.5 μg) isolated from the samples used for the Western analysis in Fig. 2B was separated alongside in vitro synthesized RNA (0.1, 0.3, 1 and 3 fmol) on 1.2% agarose gels. The cellular levels of symE mRNA and SymR RNA were determined from the ratio of the signals of control RNAs to the cellular RNAs. B. SymR RNA expressed in trans can repress SymE synthesis. The symE-SPA −10 mutant carrying pACYC and pACYC-SymR was grown to OD600∼0.3 in LB medium containing tetracycline at 37°C. Cell lysates prepared at 0, 30, 60, 90, 150 and 300 min after treatment with 1 μg ml−1 mitomycin C were analysed by immunoblot assays using monoclonal anti-FLAG M2-AP antibodies. C. Expression of an anti-antisense RNA leads to increased SymE-SPA synthesis. MG1655 PCP18-araE symE-SPA and MG1655 PCP18-araE symE-SPA −10 mutant strains carrying pAZ3-anti-SymR were grown to OD600∼0.2 in LB medium containing chloramphenicol at 37°C. The cultures were split and half of each culture was treated with 0.02% arabinose for 30 min (0 min). All cultures were then treated with 1 μg ml−1 mitomycin C for 90 min (90 min). Cell extracts prepared at the 0 and 90 min time points were analysed by immunoblot assays using monoclonal anti-FLAG M2-AP antibodies.
SymR expressed in trans represses SymE synthesis and anti-SymR expressed in trans activates SymE synthesis
To test whether SymR expressed in trans could also repress SymE-SPA synthesis, we moved a pACYC184 plasmid expressing symR from its own promoter into the −10 promoter mutant strain (Fig. 2B). In cells treated with mitomycin C, SymE-SPA was detected in the control strain carrying the vector control, similar to the wild-type strain. However, for cells harbouring pACYC-SymR, no SymE-SPA protein was detected at any time points. Thus, SymR expressed from a plasmid can also repress SymE-SPA synthesis. In the pACYC-SymR containing strains, the SymR RNA was present at 8–12 fmol μg−1, more than 40-fold higher than the levels expressed from the chromosome (Fig. 2A). At these high levels of SymR RNA no symE-SPA mRNA was detected even 90 min after mitomycin C treatment. This decrease must be resulting from effects on symE mRNA stability because assays of a symE–lacZ fusion showed that pACYC-SymR did not impact transcription from the symE promoter (data not shown).
We also constructed a plasmid expressing an RNA that is complementary to SymR and thus was predicted to interfere with SymR base-pairing with the symE mRNA. In this construct the anti-SymR RNA is under the control of the PBAD promoter of pAZ3. When the anti-SymR RNA was induced by arabinose for 30 min in the wild-type background, SymE-SPA synthesis was observed even in the absence of DNA damage and was further increased by mitomycin C treatment for 90 min (Fig. 2C). These SymE-SPA levels were comparable to the levels observed in the −10 mutant in the absence of arabinose induction. SymE-SPA levels were not further elevated in the −10 mutant when anti-SymR expression was induced by arabinose. Our interpretation of these results is that anti-SymR is exerting its effects by preventing SymR base-pairing with the symE mRNA, and this regulation is abolished when SymR is not expressed in the −10 mutant.
SymE protein levels are controlled by the Lon protease
For many well-characterized plasmid sRNAs that base-pair with cis-encoded mRNAs, the antisense RNA-target RNA duplexes are degraded by RNase III, an endoribonuclease specific for double-stranded RNA (Blomberg et al., 1990; Gerdes et al., 1992; Malmgren et al., 1997). In addition, all characterized E. coli sRNAs that base-pair with trans-encoded mRNA targets have been found to require the function of the Hfq RNA chaperone protein (Storz and Gottesman, 2006). To determine whether SymR regulation of SymE synthesis required either of these proteins, we moved the corresponding mutant alleles into the SymE-SPA strain and examined SymE-SPA synthesis at different time points after mitomycin C treatment (Fig. 3A). In the rnc mutant, the pattern of SymE-SPA synthesis was virtually identical to the wild-type strain. For the hfq mutant strain, we also did not detect SymE-SPA protein at 0 min. The peak of SymE-SPA synthesis was later than for the wild-type strain, but we attribute this delay to the slower growth of this mutant strain. In general, neither RNase III nor Hfq appears to be required for SymR RNA repression of SymE synthesis.
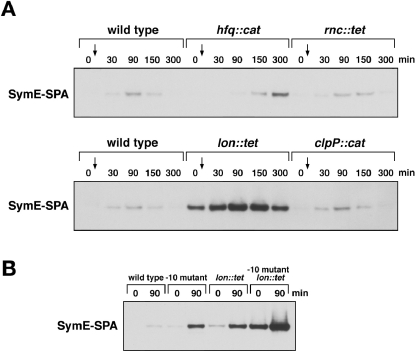
Fig. 3.
SymE protein is degraded by the Lon protease. A. SymE-SPA synthesis in hfq, rnc, lon and clpP mutant strains. The MG1655 symE-SPA strain and the corresponding mutant derivatives were grown to OD600∼0.3 in LB medium at 37°C. Cell lysates prepared at 0, 30, 90, 150 and 300 min after treatment with 1 μg ml−1 mitomycin C were analysed by Western hybridization using monoclonal anti-FLAG M2-AP antibodies. B. Relative SymE-SPA levels in the −10 symR promoter and lon mutant backgrounds. The MG1655 symE-SPA, symE-SPA −10 mutant, symE-SPA lon mutant, symE-SPA −10 lon mutant strains were grown to OD600∼0.5 in LB medium at 37°C. Cell lysates prepared at 0 and 90 min after treatment with 1 μg ml−1 mitomycin C were analysed by immunoblot assays using monoclonal anti-FLAG M2-AP antibodies.
Numerous SOS-induced proteins have been found to be unusually labile to proteolysis; a feature which should allow rapid return to a non-stressed state once the DNA damage has been eliminated (Neher et al., 2006). Thus, we examined the effects of mutations that eliminate the expression of the ClpP and Lon proteins on SymE-SPA levels (Fig. 3A). The clpP mutant also was virtually identical to the wild-type strain. For the lon mutant strain however, the levels of the SymE-SPA protein, but not other control SPA-tagged proteins (data not shown) were significantly elevated at every time point, indicating that the SymE protein is a target of the Lon protease.
Together our results showed that synthesis of the SymE is tightly repressed at multiple levels; at the transcriptional level by the LexA repressor, at the level of mRNA stability and translation by the SymR RNA and at the level of protein stability by the Lon protease. To examine the relative contributions of each of these three regulators on SymE synthesis, we compared SymE-SPA levels in strains carrying the −10 mutant, the lon mutation and these two mutations in combination in the presence and absence of DNA damage (Fig. 3B). In the absence of mitomycin C treatment, SymE-SPA levels were slightly elevated in both single mutant strains, similar to levels observed in a LexA mutant strain (lexA51; data not shown). The SymE-SPA levels were significantly elevated in the double mutant and were induced even further upon deactivation of the LexA repressor with mitomycin C treatment indicating that the contributions of LexA, SymR and Lon to SymE repression are additive.
SymE protein co-purifies with ribosomal proteins
To learn about the function of SymE, we purified plasmid-expressed, SPA-tagged and His-tagged SymE protein and identified co-purifying proteins (Fig. 4A and data not shown). A number of protein bands are observed for the SymE-SPA sample that are not in the vector control sample. Several of the identified bands correspond to ribosomal subunits (30S subunits S1, S3 and 50S subunits L1, L2, L10 and L13). Two other identified proteins (CsdA and trigger factor) are known to be associated with ribosomes. We also see an abundance of lower bands, which are likely to be small ribosomal proteins.
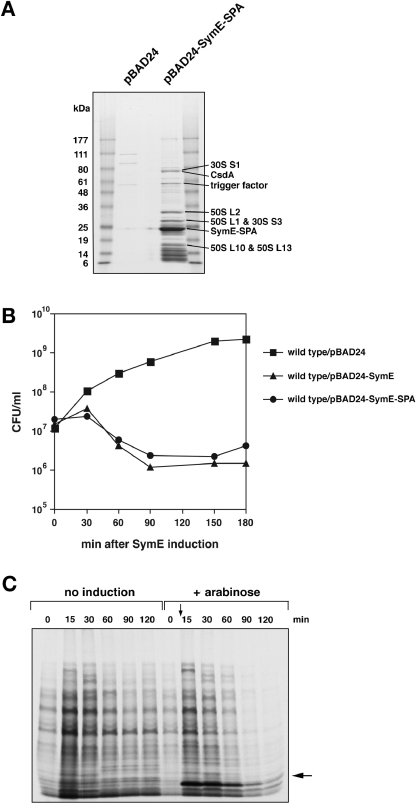
Fig. 4.
Overexpression of ribosome-associated SymE leads to reduced colony formation and decreased protein synthesis. A. Purification of SymE-SPA. Extracts prepared from MG1655 kan-PCP18-araE carrying either pBAD24 or pBAD-SymE-SPA were subjected to affinity purification as described in Experimental procedures. The samples then were analysed by 4–20% SDS-PAGE and visualized using GelCode Blue Stain Reagent. The identities of the indicated protein bands were determined by LC/MS/MS mass spectrometry. B. SymE overexpression results in reduced colony-forming ability. MG1655 PCP18-araE carrying pBAD24, pBAD-SymE and pBAD-SymE-SPA were grown to OD600∼0.3 at 37°C in LB medium containing ampicillin. Two min after time 0, 0.02% arabinose was added to induce transcription of symE or symE-SPA. At the indicated time points, cells were diluted and plated on LB solid medium containing ampicillin. C. SymE overexpression results in reduced protein synthesis. SDS-PAGE analysis of in vivo total protein synthesis after the induction of SymE. GSO120/pBAD-SymE cells were grown in M9 medium with glycerol, casamino acids and ampicillin with or without arabinose. Aliquots (0.5 ml) of the cell culture were taken at the indicated time points and added to 20 μCi of Tran35S-label containing 35S-methionine and 35S-cysteine as described in Experimental procedures. The band indicated with an arrow is SymE (∼12 kDa). This experiment was performed multiple times. The figure is a representative experiment.
SymE protein overexpression affects the colony-forming ability and protein synthesis
We did not observe any growth defects associated with the induced levels of SymE in either the −10 mutant strain or the strain carrying pACYC-SymR (data not shown). However, growth was markedly affected when SymE was overexpressed from the arabinose-inducible PBAD promoter on a multicopy plasmid (Fig. 4B). While the number of colony-forming cells steadily increased for the vector control strain, there was a marked decrease for cells overexpressing SymE. The effects of overexpressing SymE-SPA were slightly less, but the tagged protein still clearly affected growth indicating that the SPA-tag does not dramatically alter the activity of the protein.
To further characterize the detrimental effects of SymE overexpression, we examined protein synthesis at different times after SymE induction from the PBAD promoter plasmid (Fig. 4C). For each time point cells were labelled with 35S-methionine and 35S-cysteine for 1 min. Without SymE induction protein synthesis, as indicated by label incorporation, was detected at all time points. This was not the case upon SymE overproduction, where almost no protein labelling was observed at 90 and 120 min after arabinose induction. Both of these SymE overproduction phenotypes, the reduction in colony-forming ability and protein synthesis was reminiscent of phenotypes observed upon overexpression of the RelE and MazF-like toxins (Christensen and Gerdes, 2003; Christensen et al., 2003; Zhang et al., 2003). Like SymE, the RelE protein also has been shown to be associated with ribosomes (Galvani et al., 2001).
SymE overexpression leads to RNA cleavage
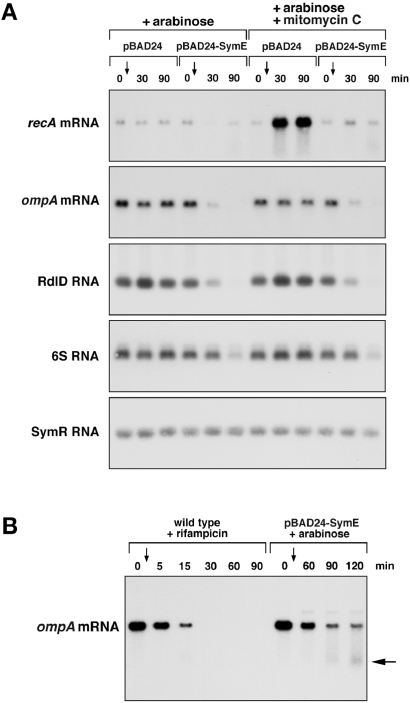
Overproduction of RelE and MazF as well as other related toxins leads to the degradation of mRNA molecules (Christensen and Gerdes, 2003; Zhang et al., 2003; 2005). To test whether the same was observed upon SymE overexpression, total RNA was isolated at different times after arabinose induction with and without mitomycin C treatment, and the levels of different mRNAs and sRNAs was examined by Northern analysis (Fig. 5A). The levels of the recA and ompA mRNAs were dramatically decreased upon SymE overexpression. The levels of two sRNAs, RdlD and 6S, also were affected, particularly at 90 min. In contrast the levels of the SymR RNA itself were not changed. For all of the RNAs, similar effects were observed with and without mitomycin C treatment. We suggest that the decrease in the recA, ompA, RdlD and 6S RNA levels most likely is resulting from SymE-promoted cleavage of these RNAs rather than general inhibition of transcription because distinct shorter ompA mRNA fragments were observed at 90 and 120 min after SymE induction which were not detected when transcription was inhibited with rifampicin (Fig. 5B).
Fig. 5.
SymE exhibits ribonuclease activity. A. SymE overexpression leads to reduced levels of some RNAs. MG1655 PCP18-araE carrying either pBAD24 or pBAD-SymE were grown to OD600∼0.5 at 37°C in M9 minimal medium supplemented with glycerol and casamino acids. Total RNA isolated at 0, 30, and 90 min after treatment with 0.02% arabinose and 0.02% arabinose plus 1 μg ml−1 mitomycin C. Samples (5 μg) were analysed by Northern hybridization using oligonucleotide probes specific to recA, ompA, RdlD, 6S and SymR. B. Detection of degradation products upon SymE overexpression. MG1655 cells were grown to OD600∼0.5 at 37°C in LB medium, and total RNA was isolated at 0, 5, 15, 30, 60 and 90 min after treatment with 300 μg ml−1 rifampicin. MG1655 PCP18-araE cells carrying pBAD-SymE were grown to OD600∼0.25 at 37°C in LB medium, and total RNA was isolated at 0, 60, 90, and 120 min after treatment with 0.02% arabinose. Samples (5 μg) were analysed by Northern hybridization using an oligonucleotide probe specific to ompA. Degradation products are indicated by the larger arrow.
The fact that RdlD and 6S RNA levels are decreased upon SymE overproduction suggests that the RNAs do not need to be actively translated in order to be targets of SymE-mediated degradation. This is supported by the observation that a lacI mRNA with and without a start codon mutation appear to be degraded at the same rate (Fig. S1) and is similar to what is found for MazF, which can cleave mRNAs independent of the ribosome (Zhang et al., 2003).
SymE protein belongs to the AbrB superfamily
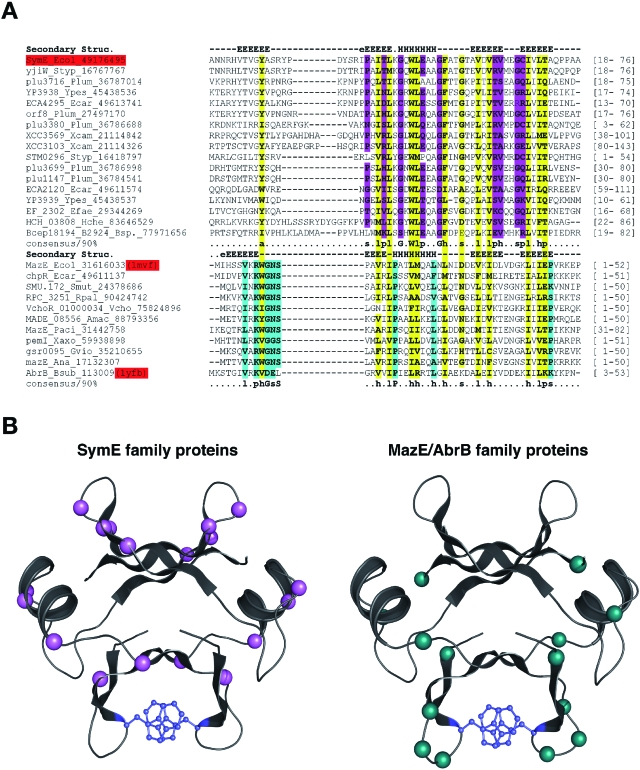
Given the functional analogies between the SymE-SymR and the toxin-antitoxin modules, we sought to understand better the evolutionary relationships between SymE and previously known protein components of toxin-antitoxin modules. A psi-blast search initiated with SymE recovers statistically significant hits (e-value = 10−3−10−26) to over 60 proteins from a wide range of species of different γ- and β-proteobacterial genera. In all of these proteins, the region of similarity spanned a globular domain of approximately 60 amino acids. In psi-blast searches with these globular regions, there were hits, albeit with border-line statistical significance (e-value ∼0.05–0.1) to proteins of the AbrB superfamily such as MazE, which function as transcription factors and antitoxins in various toxin-antitoxin modules. We also found that sensitive sequence profiles (PSSMs) and hidden Markov models (HMMs) of the AbrB domain superfamily recovered highly significant hits to SymE and its homologues (e-value = 10−6−10−10). In addition, sequence-structure threading of SymE using the 3PSSM program recovered the structure of the antitoxin MazE (1UB4; 1MVF).
We prepared a multiple sequence alignment of the conserved globular domain of SymE and all its homologues (denoted SymE family) (Fig. 6A) and used the jpred program, which combines the information from a PSSM, HMM and amino-acid frequencies in the alignment to predict the secondary structure of SymE (Cuff and Barton, 2000). The predicted structure was entirely congruent to that of the AbrB fold (Fig. 6B). Comparison of multiple alignments of the SymE family with classical members of the AbrB superfamily indicated that they shared all the key hydrophobic residues, which constitute the core of this folding. These include a highly conserved hydrophobic residue in the middle of the central α-helix, and an aromatic residue in β strand-1 that forms a π–π stacking interaction critical for the characteristic dimerization of these domains. Taken together these observations strongly indicate that SymE and its homologues contain an AbrB fold and constitute a distinct family within this superfamily.
Fig. 6.
SymE is an AbrB superfamily member that has acquired a toxin-like function. A. Multiple alignment of the SymE family and other members of the AbrB superfamily. Proteins are denoted by gene name, species abbreviation, and GenBank Identifier (gi) number; separated by underscores. Positions strongly conserved at or above the 90% applied on the entire family of proteins only among the classic SymE family proteins are shaded pink, whereas those similarly conserved only in the classic MazE/AbrB family are shaded aqua and those conserved between both are shaded yellow. Consensus similarity designations are as follows: h, hydrophobic residues (ACFILMVWY); s, small residues (AGSVCDN); p, polar residues. Secondary structure assignments obtained from the jpred prediction for the SymE family and from the crystal structures (like PDB: 1mvf and 1ub4) for the rest of the AbrB superfamily are shown above the alignment where E represents a strand and H represents a helix. The region shown in the alignment spans the entire length of the DNA binding domain of the classical AbrB proteins. The boundaries are shown to the right. Species abbreviations are as given for the full alignment in Fig. S2. B. Models of the classic SymE family and classic MazE/AbrB family proteins. An idealized version of the AbrB fold was constructed using the consensus sequence derived from the hidden Markov model for the entire fold using SWISSMODEL server of SWISSPDB and the 1mvf structure as a template; in both cases the structure is depicted as a dimer formed by two interlocking monomers, which was achieved using the oligomer mode in SWISSMODEL (Guex and Peitsch, 1997). The positions that are conserved in the SymE family at the 90% consensus are coloured pink as in Fig. 6A. The majority of them line a groove on one face of the protein. This surface faces away from the surface that contains the most conserved positions unique to the classical members of the MazE/AbrB superfamily (coloured aqua). The uniquely conserved regions include polar residues that could potentially mediate the key interactions of SymE with the ribosome or target RNA. The aromatic residues involved in stabilizing the dimer through a π–π stacking interaction are shown in blue.
This relationship between SymE and the AbrB family was astonishing, because all previously characterized AbrB members of the toxin-antitoxin modules were transcription factors that act as antitoxins. In contrast, SymE exhibited properties that were hitherto only exhibited by toxins. A careful examination of the sequence conservation patterns and their distribution on the three-dimensional structure suggested that beyond the residues those stabilize the structural core the AbrB fold, the SymE family had acquired a novel constellation of residues (Fig. 6). These were predicted to map to regions involved in potential nucleic acid interactions and included some polar residues that might have a role in potential cleavage of RNAs. It was conceivable that SymE might antagonize the functions of other AbrB family members thereby releasing another toxin, but we observed the same decreases in colony-forming ability upon SymE overproduction in a wild-type strain and a strain lacking the five previously described toxin-antitoxin modules (data not shown). Thus, we suggest that the SymE proteins have undergone a functional shift from a transcription factor or antitoxin to an RNA-associating protein with toxin-like properties.
Gene neighbourhood analysis of the SymE systems supports a toxin-like function
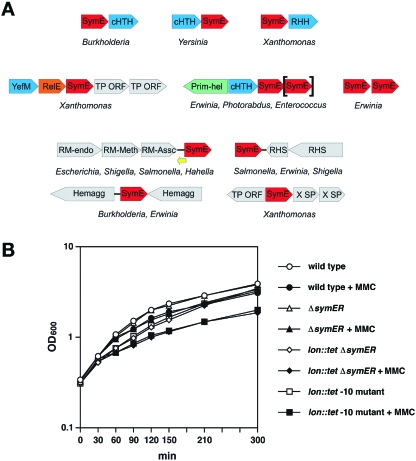
To further investigate if SymE family proteins might have indeed acquired toxin-like function, we performed a detailed analysis of the gene neighbourhoods of SymE family members (Fig. 7A). The results of this analysis showed that SymE-family genes are often found in predicted operons together with genes encoding a transcription factor. The most commonly occurring transcription factor is a Cro/cI helix–turn–helix type protein (cHTH), which is found as the repressor and antitoxin in various toxin-antitoxin modules containing toxins of the PIN, RelE and Doc superfamilies. More infrequently, SymE family genes are found in operons encoding transcription factors of the YefM or MetJ/Arc ribbon–helix–helix type (RHH) superfamilies, which are also represented in toxin-antitoxin operons. Thus, where SymE genes are embedded in predicted operons with protein coding genes, they are always accompanied by another gene that encodes a transcription factor that typically functions as an antitoxin in the characterized toxin-antitoxin modules. This observation is consistent with the inference that, despite having an AbrB fold like the MazE antitoxins, the SymE family has undergone a functional shift to a toxin-like role. In these cases, SymE family genes most likely are regulated by means of transcriptional control.
Fig. 7.
Physiological role of SymE. A. Gene neighbourhoods and predicted operons of the SymE family of genes. The orientation of the genes is denoted by the direction of the arrow. The yellow arrow denotes the SymR RNA that is transcribed in the SymE proper group. The genes are all labelled as per the gene products: SymE = SymE family; cHTH = a Cro/cI type transcriptional regulator, RHH = a MetJ/Arc-type transcription factor, YefM = a YefM-type transcription factor, RelE = a toxin homologous to the RelE toxins, Prim-Hel = the gene encoding a two module protein with a N-terminal DNAG-type primase domain and C-terminal MCM-like AAA + helicase domain. Shown in grey are other genes belonging to the larger genomic context, in the mobile islands in which the SymE family genes are inserted. These include transposons = TP ORF and restriction-modification operons = RM-endo, RM-Meth and RM-Assc for the endonuclease, methylase and accessory subunits respectively. Representative examples of neighbourhoods with the Xanthomonas-specific secreted protein (X SP) and the haemagglutinin and RHS cell-surface complex genes are also shown. The SymE enclosed in ‘[]’indicates the presence of an optional second SymE gene in some of these neighbourhoods. B. ΔsymER growth after DNA damage. Wild type, ΔsymER, lon::tetΔsymER and lon::tet−10 mutant cells were grown to OD600∼0.3 at 37°C in LB medium. Expression of SymE was induced by the addition of 1 μg ml−1 mitomycin C, and cell growth was monitored by measuring OD600 at the indicated time points.
A subset of the SymE genes, including SymE itself, did not contain any other transcription factor gene in their neighbourhood. In several of these cases there was noticeable nucleotide conservation upstream of the predicted start methionine, suggesting that members of this group are potentially all regulated by non-codings sRNAs as in the case of SymE. Thus, it appears likely that the SymE systems, like other toxin-antitoxin modules, had evolved through in situ operonic displacement of the transcription factor genes by genes for other such regulators or regulatory sRNAs that introduced a regulatory stricture an entirely different level.
SymE role in the SOS response
Various physiological roles have been proposed for the antitoxin-toxin modules including altruistic killing (Aizenman et al., 1996), reversible stasis (Pedersen et al., 2002) and long-term persistence (Lewis, 2005) as well as quality control and the reuse of resources (Gerdes et al., 2005). To begin to examine the role of SymE in the SOS response, the symER coding region was replaced by the kan gene. Wild type and ΔsymER mutant cells as well as lon mutant cells with and without the ΔsymER deletion were treated with mitomycin C, and cell growth (Fig. 7B) and colony-forming units were assays at different time points (data not shown). We did not detect any differences between the ΔsymER deletion and parent wild type or lon mutant cells in any of these assays suggesting that endogenous levels of SymE expressed after SOS induction do not play a role in altruistic killing, bacterial stasis or long-term persistence. We thus propose that a more likely role of SymE in the SOS response is the reuse of resources, in particular damaged RNAs.
Discussion
Small, non-coding RNAs have been found to have a wide variety of regulatory functions in both bacteria and eukaryotes. We sought to define the role of SymR, a cis-encoded sRNA in E. coli and found that the antisense RNA tightly controls the synthesis of SymE, a SOS-induced protein that is also subject to degradation by the Lon protease. Surprisingly, despite homology to the AbrB superfamily of proteins which previously have been characterized as antitoxins, SymE has properties of the toxin-family of proteins. Similar to other toxins, SymE co-purifies with ribosomes, and overexpression of SymE leads to reduced colony formation, decreased protein synthesis as well as significant decreases in the levels of several RNAs tested. Sequence conservation and gene neighbourhood across bacterial species support the conclusion that this AbrB-family member has evolved to have a toxin-like function. These results have led us to suggest that SymE acts as an endoribonuclease, either on its own or in association with the ribosomes. Given its induction in response to DNA damage, we also propose that SymE might play a role in recycling RNAs damaged by the agents that induce the SOS response.
Tight regulation of SymE synthesis
SymE synthesis is repressed at all levels; by the LexA repressor at the level of transcription, by the SymR RNA at the level of mRNA stability and translation, and by the Lon protease at the level of protein stability. Given the toxicity associated with SymE overexpression, we suggest that this tight, additive repression ensures endogenous SymE levels never rise to detrimental concentrations. Whether still other regulators modulate SymE synthesis remains to be seen.
We found several aspects of the SymR RNA regulation of SymE mRNA stability and translation to be surprising. First, at endogenous levels, the antisense RNA had a stronger impact on symE mRNA translation than on degradation. Because the SymR RNA is completely complementary to the 5′ end of the symE mRNA, there is extensive possibility for base-pairing and we expected this RNA-RNA duplex to be a good target for cleavage by RNase III. However, the absence of SymR only led to an approximately threefold increase in symE mRNA levels, and SymE synthesis was not affected in the rnc mutant strain. A second surprise was the high 10:1 SymR to symE ratio even after symE mRNA induction after DNA damage. All symE mRNA in the cell could be complexed with the SymR RNA. However, given that we do observe SymE protein synthesis, a subpopulation of the mRNA must not be involved in pairing or the pairing is sufficiently transient to allow occasional ribosome binding. This observation also raises the possibility that the SymR RNA has additional functions. Finally, the SymR RNA was unexpectedly stable. The levels or activities of most regulators are modulated. It could be that the SymR RNA is acting as a titrator rather than a switch. Alternatively there might be additional cellular factors that affect SymR RNA base-pairing with symE, and thus control the amount of SymR RNA that is active for regulation. The stability of SymR can probably be explained by the predicted structure of the RNA, which consists of two strong hairpins.
An intriguing difference between the SymE-SymR system and all previously characterized toxin-antitoxin modules is that in all other cases the antitoxin is rapidly degraded (for example the RelB antitoxin is a target of the Lon protease and the Sok antisense RNA is extremely unstable). In contrast, SymR antitoxin RNA is quite stable and in this case the toxin is target of Lon.
Evolution of an AbrB-fold protein into a toxin
Different independent lines of evidence, sequence divergence, operon organization and experimental results imply that the SymE protein, while originating within the AbrB fold, has evolved into a protein with toxin-like properties. AbrB-type antitoxins are widely distributed throughout the bacterial superkingdom, but the distinctive SymE family is nearly exclusively found only in proteobacteria. This suggests that evolution of the endonuclease is a recent innovation within the AbrB superfamily. While it was previously known that different toxins or antitoxin might be replaced by functionally equivalent, but evolutionarily unrelated equivalents, this appears to be the first case where we observe a possible emergence of a protein with toxin-like properties within a superfamily that contains antitoxins. The barrel-like structure of the dimers and the DNA-binding properties of the AbrB fold may have favoured evolution of an RNA cleaving activity. The presence of predicted operons with two tandem genes of distinct SymE proteins (Fig. 7A) raises the possibility that such duplications might have allowed the original diversification of the fold.
We note that an astonishing range of structurally unrelated protein folds have been recruited for cleavage or degradation of RNAs in toxin-antitoxin modules. These include members of ancient RNase folds such as the PIN domain, which contains the same catalytic fold as the 5′-to 3′ exonucleases, and predicted toxins, like HicA, which contains an RNase H fold (Takagi et al., 2005; Anantharaman and Aravind, 2006). The RelE nuclease domain was originally reported to contain a unique α-β fold with no resemblance to other previously characterized nucleases (Takagi et al., 2005). However, careful structural comparisons show that it shares a common fold, including a core α(2)-β(4) topology, with the tRNA-specific RNase domain of the colicin D family. The MazF/KiD superfamily of toxins has been derived from the ancient SH3-like β-barrel fold, which includes several ancient non-catalytic RNA-binding domains like the ribosomal proteins L24/L21e and the SM domain (Anantharaman and Aravind, 2003). Doc toxins, in contrast appear to have a distinctive all-α helical metal-binding fold (Anantharaman and Aravind, 2003). In this context, the SymE family may represent convergent innovation of catalytic activity from a protein fold, whose previously characterized members were DNA-binding proteins. All together it appears that there has been a selection for similar RNA-cleaving/destabilizing functions on at least six independent occasions in evolution.
The phyletic patterns and genomic distributions of the SymE systems are strongly suggestive of movement between species as well as intragenomic proliferation and mobility. The presence of identical copies of the SymE systems in different genomes is especially indicative of recent duplications and transpositions of these operons. For example, the Erwinia genome has at least 14 copies (including two pairs of identical copies). The isolated presence of a SymE-like system in the Gram-positive bacterium Enterococcus faecalis suggests that the operons might also be laterally transferred between phylogenetically distant bacteria. Another notable feature is their genomic association with diverse mobile elements, such as restriction-modification systems, transposons and pathogenicity islands (see Fig. 7A for examples) (Kobayashi et al., 1999; Schmidt and Hensel, 2004). Widespread lateral mobility across bacterial genomes and association with other mobile elements are observed in other toxin-antitoxin modules. For example, a relBE-type system is associated with the restriction-modification system HsdS in the Gram positive bacterium Streptococcus mutans (Lemos et al., 2005). These observations suggest that SymE and other toxin-antitoxin modules are selfish elements that probably maintain themselves through toxin-dependent cell killing, and to tend proliferate within genomes and colocate to genomic hot spots harbouring other mobile elements. However, it is known that other mobile elements, like the restriction-modification systems and some transposons, on occasion, confer selective advantages to their hosts in the form of defences against DNA viruses or as DNA repair proteins and transcriptional regulators (Kobayashi et al., 1999; Aravind et al., 2000; 2005). Thus, it is possible that some toxin-antitoxin modules were recruited to control the cleavage and degradation of transcripts under specific conditions.
Protection against RNA damage
Until now, RNA lesions have largely been ignored, primarily because damage to DNA was understood to have consequences for subsequent generations, while effects of RNA damage were transient and likely to only affect a limited number of RNA molecules. However, the universal importance of the tmRNA in bacteria and the presence of various mRNA decay mechanisms in eukaryotes, such as ‘no-go decay’ (NGD) for the degradation of mRNAs associated with stalled ribosome-mRNA complexes (Doma and Parker, 2006), indicate that damaged RNAs can also have serious consequences for the cell, in part by inhibiting ribosome function. Thus, it is perhaps not surprising that the SOS response to DNA damage would also lead to the induction of activities that could degrade damaged RNA.
Given the wide distribution of the toxin-antitoxin modules, including the SymE-like toxins, it is intriguing to ask whether other toxin-antitoxin modules are induced by and play roles in the DNA damage responses of other organisms. A transcript encoding the E. coli hokE gene, which shows homology to the plasmid-encoded hok genes, is induced during the SOS response, but it is not clear whether a functional toxin is made from this transcript (Fernández De Henestrosa et al., 2000). The synthesis of the SOS-induced TisB toxin also is regulated by an sRNA, in this case the adjacently encoded IstR RNA (Vogel et al., 2004). The function of this 29-amino acid toxin is not yet known. In addition, a LexA binding site was predicted to be downstream of the promoter of the dinJ-yafQ toxin-antitoxin operon in E. coli (Lewis et al., 1994), although there is no evidence for SOS induction of this operon (Fernández De Henestrosa et al., 2000). Because many other chromosomally encoded toxin-antitoxin genes are induced by stress conditions such as starvation that could lead to RNA damage and stalled ribosomes, we favour the model that RNA recycling is a general function of the ubiquitous toxin-antitoxin modules. Possibly many of the toxin-antitoxin-like systems of bacteria correspond to RNA degradation systems that are functionally analogous to the extensive RNA-based post-transcriptional regulatory mechanisms of eukaryotes.
Experimental procedures
Media and growth conditions
Cells were grown aerobically at 37°C in either Luria–Bertani (LB) medium or M9 minimal medium supplemented with 0.4% glycerol, 0.05% casamino acids, 1 mM MgSO4, 0.1 mM CaCl2 and 1 μg ml−1 thiamine. Antibiotics were used at the following concentrations when needed: ampicillin (50 μg ml−1), kanamycin (30 μg ml−1), chloramphenicol (25 μg ml−1), tetracycline (12.5 μg ml−1). Bacterial growth was monitored by measuring optical density at 600 nm (OD600).
Bacterial strains and plasmids
Bacterial strains and plasmids used in this study are listed in Tables S1 and S2 respectively. The sequences of all oligonucleotides used to generate the strains and plasmids are given in Table S3. The E. coli strains are derivatives of wild-type strain MG1655 (F– lambda–ilvG–rfb-50 rph-1).
SPA-tagged symE strain
The SPA tag which contains the 3 × FLAG and the calmodulin binding peptide (CBP) sequences separated by a TEV protease cleavage site was synthesized by the polymerase chain reaction (PCR) with plasmid pJL148 as a template (Zeghouf et al., 2004). The gel-purified PCR product was used to transform NM1100 (MG1655 mini-λ tet) to introduce the SPA tag at the end of symE using mini-λ Red recombination (Yu et al., 2000). The inserted symE-SPA-kan allele was moved onto MG1655 by P1 transduction. Subsequently, the kan cassette was excised by using the pCP20 plasmid (Cherepanov and Wackernagel, 1995) generating GSO114.
The −10 mutant strain
The −10 knockdown mutation of the symR promoter was constructed by using the mini-λ Red recombination method with strain NM1100 (Court et al., 2003) and single-stranded oligonucleotides with two silent mutations (CATTCT to CACTCC) in the symE open reading frame that abolish the symR promoter. The mini-λ tet prophage simultaneously was eliminated from this strain by a temperature shift from 30°C to 42°C, generating GSO115. The parent NM1100 strain for which the mini-λ tet prophage was similarly eliminated was used as the corresponding wild-type strain. To construct a −10 mutant derivative of the symE-SPA strain, the mini-λ tet allele of NM1100 was introduced into GSO114 by P1 transduction, and the −10 mutation was introduced as described above generating GSO116.
ΔsymER mutant strain
The coding region of the symE gene in strain DY330 was deleted and replaced by the kan gene, which was synthesized by PCR with the plasmid pKD4 as a template (Datsenko and Wanner, 2000; Yu et al., 2000), again using the mini-λ Red recombination method (Yu et al., 2000). The inserted kan allele was introduced into MG1655 by P1 transduction generating GSO117. Finally, the kan cassette was excised by using the pCP20 plasmid (Cherepanov and Wackernagel, 1995) generating GSO118.
Other mutant strains
To allow for more homogeneous expression from the PBAD promoter on some of the plasmids used, the native araE promoter on the chromosome was replaced by constitutive promoter PCP18 by P1 transduction of the kan-PCP18-araE allele of strain BW27750 (Khlebnikov et al., 2001) into MG1655 generating GSO119. For some experiments the kan cassette in GSO119 was excised by using the pCP20 plasmid (Cherepanov and Wackernagel, 1995) generating GSO120. The hfq-1::Ω (Tsui et al., 1997), rnc-14::ΔTn10 (Takiff et al., 1989), lon146::Tn10 (Maurizi et al., 1985) and clpP::cat (Maurizi et al., 1990) alleles were introduced into GSO114, GSO115, GSO116 or GSO118 by P1 transduction to give GSO122-GSO128.
Plasmids
The symR promoter and coding sequence were amplified by PCR and cloned into the ScaI and BstZ17I sites of pACYC184 (Chang and Cohen, 1978) to generate pACYC-SymR. To generate a pBAD18-Cm (Guzman et al., 1995) derivative in which sRNA genes could easily be cloned behind the PBAD promoter such that the sRNA would be expressed with the proper +1, the EcoRI site within cat gene was abolished by site-directed mutagenesis (GAATTC to GAATTT; Stratagene). A new EcoRI site was generated by site-directed mutagenesis downstream of PBAD such that transcription would be initiated directly downstream of the EcoRI site generating pAZ3. The symR gene amplified and cloned into the new EcoRI and the HindIII sites in a reverse orientation to generate pBAD-anti-SymR. The coding sequences of symE and symE-SPA were PCR-amplified from MG1655 and GSO114 and cloned into the filled-in NcoI site of pBAD24 (Guzman et al., 1995) to generate pBAD-SymE and pBAD-SymE-SPA respectively.
RNA analysis
In all cases, total RNA was isolated by acid hot-phenol extraction (Kawano et al., 2002).
Northern analysis
Total RNA in Formaldehyde Loading Dye (Ambion) containing 5 μg ml−1 ethidium bromide was denatured at 75°C for 5 min and separated on 1.2% agarose gel containing formaldehyde alongside radiolabelled RNA Perfect Markers (Novagen). The RNA was transferred to NYTRAN nylon transfer membranes (Schleicher and Schuell) by capillary action and subject to UV cross-linking. Membranes then were probed with 32P-labelled oligonucleotide probes (listed in Table S3) in ULTRAhyb-Oligo buffer (Ambion) at 45°C and washed as described in the manual for NYTRAN nylon transfer membranes.
Quantitative northern analysis
The quantitative northern analysis was performed essentially as described above except that different amounts of total RNA were analysed depending on the sample. The total RNA samples were run alongside in vitro synthesized control symE mRNA and SymR RNA. To synthesize the control RNA, DNA fragments for symE mRNA and SymR RNA were amplified by PCR using primers containing T7 promoter sequence (Table S3) and genomic DNA of MG1655 as a template. In vitro transcription were performed with the gel-purified PCR products and T7 polymerase, and synthesized symE mRNA and SymR were purified with MicroSpin G-50 spin columns (Amersham Biosciences) and quantified by measurement at A260.
Protein analysis
Immunoblot assays
Total cell lysate was mixed with 2 × sample buffer (Sigma-Aldrich), heated at 95°C for 5 min, a fraction equivalent to the cells in OD600 = 0.05 was separated on Novex 10–20% Tris-Glycine gel (Invitrogen), and transferred to a nitrocellulose membrane (Invitrogen). The membranes were incubated with anti-FLAG M2-AP monoclonal antibody (Sigma-Aldrich) and polyclonal anti-RecA antibody (kindly provided by D. Camerini-Otero) to detect the SPA-tagged SymE and RecA proteins respectively. Signals were visualized using the Lumi-Phos WB (Pierce) for anti-FLAG M2-AP monoclonal antibody and HRP-conjugated secondary antibody (Amersham) together with the SuperSignal West Pico Chemiluminescent Substrate (Pierce) for the anti-RecA antibody.
Affinity purification of SPA-tagged protein
The protocol used to purify SymE-SPA was adapted from (Morita et al., 2005) with some modifications. GSO119/pBAD-SymE-SPA was grown to OD600∼0.4 at 37°C in 500 ml of LB medium containing ampicillin. Expression of SymE-SPA was induced by the addition of 0.02% arabinose and 0.5 μg ml−1 mitomycin C. After 2 h, cells (450 ml) were harvested and then washed with 20 ml of STE buffer (10 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1 mM EDTA). The cell pellets were resuspended with 10 ml of IP buffer 2 (20 mM Tris-HCl [pH 8.0], 0.2 M KCl, 5 mM MgCl2, 10% glycerol, 0.1% Tween 20) containing Complete Mini protease inhibitor cocktail (Roche). The cell suspension was sonicated, and the cell debris was removed by centrifugation at 10 000 g for 1 h at 4°C. The crude extract was incubated with 500 μl of anti-FLAG M2-agarose beads (Sigma-Aldrich) overnight with rotation at 4°C. The mixture was then filtered using a poly prep chromatography column (Bio-Rad). The agarose beads were washed five times with 10 ml of IP buffer 2. Subsequently, bound proteins were eluted with 200 μl of IP buffer 2 containing 2 mg ml−1 3 × FLAG peptide (Sigma-Aldrich). The purified proteins were analysed by SDS-PAGE using Novex 4–20% Tris-Glycine gel (Invitrogen), and the gel was stained with GelCode Blue Stain Reagent (Pierce).
Identification of proteins by mass spectrometry
Stained protein bands were excised from the gel and digested with 250 ng of sequencing grade trypsin (Roche Applied Science) in 25 μl of 25 mM ammonium bicarbonate overnight at 37°C. The digested peptides were eluted with mixture of acetonitrile and trifluoroacetic acid, and concentrated to approximately 10 μl by speed vac. The samples were desalted with ZipTipc18 pipette tips (Millipore). Protein identification was accomplished by automated LC/MS/MS analysis with searches of the E. coli protein database (http://trypsin.nichd.nih.gov/mshome.htm) using the software tool Mascot (Matrix Science).
Pulse-chase labelling
GSO120/pBAD-SymE cells were grown to OD600∼0.5 at 37°C in 50 ml of M9 medium with glycerol, casamino acids and ampicillin. Cells were then treated with 0.02% arabinose to induce symE transcription. Samples (0.5 ml) were taken at the indicated time points and added to 20 μCi of Tran35S-label containing 35S-methionine and 35S-cysteine (MP Biomedicals). After 1 min, 0.6 mg of unlabelled methionine and cysteine was added to the samples. After 10 more min at 37°C, cells were precipitated with 5% trichloroacetic acid. The precipitates were collected by centrifugation, washed with acetone, dissolved in about 80 μl of 2 × sample buffer (Sigma-Aldrich). Finally the samples were analysed by SDS-PAGE using Novex 10–20% Tris-Glycine gel (Invitrogen) followed by autoradiography.
Protein sequence and structure analysis
The non-redundant (NR) database of protein sequences (National Center for Biotechnology Information, NIH, Bethesda) was searched using the blastp program (Altschul et al., 1997; Schaffer et al., 2001). Gene neighbourhoods were determined using a custom script that uses completely sequenced genomes or whole genome shot gun sequences to derive a table of gene neighbours centred on a query gene. Then the blastclust program is used to cluster the products in the neighbourhood and establish conserved co-occurring genes (http://www.ncbi.nlm.nih.gov/blast/docs/blastclust.html). These conserved gene neighbourhood are then sorted as per a ranking scheme based on occurrence in at least one other phylogenetically distinct lineage (‘phylum’ in NCBI Taxonomy database) and physical closeness (< 70 nucleotides) on the chromosome indicating sharing of regulatory −10 and −35 elements. Putative promoter regions were predicted if required by scanning for the consensus of the −10 and −35 elements in the predicted upstream regions.
Profile searches were conducted using the psi-blast program (Altschul et al., 1997; Schaffer et al., 2001) with either a single sequence or an alignment used as the query, with a default profile inclusion expectation (E) value threshold of 0.01 (unless specified otherwise), and was iterated until convergence. HMM searches were carried out using the hmm_search program of the HMMER package, after they were optimized with the HMM_caliberate program (Eddy, 1998). For all searches involving membrane-spanning domains we used a statistical correction for compositional bias to reduce false positives due to the general hydrophobicity of these proteins. Multiple alignments were constructed using MUSCLE program followed by manual adjustments based on psi-blast results (Edgar, 2004). Structural manipulations were carried out using the Swiss-PDB viewer program (Guex and Peitsch, 1997). Protein secondary structure was predicted using a multiple alignment as the input for the jpred program, with information extracted from a PSSM, HMM and the seed alignment itself (Cuff and Barton, 2000).
Acknowledgments
We thank A. Yergey for performing the mass spectrometric analysis, D. Camerini-Otero, E. Fozo, N. Majdalani, M. Maurizi, L. Van Melderen and R. Woodgate for providing advice, strains and antibodies and D. Camerini-Otero, S. Gottesman and L. Van Melderen for editorial comments. Supported by the intramural programs of the National Institute of Child Health and Human Development and National Center for Biotechnology Information and by a research fellowship from the Japan Society for the Promotion of Science (M.K.).
Supplementary material
The following supplementary material is available for this article online:
SymE overexpression leads to reduced levels of lacI mRNA with and without a start codon.
Multiple alignment of the SymE family and other members of the AbrB superfamily.
Strains used in study.
Plasmids used in study.
Oligonucleotides used in study.
This material is available as part of the online article from http://www.blackwell-synergy.com
Please note: Blackwell Publishing is not responsible for the content or functionality of any supplementary materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Aizenman E, Engelberg-Kulka H, Glaser G. An Escherichia coli chromosomal ‘addiction module’ regulated by guanosine 3′,5′-bispyrophosphate: a model for programmed bacterial cell death. Proc Natl Acad Sci USA. 1996;93:6059–6063. doi: 10.1073/pnas.93.12.6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped blast and psi-blast: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantharaman V, Aravind L. New connections in the prokaryotic toxin-antitoxin network: relationship with the eukaryotic nonsense-mediated RNA decay system. Genome Biol. 2003;4:R81. doi: 10.1186/gb-2003-4-12-r81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantharaman V, Aravind L. Novel predicted RNAses with a PIN Domain-like Fold. RNA Biol. 2006;3:18–27. doi: 10.4161/rna.3.1.2548. [DOI] [PubMed] [Google Scholar]
- Aravind L, Makarova KS, Koonin EV. Holliday junction resolvases and related nucleases: identification of new families, phyletic distribution and evolutionary trajectories. Nucleic Acids Res. 2000;28:3417–3432. doi: 10.1093/nar/28.18.3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Anantharaman V, Balaji S, Babu MM, Iyer LM. The many faces of the helix–turn–helix domain: transcription regulation and beyond. FEMS Microbiol Rev. 2005;29:231–262. doi: 10.1016/j.femsre.2004.12.008. [DOI] [PubMed] [Google Scholar]
- Blomberg P, Wagner EGH, Nordstrom K. Control of replication of plasmid R1: the duplex between the antisense RNA, CopA, and its target, CopT, is processed specifically in vivo and in vitro by RNase III. EMBO J. 1990;9:2331–2340. doi: 10.1002/j.1460-2075.1990.tb07405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buts L, Lah J, Dao-Thi M-H, Wyns L, Loris R. Toxin-antitoxin modules as bacterial metabolic stress managers. Trends Biochem Sci. 2005;30:672–679. doi: 10.1016/j.tibs.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Chang AC, Cohen SN. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanov PP, Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- Christensen SK, Gerdes K. RelE toxins from bacteria and Archaea cleave mRNAs on translating ribosomes, which are rescued by tmRNA. Mol Microbiol. 2003;48:1389–1400. doi: 10.1046/j.1365-2958.2003.03512.x. [DOI] [PubMed] [Google Scholar]
- Christensen SK, Pedersen K, Hansen FG, Gerdes K. Toxin-antitoxin loci as stress-response-elements: ChpAK/MazF and ChpBK cleave translated RNAs and are counteracted by tmRNA. J Mol Biol. 2003;332:809–819. doi: 10.1016/s0022-2836(03)00922-7. [DOI] [PubMed] [Google Scholar]
- Condon C. Shutdown decay of mRNA. Mol Microbiol. 2006;61:573–583. doi: 10.1111/j.1365-2958.2006.05270.x. [DOI] [PubMed] [Google Scholar]
- Court D, Swaminathan S, Yu D, Wilson H, Baker T, Bubunenko M, et al. Mini-lambda: a tractable system for chromosome and BAC engineering. Gene. 2003;315:63–69. doi: 10.1016/s0378-1119(03)00728-5. [DOI] [PubMed] [Google Scholar]
- Cuff JA, Barton GJ. Application of multiple sequence alignment profiles to improve protein secondary structure prediction. Proteins. 2000;40:502–511. doi: 10.1002/1097-0134(20000815)40:3<502::aid-prot170>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doma MK, Parker R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 2006;440:561–564. doi: 10.1038/nature04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelberg-Kulka H, Glaser G. Addiction modules and programmed cell death and antideath in bacterial cultures. Annu Rev Microbiol. 1999;53:43–70. doi: 10.1146/annurev.micro.53.1.43. [DOI] [PubMed] [Google Scholar]
- Fernández De Henestrosa AR, Ogi T, Aoyagi S, Chafin D, Hayes JJ, Ohmori H, Woodgate R. Identification of additional genes belonging to the LexA regulon in Escherichia coli. Mol Microbiol. 2000;35:1560–1572. doi: 10.1046/j.1365-2958.2000.01826.x. [DOI] [PubMed] [Google Scholar]
- Galvani C, Terry J, Ishiguro EE. Purification of the RelB and RelE proteins of Escherichia coli: RelE binds to RelB and to ribosomes. J Bacteriol. 2001;183:2700–2703. doi: 10.1128/JB.183.8.2700-2703.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K, Nielsen A, Thorsted P, Wagner EGH. Mechanism of killer gene activation. Antisense RNA-dependent RNase III cleavage ensures rapid turn-over of the stable Hok SrnB and PndA effector messenger RNAs. J Mol Biol. 1992;226:637–649. doi: 10.1016/0022-2836(92)90621-p. [DOI] [PubMed] [Google Scholar]
- Gerdes K, Gultyaev AP, Franch T, Pedersen K, Mikkelsen ND. Antisense RNA-regulated programmed cell death. Annu Rev Genet. 1997;31:1–31. doi: 10.1146/annurev.genet.31.1.1. [DOI] [PubMed] [Google Scholar]
- Gerdes K, Christensen SK, Løbner-Olesen A. Prokaryotic toxin-antitoxin stress response loci. Nat Rev Microbiol. 2005;3:371–382. doi: 10.1038/nrmicro1147. [DOI] [PubMed] [Google Scholar]
- Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano M, Oshima T, Kasai H, Mori H. Molecular characterization of long direct repeat (LDR) sequences expressing a stable mRNA encoding for a 35-amino-acid cell-killing peptide and a cis-encoded small antisense RNA in Escherichia coli. Mol Microbiol. 2002;45:333–349. doi: 10.1046/j.1365-2958.2002.03042.x. [DOI] [PubMed] [Google Scholar]
- Kawano M, Reynolds AA, Miranda-Rios J, Storz G. Detection of 5′-and 3′ UTR-derived small RNAs and cis-encoded antisense RNAs in Escherichia coli. Nucleic Acid Res. 2005;33:1040–1050. doi: 10.1093/nar/gki256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khlebnikov A, Datsenko KA, Skaug T, Wanner BL, Keasling JD. Homogeneous expression of the PBAD promoter in Escherichia coli by constitutive expression of the low-affinity high-capacity AraE transporter. Microbiology. 2001;147:3241–3217. doi: 10.1099/00221287-147-12-3241. [DOI] [PubMed] [Google Scholar]
- Kobayashi I, Nobusato A, Kobayashi-Takahashi N, Uchiyama I. Shaping the genome – restriction-modification systems as mobile genetic elements. Curr Opin Genet Dev. 1999;9:649–656. doi: 10.1016/s0959-437x(99)00026-x. [DOI] [PubMed] [Google Scholar]
- Lemos JA, Brown TA, Abranches J, Burne RA. Characteristics of Streptococcus mutans strains lacking the MazEF and RelBE toxin-antitoxin modules. FEMS Microbiol Lett. 2005;253:251–257. doi: 10.1016/j.femsle.2005.09.045. [DOI] [PubMed] [Google Scholar]
- Lewis K. Persister cells and the riddle of biofilm survival. Biochemistry (Mosc) 2005;70:267–274. doi: 10.1007/s10541-005-0111-6. [DOI] [PubMed] [Google Scholar]
- Lewis LK, Harlow GR, Gregg-Jolly LA, Mount DW. Identification of high affinity binding sites for LexA which define new DNA damage-inducible genes in Escherichia coli. J Mol Biol. 1994;241:507–523. doi: 10.1006/jmbi.1994.1528. [DOI] [PubMed] [Google Scholar]
- Malmgren C, Wagner EGH, Ehresmann C, Ehresmann B, Romby P. Antisense RNA control of plasmid R1 replication. The dominant product of the antisense rna-mrna binding is not a full RNA duplex. J Biol Chem. 1997;272:12508–12512. doi: 10.1074/jbc.272.19.12508. [DOI] [PubMed] [Google Scholar]
- Maurizi MR, Trisler P, Gottesman S. Insertional mutagenesis of the lon gene in Escherichia coli: lon is dispensable. J Bacteriol. 1985;164:1124–1135. doi: 10.1128/jb.164.3.1124-1135.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurizi MR, Clark WP, Katayama Y, Rudikoff S, Pumphrey J, Bowers B, Gottesman S. Sequence and structure of Clp P, the proteolytic component of the ATP-dependent Clp protease of Escherichia coli. J Biol Chem. 1990;265:12536–12545. [PubMed] [Google Scholar]
- Morita T, Maki K, Aiba H. RNase E-based ribonucleoprotein complexes: mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes Dev. 2005;19:2176–2186. doi: 10.1101/gad.1330405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neher SB, Villen J, Oakes EC, Bakalarski CE, Sauer RT, Gygi SP, Baker TA. Proteomic profiling of ClpXP substrates after DNA damage reveals extensive instability within SOS regulon. Mol Cell. 2006;22:193–204. doi: 10.1016/j.molcel.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Pandey DP, Gerdes K. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 2005;33:966–976. doi: 10.1093/nar/gki201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen K, Christensen SK, Gerdes K. Rapid induction and reversal of a bacteriostatic condition by controlled expression of toxins and antitoxins. Mol Microbiol. 2002;45:501–510. doi: 10.1046/j.1365-2958.2002.03027.x. [DOI] [PubMed] [Google Scholar]
- Schaffer AA, Aravind L, Madden TL, Shavirin S, Spouge JL, Wolf YI, et al. Improving the accuracy of psi-blast protein database searches with composition-based statistics and other refinements. Nucleic Acids Res. 2001;29:2994–3005. doi: 10.1093/nar/29.14.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H, Hensel M. Pathogenicity islands in bacterial pathogenesis. Clin Microbiol Rev. 2004;17:14–56. doi: 10.1128/CMR.17.1.14-56.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz G, Gottesman S. Versatile roles of small RNA regulators in bacteria. In: Gesteland RF, Cech TR, Atkins JF, editors. The RNA World. 3. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2006. pp. 567–594. [Google Scholar]
- Takagi H, Kakuta Y, Okada T, Yao M, Tanaka I, Kimura M. Crystal structure of archaeal toxin-antitoxin RelE-RelB complex with implications for toxin activity and antitoxin effects. Nat Struct Mol Biol. 2005;12:327–331. doi: 10.1038/nsmb911. [DOI] [PubMed] [Google Scholar]
- Takiff HE, Chen SM, Court DL. Genetic analysis of the rnc operon of Escherichia coli. J Bacteriol. 1989;171:2581–2590. doi: 10.1128/jb.171.5.2581-2590.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui HC, Feng G, Winkler ME. Negative regulation of mutS and mutH repair gene expression by the Hfq and RpoS global regulators of Escherichia coli K-12. J Bacteriol. 1997;179:7476–7487. doi: 10.1128/jb.179.23.7476-7487.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J, Argaman L, Wagner EGH, Altuvia S. The small RNA IstR inhibits synthesis of an SOS-induced toxic peptide. Curr Biol. 2004;14:2271–2276. doi: 10.1016/j.cub.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Wagner EG, Altuvia S, Romby P. Antisense RNAs in bacteria and their genetic elements. Adv Genet. 2002;46:361–398. doi: 10.1016/s0065-2660(02)46013-0. [DOI] [PubMed] [Google Scholar]
- Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci USA. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeghouf M, Li J, Butland G, Borkowska A, Canadien V, Richards D, et al. Sequential peptide affinity (SPA) system for the identification of mammalian and bacterial protein complexes. J Proteome Res. 2004;3:463–468. doi: 10.1021/pr034084x. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhang J, Hoeflich KP, Ikura M, Qing G, Inouye M. MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Mol Cell. 2003;12:913–923. doi: 10.1016/s1097-2765(03)00402-7. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhu L, Zhang J, Inouye M. Characterization of ChpBK, an mRNA interferase from Escherichia coli. J Biol Chem. 2005;280:26080–26088. doi: 10.1074/jbc.M502050200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SymE overexpression leads to reduced levels of lacI mRNA with and without a start codon.
Multiple alignment of the SymE family and other members of the AbrB superfamily.
Strains used in study.
Plasmids used in study.
Oligonucleotides used in study.