Abstract
The mutation rates of cancer cells to drug and multidrug resistance are paradoxically high, i.e., 10−3 to 10−6, compared with those altering phenotypes of recessive genes in normal diploid cells of about 10−12. Here the hypothesis was investigated that these mutations are due to chromosome reassortments that are catalyzed by aneuploidy. Aneuploidy, an abnormal number of chromosomes, is the most common genetic abnormality of cancer cells and is known to change phenotypes (e.g., Down's syndrome). Moreover, we have shown recently that aneuploidy autocatalyzes reassortments of up to 2% per chromosome per mitosis because it unbalances spindle proteins, even centrosome numbers, via gene dosage. The hypothesis predicts that a selected phenotype is associated with multiple unselected ones, because chromosome reassortments unbalance simultaneously thousands of regulatory and structural genes. It also predicts variants of a selected phenotype based on variant reassortments. To test our hypothesis we have investigated in parallel the mutation rates of highly aneuploid and of normal diploid Chinese hamster cells to resistance against puromycin, cytosine arabinoside, colcemid, and methotrexate. The mutation rates of aneuploid cells ranged from 10−4 to 10−6, but no drug-resistant mutants were obtained from diploid cells in our conditions. Further selection increased drug resistance at similar mutation rates. Mutants selected from cloned cells for resistance against one drug displayed different unselected phenotypes, e.g., polygonal or fusiform cellular morphology, flat or three-dimensional colonies, and resistances against other unrelated drugs. Thus our hypothesis offers a unifying explanation for the high mutation rates of aneuploid cancer cells and for the association of selected with unselected phenotypes, e.g., multidrug resistance. It also predicts drug-specific chromosome combinations that could become a basis for selecting alternative chemotherapy against drug-resistant cancer.
The spontaneous emergence of drug- and multidrug-resistant mutants is the nemesis of cancer chemotherapy (1–5). Therefore it has been studied for half a century now (6), ever since cytotoxic drugs were first used for cancer therapy (5), but its mechanism is still unclear (7, 8). The large body of work on the subject shows that the mutation of cancer cells, and of tumorigenic cells in culture, is strikingly different from mutations of normal diploid cells, based on the following five criteria.
(i) Paradoxically High Rates of Mutation to Drug Resistance.
Resistance to cytotoxic drugs reflects the loss of some normal metabolic function. Therefore, both alleles of normal diploid cells must be mutated to manifest the resistant phenotype. According to the spontaneous mutation rates of haploid human genes, which are about 10−6 (9), the mutation rates of diploid cells to drug resistance by gene mutation are predictably very low, i.e., only about 10−12 (7, 8, 10, 11). But numerous investigators (7, 8, 10–13) have observed that the mutation rates to drug resistance of cancer cells and of tumorigenic cell lines “are much higher than expected on the basis of germinal eukaryotic mutation rates” (14). The frequencies of these mutations range between 10−3 and 10−6 per mitosis (10, 11, 15–19). For example, one of 106 human leukemic cells is resistant to amethopterin (1), and one of 105 cells from a mouse cancer is metastatic (20). According to Gartler and Pious, “resistance to high drug levels on a classical genetic scheme would involve at least two mutational steps. Yet the mutation rate estimates for their resistance are of the same order of magnitude or much higher than are reported for single events in germinally occurring mutants. On this interpretation the rate of occurrence of somatic cell variants is thousands of times higher than for germinal mutations” (10). This difference in the rate of occurrence, according to Henry Harris, defines a “major conceptual difficulty,” i.e., “to reconcile the very high mutational frequency with genetic theory if two functional alleles are present in the same cells” (8).
(ii) High Rates of Reversion of Drug-Resistant Phenotypes.
Conventional gene mutations are genetically just as stable as their parents (9). But drug resistance of cancer cells and tumorigenic cell lines is significantly less stable than conventional mutations, some reverting to the original or other phenotypes at the same rates with which they were generated (2, 7, 12, 18, 20–22).
(iii) A Continuum of Homologous Phenotypes.
Mutations of recessive genes are typically all or nothing (9). But the drug resistance of cancer cells displays a continuum of transitory phenotypes shifting to increasing resistance on consecutive rounds of selection (2, 6, 23, 24). Regarding the mechanism, one investigator proposed that “step-wise drug selection provides strong, indirect evidence that an unstable gene dosage mechanism” is at work (2). Another suggests that “multiple events contribute to resistance” of the same drug (25). Likewise, Koski et al. have recently concluded that the generation of drug-resistant human leukemic cells is “a complex rather than a simple molecular mechanism” (26).
(iv) Mutation Rates Independent of Ploidy.
Based on gene mutation, “the frequency of mutants with a recessive phenotype should be exponentially related to the number of functional genes per cell” (27). In contrast, the mutation frequency of tumorigenic cells to drug resistance is independent, or practically independent, of ploidy levels ranging from pseudohaploid (numerically haploid, but structurally aneuploid) to pseudooctaploid (8, 13, 14, 28).
(v) Multidrug Resistance.
Selection of a mutant phenotype from normal diploid organisms typically yields variants with just one mutated gene or operon (9). But “[w]hen cultured cells are exposed to … a chemotherapeutic drug, individual clones can be selected that express … resistance to multiple drugs that may be structurally and functionally unrelated. Such cross-resistance occurs frequently in cultured cell lines and is termed the multidrug resistance (MDR) phenotype. The MDR phenotype is also encountered in the clinical setting where many human cancers are refractory to multiagent chemotherapy” (2). Therefore, a recent reviewer concluded that multidrug resistance “had to be understood as the net effect of a multifactorial process” (4).
In view of these peculiarities, some have tried to reconcile the high mutation rates of cancer cells and cell lines with gene mutation by postulating “mutator genes” (11) or a “mutator phenotype” (29), although, according to Breslow and Goldsby, the “suggestion of a mutator gene really begs the question. … [because] one is left with the problem of accounting for such a high frequency of mutator genes” (11). Indeed, mutator genes are not commonly found in cancer cells (30–35). Others have postulated multidrug resistance genes (2, 4). But this generates a new paradox because such genes never seem to protect normal cells against chemotherapy, e.g., no drug-resistant normal lymphocytes are ever observed in chemotherapy of leukemia. Even “nonmutational” (13) and “epigenetic” mechanisms (7, 12, 14, 17, 22) have been postulated to explain the frequent mutation of cancer cells to drug resistance. And “additional degrees of freedom which appear to exist in tumors and established cell systems in vitro ” were proposed by Morgan Harris, specifically to explain the ploidy independence of mutation (28).
Here we propose that mutation of cancer cells and of tumorigenic cell lines in culture is due to chromosome reassortments and that reassortments at high rates are catalyzed by aneuploidy, an abnormal balance of chromosomes. Aneuploidy is the most common genetic abnormality of cancer cells and tumorigenic cell lines (8, 36–38) and is known to change phenotypes by changing the expression of hundreds to thousands of genes (e.g., Down's syndrome) (39–43). Moreover, we have shown recently that aneuploidy destabilizes the karyotype, because it unbalances the many balance-sensitive components of the spindle apparatus including even the numbers of centrosomes (44–46), autocatalytically by unbalancing the dosage of the corresponding genes (42, 47, 48). The risk of gaining or losing a given chromosome per mitosis in a highly aneuploid cancer cell was found to be about 2%, which corresponds to a 46% risk for a highly aneuploid human cell to gain or lose one chromosome per mitosis (48, 49). The more aneuploid the karyotype, the more unstable it will be (42, 48, 50). In contrast, the risk of normal diploid cells of gaining or losing a single chromosome during mitosis is very low, ranging from 0% in human embryos (51) and adolescents (52) to 0.4% in adults, based on finding trisomic chromosomes in 0.2% (53).
Thus chromosome reassortment catalyzed by aneuploidy is an “additional degree of freedom” of mutation available only to aneuploid cancer cells and cell lines—the probable basis of the notorious genetic instability of cancer cells (42, 48). The high mutation rates of influenza virus via reassortments of subgenomic RNA segments (54, 55) versus the extremely low mutation rates of viruses with singular genomic RNAs (56) are an exact precedent for our model.
Chromosome reassortment can alter phenotypes by two distinct mechanisms that make different predictions. (i) Suppose there are alternative pathways to the same metabolic goal. This must be expected because alternative biochemical mechanisms have already been identified that generate the same drug-resistance phenotypes (4, 7, 22, 57, 58). In this case chromosome reassortment could switch on an alternative pathway for drug resistance by amplifying the chromosomes that contain the corresponding structural and/or regulatory genes. The neoantigens and newly expressed RNAs of cancer cells are predictable consequences (3, 42, 43, 59). The reversibility of this process would also explain the reversibility of many drug-resistant phenotypes described above. (ii) Suppose a cell is heterozygous for a recessive drug resistance gene. Such a cell could become drug resistant if the chromosome with the intact gene is lost by reassortment. This reassortment would create an irreversible mutation, even if the respective chromosome is subsequently doubled again.
Chromosome reassortment can also explain the transitory phenotypes described above as distinct chromosome combinations, i.e., phenotype-specific karyotypes. Chromosome reassortment further predicts new unselected phenotypes in addition to selected ones, because chromosome reassortments alter the dosage of thousands of regulatory and structural genes. Moreover, the hypothesis predicts that mutations by chromosome reassortment are not, or only little, dependent on the “ploidy” of the respective aneuploid karyotype.
To test this proposal we have compared here directly the mutation rates of aneuploid tumorigenic cells with those of normal, diploid cells from the same inbred line of Chinese hamsters (CHs) (60). Mutants were selected for resistance to cytotoxic chemotherapeutic drugs such as methotrexate, cytosine arabinonucleoside (araC), colcemid, and puromycin.
Materials and Methods
Cells.
Embryo cells from a single 18-day-old male of an inbred line of CHs were prepared as described previously (60). The preparation and clonal isolation of three chemically transformed CH cell lines, termed D 313, M 853, and B 644, has also been described previously (48). The cells were propagated and maintained in DMEM supplemented with 10% FCS and antibiotics following published procedures (47).
Cytogenetic Analysis.
Cell cultures that were about 75% confluent were incubated with 300 μl Karymax (GIBCO/BRL) (i.e., 3 μg colcemid) for about 3 h. The cells were then rinsed once with PBS, dissociated by trypsin, harvested before spontaneous clumping set in, mixed with a small volume of complete medium, and centrifuged for 4 min at 750 rpm at room temperature, following a protocol of GIBCO/BRL. The cells were resuspended in 100–150 μl of the supernatant, mixed with 500 μl 0.075 M KCl (GIBCO/BRL), and 1 min later were mixed with another 3.5 ml of the hypotonic KCl solution. After a total of 6 min at room temperature the cells were centrifuged for 6 min as described above. The cells were then again resuspended in 500 μl of the supernatant and first mixed with 0.5 ml and, 1 min later, with another 3.5 ml of a solution of 3 vol of ethanol and 1 vol of acetic acid. After incubation at room temperature for 15 min the cells were centrifuged for 6 min as above. This procedure was repeated once more; the cells were then suspended in 0.5–1 ml of the ethanol–acetic acid solution, and aliquots were pipetted on the upper, long edge of a tilted microscope slide with a 10-μl Eppendorf pipette tip. The chromosomes are then directly visible with a phase-contrast microscope as previously described (47).
Treatments of Cultured Cells with Cytotoxic Drugs.
Between 2 million and 4 million cells were seeded on a 10-cm Petri dish in 7 to 10 ml of medium. One or more of the cytotoxic drugs puromycin (Sigma), araC (Mack–Pfizer, Illertissen, Germany), colcemid (GIBCO/BRL), and methotrexate (Lederle Laboratories, Pearl River, NY) were added to the culture medium either when the cells were seeded or less than 24 h later. The concentration of drugs is reported in micrograms per Petri dish (rather than in molarities, which depend on the volume of medium and on uptake).
Results
Effect of Aneuploidy on Mutation Rates to Drug Resistance.
To determine whether the high mutation rates of cancer cells and tumorigenic cells to drug resistance are due to gene mutation or to chromosome reassortments catalyzed by aneuploidy, we have compared mutation rates in parallel cultures of normal diploid and aneuploid cells. To minimize other genetic differences, aneuploid and diploid cells were derived from the same inbred line of CHs (60).
The diploid CH embryo (CHE) cells were prepared from animals and had been propagated in vitro for a few generations before use in our experiments (Materials and Methods). The aneuploid CH cells studied included three chemically transformed cell lines that had been generated with dimethylbenzanthracene, methylcholanthrene, and benzpyrene (hence termed D 313, M 853, and B 644, respectively) and had subsequently been cloned as described previously (47, 48). The chromosome distribution of each of these clonally derived, aneuploid lines is very heterogeneous, as is characteristic of aneuploid cells (12), and the modal (i.e., most common) chromosome number of D 313 is 31, that of M 853 is 38, and that of B 644 is 37 (48). Thus based on 22, the normal diploid chromosome number of the CH, each of these lines is highly aneuploid, roughly in the triploid range. Because all cells studied have an identical genetic background, except for the presence or absence of aneuploidy, different mutation rates to drug resistance would have to be due to aneuploidy.
For the selection of drug-resistant mutants, 2 million to 4 million cells were treated in 10-cm plastic Petri dishes with one of the four cytotoxic drugs puromycin, araC, colcemid, or methotrexate, as described in Materials and Methods. Three of these drugs, i.e., puromycin, methotrexate, and colcemid, were chosen because they are not genotoxic and thus do not alter the spontaneous mutation rates of cells.
Within 10–40 days after the initiation of drug treatments all three aneuploid cell lines had generated drug-resistant colonies at frequencies of 1–200 per 106 unselected cells, whereas under the same conditions, normal diploid CHE cells failed to generate any resistant colonies (See Table 1). Thus our results confirm the hypothesis that chromosome reassortments catalyzed by aneuploidy may be the mechanism of mutation to drug resistance.
Table 1.
Frequency of drug-resistant colonies among cell cultures exposed to toxic drugs
| Drug | Cells | Time until resistant colonies, days | Colonies per 2 to 4 × 106 cells |
|---|---|---|---|
| Puromycin | CHE* | — | 0 (3 expts) |
| (1–20 μg per 10-cm | B 644† | 12, 18, 12 | 13, >80, >50‡ |
| dish) | D 313 | —, 11, 21 | 0, 3, 20 |
| M 853 | 14, 18 | 11, >100* | |
| SpoT 1 | 17, 10 | 2, >200* | |
| SpoT 2 | 10 | >200* | |
| Cytosine arabinoside | CHE | — | 0 (3 expts) |
| (0.25–10 μg per 10-cm | B 644 | 14, 15 | 40, >200 |
| dish) | D 313 | —, 15 | 0, 3 |
| M 853 | —, 15 | 0, >100 | |
| SpoT 1 | —, 10 | 0, >50 | |
| SpoT 2 | 30 | >200 | |
| Colcemid | CHE | — | 0, 0 |
| (0.1 μg per 10-cm dish) | B 644 | 26, 40 | 1, >200 |
| D 313 | —, 26 | 0, 3 | |
| M 853 | 26, 40 | 9, >100 | |
| (0.2 μg per 10-cm dish) | CHE | — | 0, 0 |
| B-01col† | 16, 9 | 10/7, 2 | |
| M-01col | 16, 9 | 28/75, 27 | |
| Methotrexate | CHE | — | 0 |
| (1.25–2.5 μg per 10-cm | B 644 | 11 | 82/17 |
| dish) | D 313 | 9 | 135 |
| M 853 | 16 | 91 | |
| SpoT 1 | 35 | 164 | |
| SpoT 2 | 35 | 68 |
CHE, diploid Chinese hamster embryo cells.
B 644, D 313, and M 854 are highly aneuploid, clonal lines of benzpyrene-, dimthylbenzanthracene-, and methylcholanthrene-transformed Chinese hamster cells; B-01col and M-01col are B 644 and M 854 cells resistant to 0.1 μg colcemid; and SpoT 1 and SpoT 2 are highly aneuploid spontaneously transformed Chinese hamster cells.
Experiments in which, in addition to large colonies of cells, multiple small ones survived drug treatment, which was typically observed when selection was initiated at the lowest concentration listed in the table.
It may be argued, however, that a hypothetical mutation induced by the carcinogenic, polycyclic aromatic hydrocarbons, used to generate the transformed cells (47), has generated the drug-resistant mutants directly or indirectly. But this is considered unlikely because mutation, particularly by polycyclic aromatic hydrocarbons, is inefficient, at most <10−6 (19). Thus only one in about 106 clonal cultures derived from chemically transformed cells, such as the three studied here, would have been heterozygous for drug resistance by such a mutation.
Spontaneously Transformed CH Embryo Cells Also Generate Drug-Resistant Variants at High Rates.
The argument that gene mutations of our aneuploid CH cell lines acquired from treatments with carcinogens before their clonal isolation may be responsible for their high mutation rates was further investigated by studying two spontaneously transformed aneuploid CH cell lines, SpoT 1 and 2. These lines were derived from two independent, spontaneous foci of transformed cells that appeared in different dishes of untreated control cultures of CHE cells that had been maintained in vitro for 2 and 3 months, respectively. Thus their odds for mutation, which is not necessary for transformation, are the same as those for untransformed control cells and thus are undetectably low in our system (see Table 1).
As expected from exact correlations between malignant transformation and aneuploidy described previously by us and others (47, 61–64), both of these cell lines were aneuploid. The modal chromosome number of SpoT 1 was 26/29 and that of SpoT 2 was 29, and both lines displayed the wide distribution of chromosomes that are characteristic of aneuploid cells (Table 2).
Table 2.
Chromosome distribution in spontaneously transformed CH cells
| Cell type | Chromosomes per
cell
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 22 = 2n | 24 | 25 | 26 | 27 | 28 | 29 | 30 | 31 | 33 | 54 | |
| SpoT 1* | — | — | 1 | 7 | 5 | 6 | 7 | — | 1 | 1 | — |
| SpoT 2 | — | 1 | — | 1 | — | 7 | 15 | 3 | — | — | 1 |
SpoT 1 and SpoT 2 are spontaneously transformed Chinese hamster cells.
It is documented in Table 1 that the mutation rates to resistance against puromycin, araC, and methotrexate of both of these spontaneously transformed lines were similar to those of the three polycyclic aromatic hydrocarbon-transformed aneuploid cells. Thus the high mutation rates of both polycyclic aromatic hydrocarbon-transformed and spontaneously transformed cells, compared with those of normal diploid cells, correlated exactly with aneuploidy.
Stepwise Increase in Drug Resistance During Selection.
Following the large literature on the subject, drug treatments were initiated at low micromolar concentrations and then increased in subsequent cultures to higher concentrations to obtain more and more resistant variants (6, 8, 18, 23, 25, 65). According to one investigator, “Typically, in these studies, cultured cells are selected for resistance to a single anti-cancer drug with a ‘classic’ step-wise selection protocol” (2). This protocol implies that a given phenotype is improved by multiple rounds of mutation.
Our experiments have confirmed these observations both positively and negatively. For example, colonies initially selected for resistance to 1 μg of puromycin per dish could be rendered resistant to 20 μg in subsequent rounds of selection (Table 1). In contrast, if 20 μg of puromycin was chosen for the initial round of selection of drug resistance from the same cells, no survivors were obtained.
In most cases a majority of cells resistant to a given concentration of a drug survived 2- to 5-fold increases of the selective drug if puromycin and araC were used within the limits stated in Table 1. However, in the case of colcemid only about 1 in 105 B 644 and 3 in 105 M 853 cells resistant to 0.1 μg colcemid per dish (termed B-01col and M-01col in Table 1) survived a 2-fold increase of colcemid to 0.2 μg per dish (Table 1).
It follows that the most sensitive method to detect mutation to drug resistance is to initiate selection at a very low concentration of a cytotoxic drug. But if the concentration of a toxic drug was too low to kill the majority of sensitive cells, the culture initially appeared resistant until it was dissociated into single cells with trypsin and then propagated under the same (or more stringent) selective conditions. Eventually all cells that are able to survive but unable to grow at a given drug concentration are eliminated by this method.
These experiments suggest that a multiplicity of mutants with various degrees of resistance existed in each group of cells selected for resistance to a specific drug. Thus multiple steps or mutational events appear to generate a continuum of mutants with different degrees of resistance against the same drug. Nevertheless, these events must be relatively specific because their frequency varies with the drug used, e.g., events altering the resistance to puromycin appear to be relatively common, generating tolerance to large increases of selective drugs, and events altering resistance to colcemid appear to be relatively rare, restricting the tolerance to increased drug concentration.
Selected Drug Resistance Is Associated with Different Unselected Morphologic Phenotypes.
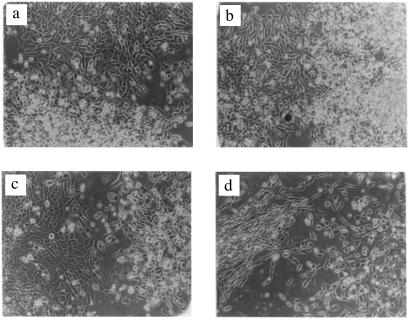
The hypothesis that drug resistance among aneuploid cells is generated by chromosome reassortments also predicts that cells with identical drug resistance markers may differ from each other in new, unselected phenotypes, depending on their particular chromosome combination. Cell morphology proved to be one such gratuitous phenotype of drug resistant cells. For example, different puromycin-resistant B 644 cells or different araC-resistant D 313 cells on the same Petri dish were either fusiform or polygonal (Fig. 1). This observation confirms the “morphologic innovations” among cells resistant to the same cytotoxic drugs observed by others over 40 years ago (66, 67).
Figure 1.
Different morphologies of drug-resistant cells derived from the same clone of aneuploid CH cells. (a) Polygonal and spherical, puromycin-resistant cells derived from the aneuploid CH clone D 313. (b) Spindle-shaped and spherical, cytosine arabinoside-resistant cells derived from the aneuploid CH clone B 644. (c) Polygonal and spindle-shaped refractile, puromycin-resistant cells derived from the aneuploid CH clone M 853. (d) Fusiform and polygonal, puromycin-resistant cells derived from the aneuploid CH clone D 313.
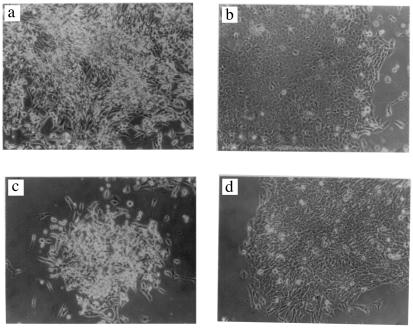
Colony morphology proved to be another gratuitous phenotype of drug-resistant cells. Two examples, a three-dimensional and a flat colony of puromycin-resistant SpoT 2 cells, and one each of puromycin plus araC-resistant M 853 cells, are shown in Fig. 2.
Figure 2.
Different colony morphologies of drug-resistant cells derived from the same clone of aneuploid CH cells. (a and b) A three-dimensional (a) and a flat (b) colony of puromycin-resistant SpoT 2 cells. (c and d) A three-dimensional (c) and a flat (d) colony of puromycin plus araC-resistant M 853 cells.
Multidrug Resistance.
Resistance to more drugs than selected for is also predicted by the hypothesis that drug resistance is achieved by chromosome reassortments that simultaneously vary thousands of genes. Several such cross-resistances have also been confirmed for the cells studied here (Table 3).
Table 3.
Multidrug resistance profiles of cells originally selected for resistance to a specific drug
| Cells resistant to a given drug, μg per 10-cm Petri dish | Cross-resistance to unselected drugs, %* |
|---|---|
| B 654*—puromycin 20 | 50% at 5 μg puromycin + 5 μg araC |
| M 853—puromycin 20 | 20% at 5 μg puromycin + 5 μg araC |
| D 313—puromycin 20 | 5% at 10 μg puromycin + 5 μg araC |
| B 654—araC 5 | 75% at 5 μg araC + 5 μg puromycin |
| D 313—araC 5 | 50% at 5 μg araC + 1 μg puromycin |
| 30% at 5 μg araC + 5 μg puromycin | |
| M 853—araC 5 | 50% at 5 μg araC + 1 μg puromycin |
| 50% at 5 μg araC + 5 μg puromycin | |
| B 654—araC 5 | 75% at 5 μg araC + 0.1 μg colchicine |
| <5% at 5 μg araC + 0.2 μg colchicine | |
| B 654—araC 5 + puromycin 5 | 50% at 5 μg araC, 5 μg puromycin + 0.1 μg colchicine |
| <5% at 5 μg araC, 5 μg puromycin + 0.2 μg colchicine | |
| D 313—puromycin 20 | 90% at 5 μg puromycin + 0.1 μg colchicine |
| 75% at 5 μg puromycin + 0.2 μg colchicine |
See text or Table 1 for a description of cells.
Cross-resistance of cells, with resistance to previously selected drugs, to further drugs was determined from the percentage of confluence a culture had reached by the time a parallel culture in the absence of further drugs was confluent.
Cross-resistance was determined from the percentage of confluence a culture resistant to a given drug had reached in the presence of further drugs by the time a parallel culture in the absence of further drugs was confluent. For this purpose 1 million to 2 million cells were challenged simultaneously with the original selective drug(s) together with other cytotoxic drugs. In parallel, 1 million to 2 million cells were grown only in the presence of the original selective drug(s). If the culture challenged with new drugs grew to confluence at the same or nearly the same rate as the unchallenged control culture, the cells of both cultures were diluted 4-fold and reseeded in the presence of the same drugs.
The results are reported in Table 3. For example, puromycin-resistant B 654 cells reached 50% confluence in puromycin and araC by the time a parallel culture, treated only with puromycin, was confluent (Table 3). In contrast, only about 1–10 of 106 unselected B 654 cells were resistant to araC (Table 1). Likewise, araC-resistant B 644 cells reached 75% confluence in the presence of puromycin by the time a parallel culture, treated only with araC, was confluent (Table 3). In contrast, only about 1 of 105 unselected B 644 cells were resistant to puromycin (Table 1). We conclude that mutants, selected for resistance against only one drug, are also resistant to lesser degrees to various other unrelated drugs.
Preliminary Evidence for Reversibility of Drug Resistance.
We have observed in many cultures of drug-resistant cells growing in the presence of a selective cytotoxic drug an excess of unattached, dead cells, compared with parallel cultures grown in the absence of the drug. The death of these cells occurred while the majority of cells in the culture were firmly attached to the dish and growing, and is thus not due to the lack of nutrients. Moreover, cell death depended on the nature of the selective drug. For example, more cells died in araC and puromycin-resistant cultures than in colcemid-resistant cultures treated with the respective drugs. Furthermore, the mortality of multidrug-resistant cultures in the presence of multiple drugs seemed to be higher than in single drug-resistant cultures in the presence of a single selective drug. These observations are preliminary evidence for the reversibility of drug resistance via chromosome reassortment. However, further work is needed to confirm this interpretation.
Discussion
(i) Chromosome Reassortment Versus Gene Mutation as a Mechanism of Drug Resistance.
The experiments described here were designed to distinguish between gene mutation and chromosome reassortment as the mechanism for the generation of drug-resistant mutants of cancer cells and tumorigenic cell lines. In contrast to the experiments conducted by others, we have investigated this question here by comparing directly the mutation rates of normal, diploid cells with those of highly aneuploid cells from the same inbred line of CHs. This comparison revealed that aneuploid, but not diploid, cells (a) mutate to drug resistance at the high rates of 10−4 to 10−6, which are paradoxical in view of conventional mutation rates of recessive genes of diploid cells (see above); (b) undergo further mutation at high rates in subsequent rounds of selection that enhances the drug-resistant phenotype; and (c) display, in addition to the selected phenotype, different unselected morphological and drug-resistant phenotypes.
Each of these results can be explained on the basis of chromosome reassortments that are autocatalyzed by aneuploidy as proposed above (Introduction). With regard to the question of whether chromosome reassortment achieves drug resistance by activating alternative biochemical pathways or by deleting chromosomes with drug-sensitive alleles from heterozygotes, our data support the first alternative. The chromosome deletion hypothesis is hard to reconcile with (a) the wide spectrum of degrees of drug resistance observed for a given resistant phenotype, (b) preliminary evidence for reversibility, and (c) the low probability that each of the five clonal cell lines from which mutants were derived would have been heterozygous for resistance genes against all of the drugs tested here. Thus based on the accuracy of its predictions, chromosome reassortment, rather than gene mutation and chromosome loss, is the mechanism of the high mutation rates of aneuploid cells.
Like us, others have previously considered chromosome loss as a possible genetic basis for drug resistance (7, 12, 14, 18, 23, 67–70). But others have not previously considered chromosome reassortment, autocatalyzed by aneuploidy, as the mechanism of the high mutation rates of aneuploid cells. This may be why a direct comparison of the mutation rates of diploid and aneuploid cells was not performed previously. Indeed, many investigators assumed that cell lines with normal or near-normal chromosome numbers were also diploid or near-diploid (7, 8, 19, 28). However, cytogenetic studies show that the cell lines that were considered diploid or near-diploid were pseudodiploid, with numerous aneuploid chromosomes or segments of chromosomes, despite diploid or near-diploid chromosome numbers. The CH lines used for most of the studies on mutation to drug resistance are a case in point (71, 72). According to Terzi, “[e]ven when the cells have the same total number of chromosomes, they usually possess a variable number of homologues” (12).
(ii) Phenotype-Specific Karyotypes.
Our hypothesis predicts phenotype-specific karyotypes that we have not identified. Indeed, the phenotype-specific karyotypes will be difficult to identify for two reasons: (a) Different chromosome combinations are likely to generate identical drug resistance phenotypes because multiple biochemical mechanisms can generate the same drug resistance phenotypes (4, 7, 22, 57, 58). (b) Collateral reassortments, resulting from the inherent instability of the aneuploid karyotype, that are irrelevant to the selected phenotype will mask the specific karyotype of a given drug-resistant cell.
Nevertheless, it should not be impossible to identify phenotype-specific chromosome combinations, by focusing on resistances with narrow tolerance to a given drug concentration and thus possibly without alternative pathways, and by studying near-diploid aneuploid cells, in which only a minimal number of chromosomes are aneuploid. An example of such an analysis, i.e., resistance to platinum compounds, has recently been described (69). Work along these lines could identify chromosome combinations that generate specific phenotypes or groups of phenotypes and thus become a basis for selecting alternative chemotherapy against drug-resistant cancer. Alternatively, it is even conceivable that substances that destabilize aneuploid chromosome combinations beyond their inherent levels by furthering chromosome reassortments, such as tumor promoters (77, 82, 83), could prove useful against drug-resistant cancer.
(iii) Chromosome Reassortment May Lead to Chromosome Rearrangements.
Next to aneuploidy, chromosome rearrangements are the second most common chromosome abnormality in cancer cells (36, 78). By upsetting the normal balance of enzymes involved in DNA breakage and reunion that are necessary for DNA replication, chromosome reassortment could also generate aneuploidy-specific chromosome rearrangements, as for example the multiple rearrangements of chromosomes in cancer cells that have been recently detected with fluorescent chromosome-specific probes (79, 80).
(iv) Mutation to Drug Resistance as a Functional Test of Preneoplastic Aneuploidy?
Aneuploidy has been investigated as a marker of incipient malignancy in benign lesions (see refs. 73–76 and 81, and literature reviewed in refs. 43 and 64). But the sensitivity of such tests is limited by the percentage of aneuploid cells in a sample (e.g., a Pap smear). However, if enough aneuploid cells were present to generate a drug-resistant mutant, i.e., between 103 and 106, they could be picked up via selection for drug-resistant mutants.
Acknowledgments
We thank Athel Cornish-Bowden (Laboratoire de Bioenergetique et Ingenierie des Proteines, Centre National de la Recherche Scientifique, Marseille), Morgan Harris (Molecular and Cell Biology, University of California, Berkeley, retired), and Heinz Schaller (Zentrum für Molekulare Biologie, University of Heidelberg, Heidelberg) for critical reviews of the manuscript, and David Rasnick (University of California Berkeley) for critical information. Robert Leppo (philanthropist, San Francisco), an American foundation that prefers to be anonymous, the Abraham J. and Phyllis Katz Foundation (New York), other private sources, and the Forschungsfonds der Fakultät für Klinische Medizin Mannheim are gratefully acknowledged for support. R.S. thanks Robert Leppo for a research fellowship. P.D. thanks the Mildred Scheel Stiftung of the Deutsche Krebshilfe for a guest professorship at the III Medizinische Klinik of the University of Heidelberg at Mannheim, and Siggi and Max for waiting at home in Berkeley.
Abbreviations
- CH
Chinese hamster
- araC
cytosine arabinonucleoside
- CHE
CH embryo
References
- 1.Skipper H E. Cancer Res. 1965;25:1544–1550. [PubMed] [Google Scholar]
- 2.Schoenlein P V. Cytotechnology. 1993;12:63–89. doi: 10.1007/BF00744658. [DOI] [PubMed] [Google Scholar]
- 3.Pitot H C. Fundamentals of Oncology. New York: Dekker; 1986. [Google Scholar]
- 4.Stein U. Onkologie. 2000;23:316–317. [Google Scholar]
- 5.DeVita V T., Jr . In: Cancer, Principles & Practice of Oncology. DeVita V T Jr, Hellman S, Rosenberg S A, editors. Philadelphia: Lippincott; 1993. pp. 276–292. [Google Scholar]
- 6.Law L W. Nature (London) 1952;169:628–629. doi: 10.1038/169628a0. [DOI] [PubMed] [Google Scholar]
- 7.Siminovitch L. Cell. 1976;7:1–11. doi: 10.1016/0092-8674(76)90249-x. [DOI] [PubMed] [Google Scholar]
- 8.Harris H. The Cells of the Body: A History of Somatic Cell Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1995. [Google Scholar]
- 9.Vogel F, Motulsky A G. Human Genetics: Problems and Approaches. Berlin: Springer; 1986. [Google Scholar]
- 10.Gartler S M, Pious D E. Humangenetik. 1966;2:83–114. [Google Scholar]
- 11.Breslow R E, Goldsby R A. Exp Cell Res. 1969;55:339–346. doi: 10.1016/0014-4827(69)90567-9. [DOI] [PubMed] [Google Scholar]
- 12.Terzi M. Proc Natl Acad Sci USA. 1974;71:5027–5031. doi: 10.1073/pnas.71.12.5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mezger-Freed L. Nat New Biol. 1972;235:245–246. doi: 10.1038/newbio235245a0. [DOI] [PubMed] [Google Scholar]
- 14.Chasin L A. J Cell Physiol. 1973;82:299–308. doi: 10.1002/jcp.1040820218. [DOI] [PubMed] [Google Scholar]
- 15.Szybalski W. Exp Cell Res. 1960;18:588–591. doi: 10.1016/0014-4827(59)90327-1. [DOI] [PubMed] [Google Scholar]
- 16.Lieberman I, Ove P. Proc Natl Acad Sci USA. 1959;45:872–877. doi: 10.1073/pnas.45.6.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coffino P, Scharff M D. Proc Natl Acad Sci USA. 1971;68:219–223. doi: 10.1073/pnas.68.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Littlefield J W. Biochim Biophys Acta. 1965;95:14–22. doi: 10.1016/0005-2787(65)90206-6. [DOI] [PubMed] [Google Scholar]
- 19.Bradley M O, Bhuyan B, Francis M C, Langenbach R, Peterson A, Huberman E. Mutat Res. 1981;87:81–142. doi: 10.1016/0165-1110(81)90029-4. [DOI] [PubMed] [Google Scholar]
- 20.Harris J F, Chambers A F, Hill R P, Ling V. Proc Natl Acad Sci USA. 1982;79:5547–5551. doi: 10.1073/pnas.79.18.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peterson A R, Peterson H, Heidelberger C. Mutat Res. 1975;29:127–137. doi: 10.1016/0027-5107(75)90026-3. [DOI] [PubMed] [Google Scholar]
- 22.Jones G E, Sargent P A. Cell. 1974;2:43–54. doi: 10.1016/0092-8674(74)90007-5. [DOI] [PubMed] [Google Scholar]
- 23.Lieberman I, Ove P. Proc Natl Acad Sci USA. 1959;45:867–877. doi: 10.1073/pnas.45.6.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Littlefield J W, Basilico C. Nature (London) 1966;211:250–252. doi: 10.1038/211250a0. [DOI] [PubMed] [Google Scholar]
- 25.Blade K, Menick D R, Cabral F. J Cell Sci. 1999;112:2213–2221. doi: 10.1242/jcs.112.13.2213. [DOI] [PubMed] [Google Scholar]
- 26.Koski T, Karhu R, Visakorpi T, Vilpo L, Knuutila S, Vilpo J. Eur J Haematol. 2000;65:32–39. doi: 10.1034/j.1600-0609.2000.9c153.x. [DOI] [PubMed] [Google Scholar]
- 27.Chasin L A. Cell. 1974;2:37–41. doi: 10.1016/0092-8674(74)90006-3. [DOI] [PubMed] [Google Scholar]
- 28.Harris M. J Cell Physiol. 1971;78:177–184. doi: 10.1002/jcp.1040780204. [DOI] [PubMed] [Google Scholar]
- 29.Loeb K R, Loeb L A. Am J Pathol. 1999;154:1621–1626. doi: 10.1016/S0002-9440(10)65415-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strauss B S. Cancer Res. 1992;52:249–253. [PubMed] [Google Scholar]
- 31.Kinzler K, Vogelstein B. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 32.Heppner G, Miller F R. Int Rev Cytol. 1998;177:1–56. doi: 10.1016/s0074-7696(08)62230-5. [DOI] [PubMed] [Google Scholar]
- 33.Harris C C. Cancer Res. 1991;51:5023s–5044s. [PubMed] [Google Scholar]
- 34.Barrett J C, Tsutsui T, Tsly T, Oshimura M. In: Genetic Mechanisms in Carcinogenesis and Tumor Progression. Harris C C, Liotta L A, editors. New York: Wiley–Liss; 1990. pp. 97–114. [Google Scholar]
- 35.Jakubezak R J, Merlino G, French J E, Muller W J, Paul B, Adhya S, Garges S. Proc Natl Acad Sci USA. 1996;93:9073–9078. doi: 10.1073/pnas.93.17.9073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heim S, Mitelman F. Cancer Cytogenetics. New York: Wiley–Liss; 1995. [Google Scholar]
- 37.Sandberg A A. The Chromosomes in Human Cancer and Leukemia. New York: Elsevier; 1990. [Google Scholar]
- 38.Gebhart E, Liehr T. Int J Oncol. 2000;16:383–399. doi: 10.3892/ijo.16.2.383. [DOI] [PubMed] [Google Scholar]
- 39.Boveri, T. (1902) reprinted in Willier, B. H. & Oppenheimer J. M. (1964) Foundations of Experimental Embryology (Prentice–Hall, Englewood Cliffs, NJ), pp. 74–97.
- 40.Lejeune J, Turpin R, Gautier M. Ann Genet. 1959;2:41–49. [Google Scholar]
- 41.Lindsley D L, Sandler L, Baker B S, Carpenter A T C, Denell R E, Hall J C, Jacobs P A, Gabor Miklos G L, Davis B K, Gethmann R C, et al. Genetics. 1972;71:157–184. doi: 10.1093/genetics/71.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rasnick D, Duesberg P. Biochem J. 1999;340:621–630. [PMC free article] [PubMed] [Google Scholar]
- 43.Duesberg P, Rasnick D. Cell Motil Cytoskeleton. 2000;47:81–107. doi: 10.1002/1097-0169(200010)47:2<81::AID-CM1>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 44.Brinkley B R, Goepfert T M. Cell Motil Cytoskeleton. 1998;41:281–288. doi: 10.1002/(SICI)1097-0169(1998)41:4<281::AID-CM1>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 45.Pihan G A, Purohit A, Wallace J, Knecht H, Woda B, Quesenberry P, Doxsey S J. Cancer Res. 1998;58:3974–3985. [PubMed] [Google Scholar]
- 46.Duesberg P. Science. 1999;284:2091–2092. doi: 10.1126/science.284.5423.2089f. [DOI] [PubMed] [Google Scholar]
- 47.Li R, Yerganian G, Duesberg P, Kraemer A, Willer A, Rausch C, Hehlmann R. Proc Natl Acad Sci USA. 1997;94:14506–14511. doi: 10.1073/pnas.94.26.14506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Duesberg P, Rausch C, Rasnick D, Hehlmann R. Proc Natl Acad Sci USA. 1998;95:13692–13697. doi: 10.1073/pnas.95.23.13692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lengauer C, Kinzler K W, Vogelstein B. Nature (London) 1997;386:623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- 50.Furuya T, Uchiyama T, Murakami T, Adachi A, Kawauchi S, Oga A, Hirano T, Sasaki K. Clin Cancer Res. 2000;6:2815–2820. [PubMed] [Google Scholar]
- 51.Harnden D G, Benn P A, Oxford J M, Taylor A M R, Webb T P. Somatic Cell Genet. 1976;2:55–62. doi: 10.1007/BF01539242. [DOI] [PubMed] [Google Scholar]
- 52.Galloway S M, Buckton K E. Cytogenet Cell Genet. 1978;20:78–96. doi: 10.1159/000130842. [DOI] [PubMed] [Google Scholar]
- 53.Petersson H, Mitelman F. Hereditas. 1985;102:33–38. doi: 10.1111/j.1601-5223.1985.tb00462.x. [DOI] [PubMed] [Google Scholar]
- 54.Duesberg P H. Proc Natl Acad Sci USA. 1968;59:930–937. doi: 10.1073/pnas.59.3.930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Duesberg P H. J Mol Biol. 1969;42:485–499. doi: 10.1016/0022-2836(69)90237-x. [DOI] [PubMed] [Google Scholar]
- 56.Fenner F, McAuslan B R, Mims C A, Sambrook J, White D O. The Biology of Animal Viruses. New York: Academic; 1974. [Google Scholar]
- 57.Subak-Sharpe H. Exp Cell Res. 1965;38:106–119. doi: 10.1016/0014-4827(65)90432-5. [DOI] [PubMed] [Google Scholar]
- 58.Orkin S H, Littlefield J W. Exp Cell Res. 1971;69:174–180. doi: 10.1016/0014-4827(71)90322-3. [DOI] [PubMed] [Google Scholar]
- 59.Zhang L, Zhou W, Velculescu V E, Kern S E, Hruban R H, Hamilton S R, Vogelstein B, Kinzler K W. Science. 1997;276:1268–1272. doi: 10.1126/science.276.5316.1268. [DOI] [PubMed] [Google Scholar]
- 60.Yerganian G, Leonard M. Science. 1961;133:1600–1601. doi: 10.1126/science.133.3464.1600. [DOI] [PubMed] [Google Scholar]
- 61.Saksela E, Moorhead P S. Proc Natl Acad Sci USA. 1963;50:390–396. doi: 10.1073/pnas.50.2.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cram L S, Bartholdi M F, Ray F A, Travis G L, Kraemer P M. Cancer Res. 1983;43:4828–4837. [PubMed] [Google Scholar]
- 63.Levan A, Biesele J J. Ann NY Acad Sci. 1958;71:1022–1053. doi: 10.1111/j.1749-6632.1958.tb46820.x. [DOI] [PubMed] [Google Scholar]
- 64.Duesberg P, Li R, Rasnick D, Rausch C, Willer A, Kraemer A, Yerganian G, Hehlmann R. Cancer Genet Cytogenet. 2000;119:83–93. doi: 10.1016/s0165-4608(99)00236-8. [DOI] [PubMed] [Google Scholar]
- 65.Jones P A, Taderera J V, Hawtrey A O. Eur J Cancer. 1972;8:595–599. doi: 10.1016/0014-2964(72)90138-7. [DOI] [PubMed] [Google Scholar]
- 66.Hauschka T S. J Cell Comp Physiol. 1958;52,(Suppl.):197–233. doi: 10.1002/jcp.1030520411. [DOI] [PubMed] [Google Scholar]
- 67.Vogt M. Genetics. 1959. 1257–1270. [Google Scholar]
- 68.Tew K D, Moy B C, Hartley-Asp B. Exp Cell Res. 1983;149:443–450. doi: 10.1016/0014-4827(83)90356-7. [DOI] [PubMed] [Google Scholar]
- 69.Leyland-Jones B, Kelland L R, Harrap K R, Hiorns L R. Am J Pathol. 1999;155:77–84. doi: 10.1016/S0002-9440(10)65102-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Law L W. Cancer Res. 1956;16:698–716. [PubMed] [Google Scholar]
- 71.Deaven L L, Petersen D F. Chromosoma. 1973;41:129–144. doi: 10.1007/BF00319690. [DOI] [PubMed] [Google Scholar]
- 72.Simi S, Colella C M, Mariani T, Piras A, Rainaldi G. Teratogenesis Carcinogenesis Mutagenesis. 1988;8:45–54. doi: 10.1002/tcm.1770080106. [DOI] [PubMed] [Google Scholar]
- 73.Spriggs A I. In: Chromosomes and Cancer. German J, editor. New York: Wiley; 1974. p. 423. [Google Scholar]
- 74.Rubin C E, Haggitt R C, Burmer G C, Brentnall T A, Stevens A C, Levine D S, Dean P J, Kimmey M, Perera D R, Rabinovitch P S. Gastroenterology. 1992;103:1611–1620. doi: 10.1016/0016-5085(92)91185-7. [DOI] [PubMed] [Google Scholar]
- 75.Segers P, Haesen S, Amy J-J, De Sutter P, Van Dam P, Kirsch-Volders M. Cancer Genet Cytogenet. 1994;75:120–129. doi: 10.1016/0165-4608(94)90163-5. [DOI] [PubMed] [Google Scholar]
- 76.Richter J, Jiang F, Gorog J P, Sartorius G, Egenter C, Gasser T C, Moch H, Mihatsch M J, Sauter G. Cancer Res. 1997;57:2860–2864. [PubMed] [Google Scholar]
- 77.Fusenig N E, Petrusevska R T, Pohlmann N. In: Tumor Promoters: Biological Approaches for Mechanistic Studies and Assay Systems. Langenbach R, editor. New York: Raven; 1988. pp. 259–273. [Google Scholar]
- 78.Mitelman F, Mertens F, Johansson B. Nature Genet. 1997;15,Suppl.:S417–S474. doi: 10.1038/ng0497supp-417. [DOI] [PubMed] [Google Scholar]
- 79.Gisselsson D, Pettersson L, Hoglund M, Heidenblad M, Gorunova L, Wiegant J, Mertens F, Dal Cin P, Mitelman F, Mandahl N. Proc Natl Acad Sci USA. 2000;97:5357–5362. doi: 10.1073/pnas.090013497. . (First Published April 10, 2000; 10.1073/pnas.090013497) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Saunders W S, Shuster M, Huang X, Gharaibeh B, Enyenihi A H, Petersen I, Gollin S M. Proc Natl Acad Sci USA. 2000;97:303–308. doi: 10.1073/pnas.97.1.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bocking A, Chatelain R. Anal Quant Cytol Histol. 1989;11:177–186. [PubMed] [Google Scholar]
- 82.Parry J M, Parry E M, Barrett J C. Nature (London) 1981;294:263–265. doi: 10.1038/294263a0. [DOI] [PubMed] [Google Scholar]
- 83.Dzarlieva R T, Fusenig N E. Cancer Lett. 1982;16:7–17. doi: 10.1016/0304-3835(82)90085-4. [DOI] [PubMed] [Google Scholar]