Abstract
In plants, disease resistance mediated by the gene-for-gene mechanism involves the recognition of specific effector molecules produced by the pathogen either directly or indirectly by the resistance-gene products. This recognition triggers a series of signals, thereby serving as a molecular switch in regulating defense mechanisms by the plants. To understand the mechanism of action of the barley stem rust resistance gene Rpg1, we investigated the fate of the RPG1 protein in response to infection with the stem rust fungus, Puccinia graminis f. sp. tritici. The investigations revealed that RPG1 disappears to undetectable limits only in the infected tissues in response to avirulent, but not virulent pathotypes. The RPG1 protein disappearance is rapid and appears to be due to specific protein degradation via the proteasome-mediated pathway as indicated by inhibition with the proteasomal inhibitor MG132, but not by other protease inhibitors.
Keywords: avirulence, cultivar, programmed cell death, Puccinia graminis
Plants have evolved diverse mechanisms to recognize pathogen attack and trigger defense responses. Pathogen recognition specificity is often determined by a pathogen avirulence (Avr) gene and its corresponding plant resistance (R) gene in a gene for gene manner (1). The R gene products may function directly or indirectly as receptors for the Avr gene products, providing detection of pathogen attack (2–6). Avr proteins secreted from the pathogen are recognized by the R proteins either in the intercellular spaces or after their transport into the plant cell. The Avr–R interactions lead to activation of defense responses and often result in the hypersensitive response (HR) (1), inhibiting the growth of the pathogen. In the absence of either the cognate R or Avr gene product, the pathogen colonizes the host and causes disease. Despite the cloning of several R genes and their corresponding Avr genes, direct physical interaction between matched Avr and R proteins has been shown only in a few cases. To exemplify, the Avr-Pita protein of the rice blast fungus Magnaporthe grisea encoding a metalloprotease is secreted with an N-terminal signal sequence. After delivery into the plant cell and removal of the proprotein sequence, the mature enzyme binds to the leucine-rich domain of the Pi-ta R protein and elicits the resistance response. This interaction was confirmed with the yeast two-hybrid system, with transient expression in rice seedling leaves of resistant or susceptible lines, by in vitro binding of the recombinantly synthesized Pi-ta protein to the Avr-Pita protein and by inactivation of either of the proteins through amino acid substitutions (7). Direct physical interaction has been demonstrated for the tomato Pto R protein and the AvrPto gene product (2, 4), the Arabidopsis RRS1 R and Ralstonia solanacearum PopP2 (8), and between the flax R gene L6 with the corresponding Avr-L6 of Melampsora lini (9). However, attempts with other R–Avr pairs have failed to establish a direct physical interaction. These observations led to development of the guard hypothesis. In this model, the R gene product acts as a sentinel of the cellular machinery, guarding key virulence targets inside the cell (10, 11). The guard hypothesis proposes that the Avr proteins interact with and modify non-R cellular proteins. The R gene protein then perceives the altered status of the virulence target and induces a defense response.
Support for the guard hypothesis comes from comprehensive analyses of the molecular patterns of responses leading to resistance or susceptibility in different plant bacterial and fungal pathogen interactions (12). A direct interaction of AvrRpt2 of Pseudomonas syringae pv. tomato with its cognate R gene product RPS2 of Arabidopsis could not be established. Instead, RPS2 interacts physically with the RIN4 protein of Arabidopsis. AvrRpt2 action results in degradation of RIN4 and activation of the RPS2 gene function (5, 13, 14). In this way, RPS2 senses RIN4 levels and guards the role of RIN4 in plant cells. RIN4 is targeted and modified by two additional Avr proteins, AvrRPM1 and AvrB, from P. syringae that interact with the Arabidopsis RPM1 resistance gene (14, 15). In this case, the Avr proteins induce phosphorylation of RIN4 by means of an unidentified kinase. This phosphorylation alters RIN4, and RPM1 detects the phosphorylated form of RIN4 and induces programmed cell death (PCD). RPM1 signals the downstream defense activities and is itself degraded by a yet to be identified mechanism (16).
Arabidopsis resistance to P. syringae bacteria expressing AvrPphB is mediated by RPS5, an R protein with a nucleotide-binding site and leucine-rich repeats, and the protein kinase PBS1 (17, 18). AvrPphB is a cysteine protease that autocleaves at a GDK triade and cleaves the PBS1 protein at the same triad, yielding a small C-terminal peptide (17). This cleavage of the cellular target is required for the RPS5-mediated induction of the defense response. It was speculated that the cleavage fragment binds to RPS5 and must be phosphorylated because inactivation of the PBS1 kinase activity also inactivated its ability to elicit disease resistance. Thus the avirulence protein is detected by the plant by means of its enzyme activity.
The induction of PCD in resistant plants by AvrPto and AvrPtoB depends on Pto, an S/T kinase and Prf, a CC-NBS-LRR protein (19). It was suggested that Prf may be the R gene product that responds to AvrPto/AvrptoB (20). The kinase activity of Pto is required for the Avr–Pto, and Avr–PtoB induced PCD as well as signaling through the Pti proteins (20). This suggests that the kinase activity of Pto generates a phosphorylated intermediate that could be a ligand for Prf. Recently, it was shown that Pto associates with a unique Prf N-terminal domain and resides in a high-molecular-weight recognition complex. In this complex, both Pto and Prf contribute to specific recognition of AvrPtoB (21).
The barley stem rust resistance gene Rpg1, confers resistance to many pathotypes of the rust fungus, Puccinia graminis f. sp. tritici (Pgt) (22). It was cloned and shown to be a receptor-like S/T kinase with tandem kinase domains (23). Whether Rpg1 fits into the receptor-ligand model or the guard hypothesis is not known, because the AvrRpg1 gene has not been identified. However, it is evident that the kinase activity of RPG1 is required, but not sufficient, for resistance (24). So could RPG1 phosphorylation and/or proteolysis act as an initial signal in recognizing the avirulent fungus? If so, does it underlie the resistance or the susceptibility pathway? Experiments designed from these perspectives revealed that RPG1 disappears to undetectable limits only in infected tissues by a proteasome-mediated pathway, and this proteolysis occurs exclusively in response to the avirulent Pgt pathotypes.
Results
RPG1 Protein Is Degraded in Barley Seedling Leaves upon Infection with P. graminis f. sp. tritici Pathotype MCC.
Real-time PCR experiments previously established that the Rpg1 gene providing resistance to Pgt pathotype MCC is transcribed constitutively over the test period of 36 h whether untreated, mock-inoculated, or inoculated with the pathogen (25). This time period covers germination of rust spores, growth of the germ tube, formation of appressoria, penetration pegs, substomatal vesicles, and haustoria (reviewed in ref. 25). Judging from the results in flax rust pathosystem, this series of events coincides with the delivery of the avirulence protein (9, 26). The beginning of PCD, visualized by cell necrosis, is also occurring in this time frame (27). By using a quantitative ELISA test, the amount of RPG1 protein was determined in 10-day-old uninfected seedlings for six genotypes with different levels of resistance to pathotype MCC (Table 1). RPG1 protein accumulation was greatest in the highly resistant cvs. Chevron and Q21861 and in GP/Rpg1T1, a transformant of cv. Golden Promise homozygous for one copy of Rpg1. The RPG1 protein also was present in the resistant and moderately resistant cultivars Morex and Beacon (Table 1). The susceptible, neutron-induced deletion mutant rpr1 of Morex is a suppressor of Rpg1-mediated stem rust resistance, but has an Rpg1 gene that is expressed at the mRNA and protein level (28).
Table 1.
RPG1 protein accumulation in barley genotypes with different levels of stem rust resistance and timing of RPG1 degradation upon inoculation with the P. graminis f. sp. tritici avirulent pathotype MCC
| Barley genotype | Stem rust phenotype‡ | RPG1, mole·g−1 protein (n = 3) | Absence of RPG1, h |
|---|---|---|---|
| GP/Rpg1T1* | Highly resistant | 6.05 ± 0.32 | 24 |
| Chevron | Highly resistant | 7.63 ± 0.42 | 24 |
| Q21861 | Highly resistant | 6.32 ± 0.73 | 24 |
| Morex | Resistant | 4.86 ± 0.49 | 22 |
| Beacon | Moderately resistant | 3.76 ± 0.36 | 20 |
| rpr1† | Susceptible | 2.0 ± 0.23 | 20 |
*GP/Rpg1T1 is a single-copy transgenic Rpg1 line in Golden Promise genomic background.
†rpr1 is a deletion mutant of a gene required for Rpg1 function (28).
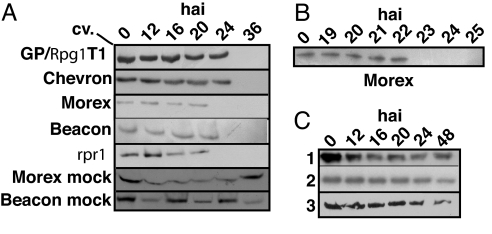
The fate of RPG1 protein after infection by the rust fungus over a period of 36 h was tested with immunoblots and RPG1-specific antibodies (Fig. 1). Surprisingly, the RPG1 protein disappeared to undetectable levels in the infected leaves between 20 and 24 h after infection (hai) in Morex and Beacon, whereas it persisted throughout the experiment in uninoculated lines (Fig. 1). RPG1 disappearance was delayed (between 24 and 36 hai) in the highly resistant lines (Chevron, Q21861, and GP/Rpg1T1). This difference in the timing of the RPG1 protein disappearance may simply be a reflection of the quantity of protein present in these lines compared with the somewhat less resistant lines. The RPG1 protein in a susceptible accession containing the suppressor mutation rpr1 also disappeared 20–24 hai. An experiment designed to determine more precisely the time of RPG1 protein disappearance in cv. Morex showed that it was rapid and between 22–23 hai (Fig. 1B; Table 1). These experiments were also repeated with quantitative RPG1 ELISA analyses, which showed similar timing of the RPG1 disappearance [supporting information (SI) Fig. 5].
Fig. 1.
RPG1 protein disappears between 20 and 24 hai with P. graminis f. sp. tritici avirulent pathotype MCC, whereas it remains stable for at least 36 h in uninoculated samples of Morex and Beacon. (A) Total proteins were extracted and RPG1 visualized on gels as described in Materials and Methods. The lines GP/Rpg1T1 and rpr1 are described in Table 1. Chevron, Q21861, Morex, and Beacon are barley cultivars. Samples were taken at the indicated hai. The RPG1 protein band is 90.2 kDa. (B) Analysis of the RPG1 protein at hourly intervals in Morex showed that it disappears between 22 and 23 h. (C) Degradation of RPG1 is not accompanied by a general proteolysis of cellular proteins after inoculation of Morex with the avirulent pathotype MCC as evidenced from immunoblots showing stability of three enzymes. glutamate-1-semialdehyde aminotransferase (1) glutamyl tRNA synthetase (2), and GST (3).
To determine whether RPG1 was degraded by a protease that might generate small, discrete degradation products, a polyclonal antibody was generated against the full-length protein and used to repeat the experiments. No discrete degradation products were detected, and comparable results were obtained whether the assays were conducted with RPG1 antibodies to the peptide or full-length protein SI Fig. 6).
Although RPG1 degradation occurred rapidly throughout the inoculated leaf, the levels of RPG1 remained unchanged in the uninfected second or subsequent leaves (data not shown). Thus, RPG1 degradation is not systemic in the sense of spreading throughout the plant. The degradation of RPG1 is specific and not accompanied by general proteolysis of cell proteins demonstrated by assays of glutamate-1-semialdehyde amino transferase, glutamyl tRNA synthetase, and GST (Fig. 1C, rows 1, 2, and 3). The minor variation in the protein band intensity could be due to nonspecific protein degradation in the cells undergoing PCD.
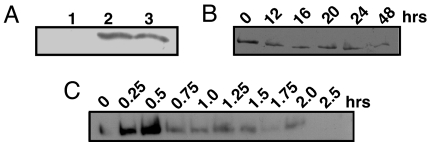
To eliminate the possibility that the RPG1 protein degradation was a spurious result by proteases released from the plant or fungus during sample preparation, rust-inoculated and -uninoculated leaves were mixed before grinding and analyzed. The results substantiated that RPG1 present in the uninoculated tissue was not degraded during processing of the samples (Fig. 2A). Another possibility considered was that, upon infection with the rust fungus, the RPG1 protein becomes associated with the membrane and is precipitated during sample preparation. This possibility was eliminated by testing the microsomal fractions after disappearance of the protein from the supernatant. The results showed that the RPG1 protein was not present in these fractions either (data not shown).
Fig. 2.
Absence of the RPG1 protein in inoculated leaves is not due to in vitro degradation during processing of the samples or inhibition of mRNA translation. (A) Morex leaves uninoculated and inoculated with Pgt avirulent pathotype MCC were harvested 28 hai and prepared either separately or mixed in equal amounts for immunoblot analysis as described in Materials and Methods. Lane 1: inoculated Morex; lane 2: uninoculated Morex; lane 3: inoculated and uninoculated leaves were mixed together before sample preparation. (B and C) Morex seedling leaves were infiltrated with cycloheximide (100 μg/ml) and incubated with nitrate under continuous light (265 μE/m2s) as described (29). Nitrate reductase has a rapid turnover rate, but it is stably maintained for at least 60 h in nitrate and under lights (29). Samples were taken at indicated time points and analyzed for the presence of either RPG1 or nitrate reductase protein with specific antibodies. (B) Cycloheximide did not affect the RPG1 protein, which was stable for at least 48 h, indicating low turnover rate. (C) Cycloheximide, an inhibitor of translation by cytosolic ribosomes, affected cytosolic nitrate reductase accumulation, which rapidly disappeared to undetectable levels after 2 h, indicating effective cycloheximide treatment. The nitrate reductase protein band is 110 kDa.
Turnover and Conditional Degradation of RPG1.
Three different types of cellular proteins can be distinguished: metabolically stable proteins, regulatory proteins that are rapidly turned over by active degradation, and conditionally degraded proteins. Although RPG1 in uninfected leaves was metabolically stable over several days, it was degraded within 24 hai with avirulent pathogens. To demonstrate that inhibition of protein synthesis is not responsible for the disappearance of the RPG1 protein, we used cycloheximide, an inhibitor of protein synthesis by cytoplasmic ribosomes. RPG1 accumulation was stable over 48 h in Morex seedling leaves vacuum infiltrated with cycloheximide (100 μg·ml−1) (Fig. 2B). The cycloheximide treatment, however, resulted in the complete disappearance of cytosolic nitrate reductase in 2 h (Fig. 2C). In the absence of cycloheximide, nitrate reductase is stably maintained for at least 60 h under the experimental conditions used (29). These comparisons classify RPG1 as an actively degraded protein after pathogen infection.
RPG1 Is Degraded in Response to Infection by Avirulent, but Not by Virulent, Stem Rust Pathotypes.
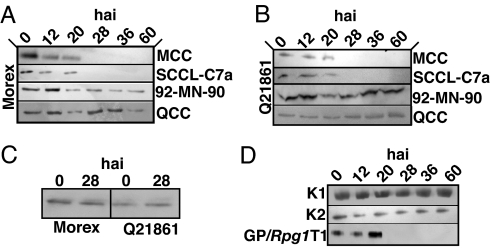
Barley is attacked by two closely related forma specialis (f. sp.) of Puccinia graminis: f. sp. tritici (the wheat stem rust fungus) and f. sp. secalis (the rye stem rust fungus) (30). Cultures of each f. sp. were used to determine their effect on the degradation of RPG1. In Q21861 and Morex, the RPG1 protein was degraded between 20 and 28 hai with avirulent Pgt pathotypes MCC or SCCL-C7a, but not upon infection with virulent Pgt pathotype QCC or virulent Pg, f. sp. secalis isolate 92-MN-90 (Fig. 3 A and B and SI Table 2). Puccinia striiformis f. sp. hordei (stripe rust), another species of Puccinia attacking barley, also was used to assess its effect on RPG1 degradation. Infection of Morex and Q21861 with the virulent stripe rust pathotype PSH-63 failed to elicit RPG1 protein degradation (Fig. 3C). The results demonstrate that RPG1 protein degradation is a specific response to the interaction of the Rpg1 gene product with the specific AvrRpg1 gene product and not with other Avr gene products carried by other stem rust or stripe rust fungi.
Fig. 3.
RPG1 disappearance is triggered by avirulent, but not virulent, rust pathotypes and is correlated with resistance to stem rust P. graminis f. sp. tritici. (A and B) The RPG1 protein in Q21861 (carrying resistance genes Rpg1, rpg4, and Rpg5) and Morex (with Rpg1) disappeared upon infection with Pgt pathotypes MCC and SCCL-C7a, avirulent on Rpg1, but not with Pgt pathotype QCC and Pgs isolate 92-MN-90, virulent on Rpg1, but avirulent on rpg4 and Rpg5. (C) RPG1 protein was not degraded upon inoculation of Morex (left lanes) or Q21861 (right lanes) with the virulent stripe rust Puccinia striiformis f. sp. hordei, pathotype, PSH-63. (D) Mutant K1 (KK151, 152NQ) and K2 (KK461, 462NQ) RPG1 protein is not degraded upon infection with the normally avirulent stem rust pathotype MCC, whereas immune transformant GP/Rpg1T1, RPG1 completely disappeared after 20 h. Mutant K1 RPG1 (in the pseudokinase domain) retains kinase activity because of the pK2 domain, whereas mutant K2 RPG1 (in the active kinase domain) is catalytically defunct (24).
RPG1 Degradation Is Correlated with Disease Resistance and Ubiquitination.
To determine whether RPG1 protein degradation is correlated with Rpg1-mediated stem rust resistance, we tested the transgenic, loss of function mutants K1 (KK152, 153NQ) and K2 (KK461, 462NQ) for RPG1 stability upon infection with the avirulent Pgt pathotype MCC. The K2 mutant has lost RPG1 kinase catalytic activity, whereas the K1 mutant retains autophosporylation activity (24). Both mutants were susceptible to infection by pathotype MCC as demonstrated (24), and the RPG1 protein was not degraded (Fig. 3D).
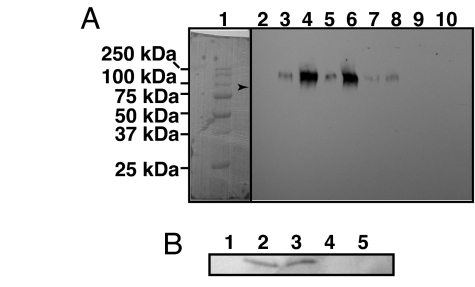
As possible mechanisms for protein degradation, we tested proteasome-mediated proteolysis and the effect of a protease inhibitor mixture. Proteasome-mediated degradation would require ubiquitination of the barley RPG1 protein. Barley, as other plants, express a gene family of the 76-aa-long ubiquitin that is highly conserved and differs from animal and yeast ubiquitin only at 3 and 2 aa, respectively (31, 32). Tagging of a protein with ubiquitin to mark it for degradation by the ubiquitin-activating, -conjugating, and -ligating enzyme complex is also highly conserved, as is the 26S proteasome organization and function. The ubiquitin enrichment kit (Pierce, Rockford, IL) was used to capture the ubiquitin-modified proteins from leaf extracts with an affinity resin containing a monoclonal antibody against ubiquitin. The eluted ubiquitinated proteins were separated by SDS/PAGE and detected with the RPG1-specific antibody on Western blots (Fig. 4A). Results showed that the RPG1 protein is polyubiquitinated in all lines tested, except the K2 mutant and Golden Promise, which does not synthesize the RPG1 protein. The amount of RPG1 polyubiquitinated is enhanced upon infection with the avirulent pathotype MCC, except in mutant K1 (Fig. 4A). Ubiquitination of the kinase-negative RPG1 mutant K2 was not detectable in uninfected or infected samples. This suggests that phosphorylation is essential for ubiquitination and protein degradation. In the mutant K1, RPG1 kinase is active, and the ubiquitination level in the uninfected samples appears to be similar to the controls, but it does not respond to infection with increased RPG1 ubiquitination. This suggests that the pK1 domain controls the response to the pathogen-induced ubiquitination of RPG1 molecules.
Fig. 4.
Disappearance of RPG1 is correlated with polyubiquitination. (A) The RPG1 protein is polyubiquitinated in all lines tested except the K2 mutant, and the amount of RPG1 polyubiquitinated is enhanced upon infection with the avirulent pathotype MCC except in mutant K1. The polyubiquitinated protein was prepared from leaves 28 hai and isolated with polyubiquitin affinity resin and separated by SDS/PAGE, and the immunoblots were decorated with RPG1 antibody. Lane 1: molecular weight markers; lane 2: Golden Promise (no RPG1 present); lanes 3, 5, 7, and 9: uninfected Morex, GP/Rpg1T1, K1 mutant, and K2 mutant, respectively; lanes 4, 6, 8, and 10: the same lines but inoculated with the avirulent pathotype MCC. These results indicate that polyubiquitination is completely blocked in the catalytically inactive kinase mutant K2, whereas it occurs at normal levels in the K1 mutant before infection, but it does not respond to the stem rust infection. The polyubiquitinated RPG1 band is of higher molecular mass than the unubiquitinated RPG1 indicated by the arrow. (B) Disappearance of the RPG1 protein is blocked by the proteasome inhibitor MG132 but not by a plant protease inhibitor mixture. Lane 1: Morex infected with P. graminis f. sp. tritici pathotype MCC; lane 2: Morex infiltrated with the proteasome peptide aldehyde inhibitor MG132 in DMSO and infected with pathotype MCC; lane 3: uninfected and untreated Morex; lane 4: Morex infiltrated with plant protease inhibitor mixture, P9599 in DMSO and infected with pathotype MCC; and lane 5: Morex infiltrated with DMSO and infected with pathotype MCC. Immunoblot analysis of extracts 28 hai was carried out with antibody specific for RPG1.
To further validate ubiquitination as the mechanism underlying the degradation of RPG1, we used the synthetic proteasome substrate benzyloxycarbonyl-l-leucyl-l-leucyl-l-norvaline 4-methyl-coumaryl-7-amide (MG132) to inhibit the proteasome activity. It targets the chymotrypsin-like activity of the catalytic subunits in the 20S proteasome channel. Alternatively, inhibition of RPG1 proteolysis was analyzed with protease inhibitor mixture P9599 (Sigma–Aldrich, St. Louis, MO). The results showed that RPG1 degradation was inhibited only in the presence of the proteasome inhibitor MG132 but not by the mixtures of protease inhibitors (Fig. 4B). This is consistent with RPG1 protein degradation by the proteasome pathway.
Discussion
We have presented evidence that the Rpg1 gene product is rapidly degraded upon infection of the barley plant with avirulent pathotypes of the stem rust fungus Pgt. Infection with virulent stem rust (both Pgt and Pgs) and stripe rust cultures did not have a noticeable effect on the RPG1 protein. Thus, RPG1 degradation appears to be a specific reaction to the avirulent pathogen and presumably important in the defense signaling pathway and disease resistance. The role of RPG1 degradation in this disease resistance is underscored by the highly susceptible mutants K1 and K2 that do not degrade the RPG1 protein upon infection with the pathotype MCC, which is avirulent on wild-type Rpg1.
Two possible roles of protein degradation in disease resistance have been postulated, i.e., negative regulation of HR and removal of a negative regulator from an R protein complex. Negative regulation of HR was suggested as a possible explanation for the Arabidopsis RPM1 R protein degradation (16). RPM1 degradation was coincident with the HR response, leading us to speculate that degradation of RPM1 may be how the cell controls HR lesion size and confines it to the site of infection. The mechanism of RPM1 degradation has not been resolved. The need to control HR is evident from necrotic mutants, such as the Arabidopsis lsd1, which exhibit spreading necrotic lesions suggesting that the wild-type gene controls the extent of HR (16, 33). The observation that RPG1 degradation does not radiate to the uninfected leaves of the infected plant tends to support this argument. However, in our case, the RPG1 degradation seems to occur ≈10 h before visible HR and thus may not be the factor that limits HR (27).
Another possibility is that RPG1 degradation initiates the disease resistance-signaling pathway either by removing a negative regulator from the R protein complex or by actively initiating the signaling pathway just before degradation perhaps by phosphorylation triggered by the interaction with AvrRPG1. We know that the absence of RPG1 is not sufficient to activate the R protein complex because commercially viable barley cultivars exist that do not produce the RPG1 protein or any cross-reacting material (34). Another possibility is that the degradation of RPG1 releases a peptide that is the real initiator of the disease resistance response. In the PBS1 case, as with RPG1, degradation and phosphorylation are both correlated with disease resistance. However, we have not detected a retained peptide in the degradation process by using antibodies directed either against an RPG1 peptide or against the whole protein. Nevertheless, this possibility cannot be excluded as illustrated by the case of RIN4 degradation by the Pseudomonas syringae effector AvrRpt2, where a small (≈6.4 kDa) membrane-embedded fragment is retained, whereas the rest of the molecule is eliminated by the proteasome pathway (35). RIN4 has been implicated as a negative regulator of disease resistance by knockdown mutants that show constitutive activation of defense responses (14).
The ubiquitin–proteasome pathway mediates specific degradation of regulatory proteins and plays an important role in controlling a variety of cellular functions (36). Degradation of a protein by the ubiquitin system involves two distinct and successive steps: covalent attachment of multiple ubiquitin molecules to the target protein and degradation of the tagged protein by the 26S proteasome. Conjugation of ubiquitin to the substrate proceeds through the action of three enzymes; ubiquitin-activating enzyme E1, ubiquitin-conjugating enzyme E2, and ubiquitin–protein ligase E3 (36, 37). Ubiquitination specificity seems to be determined by the ligase E3 and by posttranslational modification, such as phosphorylation, of the target protein. The human Janus kinase JAK2 has been shown to be specifically polyubiquitinated and degraded after phosphorylation of the Y1007 residue (38). Janus kinases are relevant to our studies because, like RPG1, they are tandem kinases with one functional and one pseudokinase domain (39).
We have shown that the elimination of RPG1 is correlated with ubiquitination and the requirement of the chymotrypsin-like activity of the catalytic subunits in the 20S proteasome channel (40). Among the E3 ligases that bind the protein to be ubiquitinated, there is one family that recognizes substrates according to the end rule, preferring proteins with basic residues at the N terminus such as R, K, and H (40), often after removal of the N-terminal methionine. It is interesting that the N terminus of RPG1 is M-M-V-R. Thus, the E3 specificity may identify the family of genes to be degraded, whereas phosphorylation of specific residue(s) targets it for degradation. The K2 mutant RPG1 with KK461, 462NQ substitutions in the kinase ATP anchor is no longer an active kinase, does not provide resistance to stem rust, fails to undergo polyubiquitination, and is not degraded upon infection with an avirulent rust pathotype (Figs. 3D and 4A). The sister mutant K1 with KK152, 153NQ substitutions in the region homologous to the K2 mutant maintains kinase activity, but is not degraded and cannot provide resistance to stem rust (Figs. 3D and 4A). In uninfected controls, the K1 mutant RPG1 protein undergoes a basal level of polyubiquitination similar to the uninfected controls Morex and GP/Rpg1T1. However, it does not respond to stem rust infection by ubiquitinating increasing numbers of RPG1 molecules (Fig. 4A). These results, and similarity to the Janus kinases, suggest that the pK1 pseudokinase domain acts as a regulator of the pK2 domain and transmits signals from external sources to the pK2 domain.
The Rpg1 suppressor line rpr1 is susceptible to pathotype MCC but still degrades its RPG1 protein upon infection. Thus, the RPG1 protein degradation is not sufficient for disease resistance. The rpr1 mutant probably affects a downstream step essential for the Rpg1-mediated disease resistance signaling (28). The degradation of RPG1 occurs at about the time of haustoria establishment in the infected tissue, ≈16–30 hai. Based on the work with flax rust (9, 26), it is reasonable to believe that synthesis and delivery to the host cell of the avirulence protein occurs in the haustoria. Therefore, the degradation of RPG1 is coincidental with interaction of the hypothetical AvrRPG1 effector.
In summary, RPG1 degradation in response to avirulent rust pathotypes and ability to phosphorylate are both associated with disease resistance, and neither by itself is sufficient. The model that we currently favor is that phosphorylation targets RPG1 for degradation. The degradation process, or products, activates the signaling pathway that results in disease resistance. Alternatively an autophosphorylated RPG1 fragment survives the degradation process long enough to initiate the downstream resistance signaling cascade.
Materials and Methods
Plant Materials.
Barley lines for the quantification and turnover experiments were grown in growth chambers in plastic pots containing potting mix with a day and night temperature of 21 ± 1°C and 18 ± 1°C, respectively and with a 16-h photoperiod provided by cool fluorescent tubes (525 μE/m2s).
Antibody Production.
Polyclonal antibody was raised in rabbits by Alpha Diagnostics (Austin, TX) against an RPG1 synthetic peptide NKLTATPLEEKSRSC, representing residues 834–848 (24). Antibody against the full-length recombinant RPG1 with a C-terminal His-tag (24) was raised at Washington State University in New Zealand White rabbits with RIBI Adjuvant (Corixa, Hamilton, MT). Rabbit anti-GST antibody was purchased from Bethyl Laboratories (Montgomery, TX). Antibodies against nitrate reductase, glutamate-1-semialdehyde aminotransferase, and glutamyl tRNA synthetase have been described (29, 41, 42).
Quantification of RPG1 by ELISA.
RPG1 protein levels were quantified by ELISA (43) by using RPG1 specific polyclonal antibodies. Approximately 200 mg of 10-d-old barley leaves were ground in 400 μl of the extraction buffer and spun at 15,300 × g for 5 min. Approximately 200 μl of the supernatant was coated onto ELISA plates and incubated at 4°C for 12 h. The supernatant was discarded, and the wells were washed three times with PBS containing Tween 20 (PBST) and refilled with ≈200 μl of the cross-absorption antiserum buffer prepared from Golden Promise. To prepare the cross-absorption antiserum, ≈200 mg of Golden Promise lacking the RPG1 protein (34), leaves were ground with 400 μl of antiserum buffer and spun at 15,300 × g for 5 min. The supernatant was mixed with affinity-purified RPG1 antisera at 1:500 dilution and used as the cross-absorption antisera. The plates were incubated for 4 h at room temperature on moist paper towels. The wells were washed three times with PBST, 200 μl of the goat anti-rabbit IgG-horseradish peroxidase conjugate was added, and the wells were incubated for 2 hours. The wells were washed five times with PBST and allowed to dry on clean paper towels. One hundred microliters of TMB (3,3′,5,5′-tetramethyl benzidine) substrate was added per well, and the color development was measured at 405 nm by using an ELISA plate reader (Bio-Rad, Hercules, CA). Purified RPG1 protein or the peptide used to develop the antibody was used to construct a standard curve for ELISA quantification. Golden Promise was used as a negative control.
Rust Infection.
Seedlings were grown in a growth chamber at 21–22°C and 80–100% relative humidity for 1 week. The plants were inoculated with avirulent (MCC and SCCL-C7a) or virulent (QCC) Pgt pathotypes or with the virulent Pgs isolate 92-MN-90. Controls were grown under the same conditions but were not inoculated with rust. Plants to be assayed for rust infection were inoculated with one of the rust cultures (concentration 4.5 mg of urediniospores/0.7 ml of Soltrol oil) at a rate of 0.25 mg per plant. Tissue samples were collected as indicated in the figures. Leaves were preserved in RNA Later (Ambion, Woodward, TX) for protein analysis. Twelve days after inoculation, the infection types (IT) were assessed based on a 0–4 rating scale as described (34). On this scale, IT 0 is characterized by no visible symptoms; IT 0; is characterized by hypersensitive “flecks” (small necrotic areas) and no uredinia (infection sites with pathogen sporulation); IT 1 is characterized by minute uredinia surrounded by distinct necrotic areas; IT 2 is characterized by small uredinia surrounded by chlorosis; IT 3 is characterized by medium-sized uredinia often surrounded by chlorosis; and IT 4 is characterized by large uredinia usually without chlorosis. ITs are divided in five general classes: highly resistant. with ITs of 0 or 0;; resistant, with ITs of 1 or 10; moderately resistant, with ITs of 12 or 21; intermediate, with ITs of 23; and susceptible, with ITs of 3 and 4.
Morex and Q21861 seedlings also were inoculated with stripe rust (P. striiformis f. sp. hordei) race PSH-63. Uredinospores were mixed with talc (Sigma, St. Louis, MO) at a 1:20 ratio and spread on the leaves. The inoculated plants were kept in a dew chamber at 10°C in the dark for 24 h and then transferred to a growth chamber at a diurnal temperature cycle gradually changing from 4°C at 2 a.m. to 20°C at 2 p.m. with a daily 16-h photoperiod. Samples were collected 28 hai and phenotyped at 21 dai.
Immunoprecipitation.
Approximately 200 μg of leaf tissue were ground in 500 μl of ice-cold extraction buffer [0.5 M sorbitol, 50 mM Tris·HCl (pH 7.5)/ 10 mM MgCl2/1 mM DTT]. Cell debris was removed by centrifugation at 15,300 × g for 10 min, and total protein remaining in the supernatant was quantified by a dye-binding assay according to the manufacturer's instruction (Bio-Rad). For immunoprecipitation, 500 μg of total protein was combined with 30 μl of affinity-purified antisera in extraction buffer and 500 μl of 2× immunoprecipitation buffer (1M KCl/0.02M EDTA/2 mM PMSF) and rotated end-over-end at 4°C for 12 h. Protein A-agarose (30 μl) was added and incubated on ice for 1 h to precipitate the immunocomplexes, which were collected at 15,300 × g. Immunocomplexes were washed four times with 1 ml of ice-cold immunoprecipitation buffer, resuspended in 30 μl of Laemmli sample buffer (44), boiled at 95°C for 3 min, and analyzed by SDS/PAGE. The immunoprecipitation of the RPG1 protein was used to concentrate the protein and enable visualization of possible breakdown products.
Proteins were electroblotted to PVDF membranes and blocked in TBST [20 mM Tris/500 mM NaCl/0.1% Tween-20 (pH 7.5)] containing 10% nonfat dry milk. The blots were reacted with the primary antibodies for 12 h at room temperature. Horseradish peroxidase-conjugated secondary antibodies were diluted 1:10,000 (Alpha Diagnostics). Bands were visualized with Nu Glo-chemiluminescent detection system according to manufacturer's directions (Alpha Diagnostics).
Enrichment of Ubiquitinated RPG1 from Barley Plants.
Morex, GP/Rpg1T1, K1, and K2 mutant seedlings were infected with Pgt pathotype MCC and sampled at 28 hai. Uninfected samples were used as controls. Extracts were prepared by grinding 200 mg of leaf tissue in 500 μl of ice-cold extraction buffer devoid of DTT [0.5 M sorbitol/50 mM Tris·HCl (pH 7.5)/10 mM MgCl2). Cell debris was removed by centrifugation at 15,300 × g for 10 min, and total protein remaining in the supernatant was quantified by a dye-binding assay (Bio-Rad). For enrichment, ≈500 μg of the total protein was suspended in TBS in a 1:1 ratio, mixed with 20 μl of polyubiquitin affinity resin (Pierce, Rockford, IL), incubated by rotation end-over-end overnight at 4°C, and washed three times with a 1:1 mixture of TBS and the extraction buffer devoid of DTT in a spin column provided by the manufacturer. The ubiquitinated proteins were then eluted by boiling in Laemmli buffer and subjected to SDS/PAGE and Western blotting. The enriched proteins were visualized by using RPG1-specific antisera by a chemiluminescent method according to the manufacturer's directions (Alpha Diagnostics).
Ubiquitin Inhibitor and Plant Protease Inhibitor Experiments.
Ten-day-old cv. Morex plants were infiltrated with either 100 μM MG132 (Sigma–Aldrich) or a mixture of plant protease inhibitors (Sigma–Aldrich) dissolved in DMSO. The protease inhibitor mixture contains the following: 4-(2-aminoethyl) benzenesulfonyl fluoride targeting serine proteases; bestatin (3-amino-2-hydroxy-4-phenylbutanoyl-l-leu) inhibiting aminopeptidases; pepstatin A targeting aspartate proteases; leupeptin (propionyl-l-leu-l-leu-arginyl) inhibiting serine and cysteine proteases; transepoxysuccinyl-l-leucylamido (4-guanidino) butane targeting cysteine proteases, and 1,10-phenanthroline inhibiting metalloproteases. The plants were dried for 4 h and then inoculated with Pgt. pathotype MCC and sampled at 28 hai and subjected to immunoprecipitation and subsequent Western blot analysis with RPG1-specific antisera.
Supplementary Material
Acknowledgments
We thank Laura Penman for technical assistance. This work was supported by National Research Initiative of the U. S. Department of Agriculture Cooperative State Research, Education, and Extension Service Grant 2004-35301-14635 (to A.K. and B.J.S.) This is scientific paper # 0301-07 from the College of Agriculture, Human, and Natural Resource Sciences, Washington State University, Project 0196.
Abbreviations
- Avr
avirulence
- cv.
cultivar
- hai
hours after infection
- R
resistance
- PCD
programmed cell death
- Pgt
Puccinia graminis f. sp. tritici
- Pgs
Puccinia graminis f. sp. secalis.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0703758104/DC1.
References
- 1.Hammond-Kosack KE, Jones JDG. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:575–607. doi: 10.1146/annurev.arplant.48.1.575. [DOI] [PubMed] [Google Scholar]
- 2.Tang X, Frederick RD, Zhou J, Halterman DA, Yia Y. Science. 1996;274:2060–2063. doi: 10.1126/science.274.5295.2060. [DOI] [PubMed] [Google Scholar]
- 3.Ellis JG, Lawrence GJ, Luck JE, Dodds PN. Plant Cell. 1999;11:495–506. doi: 10.1105/tpc.11.3.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scofield SR, Tobias CM, Rathjen JP, Chang JH, Lavelle DT, Michelmore RW, Staskawicz BJ. Science. 1996;274:2063–2065. doi: 10.1126/science.274.5295.2063. [DOI] [PubMed] [Google Scholar]
- 5.Leister RT, Katagiri F. Plant J. 2000;22:345–354. doi: 10.1046/j.1365-313x.2000.00744.x. [DOI] [PubMed] [Google Scholar]
- 6.Thomas CM, Jones DA, Parniske M, Harrison K, Balint-Kurti PJ, Hatzinxanthis K, Jones JDG. Plant Cell. 1997;9:2209–2224. doi: 10.1105/tpc.9.12.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jia J, McAdams SA, Bryan GT, Hershey HP, Valent B. EMBO J. 2000;19:4004–4014. doi: 10.1093/emboj/19.15.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deslandes L, Olivier J, Peeters N, Feng DX, Khounlotham M, Boucher C, Somssich I, Genin S, Marco Y. Proc Natl Acad Sci USA. 2003;100:8024–8029. doi: 10.1073/pnas.1230660100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dodds PN, Lawrence GJ, Catanzariti A-M, Teh T, Ching I, Wang A, Ayliffe MA, Kobe B, Ellis JG. Proc Natl Acad Sci USA. 2006;103:8888–8893. doi: 10.1073/pnas.0602577103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Der Bizen EA, Jones JDG. Trends Biochem Sci. 1998;23:454–456. doi: 10.1016/s0968-0004(98)01311-5. [DOI] [PubMed] [Google Scholar]
- 11.Dangl JL, Jones JD. Nature. 2001;411:826–833. doi: 10.1038/35081161. [DOI] [PubMed] [Google Scholar]
- 12.Chisholm ST, Coaker G, Day B, Staskawicz BJ. Cell. 2006;124:803–814. doi: 10.1016/j.cell.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 13.Axtell MJ, Staskawicz BJ. Cell. 2003;112:369–377. doi: 10.1016/s0092-8674(03)00036-9. [DOI] [PubMed] [Google Scholar]
- 14.Mackey D, Holt BF, Wiig A, Dangl JL. Cell. 2002;108:743–754. doi: 10.1016/s0092-8674(02)00661-x. [DOI] [PubMed] [Google Scholar]
- 15.Bisgrove SR, Simonich MT, Smith NM, Sattler A, Innes RW. Plant Cell. 1994;6:927–933. doi: 10.1105/tpc.6.7.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boyes DC, Nam J, Dangl JL. Proc Natl Acad Sci USA. 1998;95:15849–15854. doi: 10.1073/pnas.95.26.15849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shao F, Goldstein C, Ade J, Stioutemyer M, Dixon JE, Innes RW. Science. 2003;301:1230–1233. doi: 10.1126/science.1085671. [DOI] [PubMed] [Google Scholar]
- 18.Warren RF, Merritt PM, Holub E, Innes RW. Genetics. 1999;152:401–412. doi: 10.1093/genetics/152.1.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salmeron JM, Oldroyd GED, Rommens CMT, Scofield SR, Kim H-S, Lavelle DT, Dahlbeck D, Staskawicz BJ. Cell. 1996;86:123–133. doi: 10.1016/s0092-8674(00)80083-5. [DOI] [PubMed] [Google Scholar]
- 20.Pedley KF, Martin GB. Annu Rev Phytopathol. 2003;41:215–243. doi: 10.1146/annurev.phyto.41.121602.143032. [DOI] [PubMed] [Google Scholar]
- 21.Mucyn TS, Clemente A, Andriotis VME, Balmuth AL, Oldroyd GED, Staskawicz BJ, Rathjen JP. Plant Cell. 2006;18:2792–2806. doi: 10.1105/tpc.106.044016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steffenson BJ. Euphytica. 1992;63:153–167. [Google Scholar]
- 23.Brueggeman R, Rostoks N, Kudrna D, Kilian A, Han F, Chen J, Druka A, Steffenson B, Kleinhofs A. Proc Natl Acad Sci USA. 2002;99:9328–9333. doi: 10.1073/pnas.142284999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nirmala J, Brueggeman R, Maier C, Clay C, Rostoks N, Kannangara CG, von Wettstein D, Steffenson B, Kleinhofs A. Proc Natl Acad Sci USA. 2006;103:7518–7523. doi: 10.1073/pnas.0602379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rostoks N, Steffenson BJ, Kleinhofs A. Physiol Mol Plant Pathol. 2004;64:91–101. [Google Scholar]
- 26.Dodds PN, Lawrence GJ, Catanzariti A-M, Ayliffe M, Ellis J. Plant Cell. 2004;16:755–768. doi: 10.1105/tpc.020040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin KC, Bushnell WR, Smith AG, Szabo LJ. Physiol Mol Plant Pathol. 1998;52:95–114. [Google Scholar]
- 28.Zhang L, Fetch T, Nirmala J, Schmierer D, Brueggeman R, Steffenson B, Kleinhofs A. Theor Appl Genet. 2006;113:847–855. doi: 10.1007/s00122-006-0342-y. [DOI] [PubMed] [Google Scholar]
- 29.Somers DA, Kuo T-M, Kleinhofs A, Warner RL, Oaks A. Plant Physiol. 1983;72:949–952. doi: 10.1104/pp.72.4.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roelfs AP. In: The Cereal Rusts. Roelfs AP, Bushnell WR, editors. Vol II. New York: Harcourt Brace Jovanovich; 1985. pp. 3–37. [Google Scholar]
- 31.Gausing K, Barkardottir R. Eur J Biochem. 1986;158:57–62. doi: 10.1111/j.1432-1033.1986.tb09720.x. [DOI] [PubMed] [Google Scholar]
- 32.Vierstra RD. Plant Mol Biol. 1996;32:275–302. doi: 10.1007/BF00039386. [DOI] [PubMed] [Google Scholar]
- 33.Dietrich RA, Richberg MH, Schmidt R, Dean C, Dangl JL. Cell. 1997;88:685–694. doi: 10.1016/s0092-8674(00)81911-x. [DOI] [PubMed] [Google Scholar]
- 34.Horvath H, Rostoks N, Brueggeman R, Steffenson B, von Wettstein D, Kleinhofs A. Proc Natl Acad Sci USA. 2003;100:364–369. doi: 10.1073/pnas.0136911100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim H-S, Desveaux D, Singer AU, Patel P, Sondek J, Dangl JL. Proc Natl Acad Sci USA. 2005;102:6496–6501. doi: 10.1073/pnas.0500792102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ciechanover A. EMBO J. 1998;17:7151–7160. doi: 10.1093/emboj/17.24.7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laney JD, Hochstrasser M. Cell. 1999;97:427–430. doi: 10.1016/s0092-8674(00)80752-7. [DOI] [PubMed] [Google Scholar]
- 38.Ungureanu D, Saharinen P, Juntilla I, Hilton DJ, Silvennoinen O. Mol Cell Biol. 2002;22:3316–3326. doi: 10.1128/MCB.22.10.3316-3326.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Giordanetto F, Kroemer RT. Protein Engineering. 2002;15:727–737. doi: 10.1093/protein/15.9.727. [DOI] [PubMed] [Google Scholar]
- 40.Myung J, Kim KB, Crews CM. Med Res Rev. 2001;21:245–273. doi: 10.1002/med.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grimm B, Bull A, Welinder KG, Gough SP, Kannangara CG. Carlsberg Res Commun. 1989;54:67–79. doi: 10.1007/BF02907586. [DOI] [PubMed] [Google Scholar]
- 42.Kannangara CG, Gough SP, Bruyant P, Hoober JK, Kahn A, von Wettstein D. Trends Biochem Sci. 1988;13:139–143. doi: 10.1016/0968-0004(88)90071-0. [DOI] [PubMed] [Google Scholar]
- 43.Li Z, Jayasankar S, Gray DJ. Plant Mol Biol Rep. 2001;19:341–351. [Google Scholar]
- 44.Laemmli UK. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.