Abstract
During embryogenesis, the XIST RNA is expressed from and localizes to one X chromosome in females and induces chromosome-wide silencing. Although many changes to inactive X heterochromatin are known, the functional relationships between different modifications are not well understood, and studies of the initiation of X-inactivation have been largely confined to mouse. We now present a model system for human XIST RNA function in which induction of an XIST cDNA in somatic cells results in localized XIST RNA and transcriptional silencing. Chromatin immunoprecipitation and immunohistochemistry shows that this silencing need only be accompanied by a subset of heterochromatic marks and that these can differ between integration sites. Surprisingly, silencing is XIST-dependent, remaining reversible over extended periods. Deletion analysis demonstrates that the first exon of human XIST is sufficient for both transcript localization and the induction of silencing and that, unlike the situation in mice, the conserved repeat region is essential for both functions. In addition to providing mechanistic insights into chromosome regulation and formation of facultative heterochromatin, this work provides a tractable model system for the study of chromosome silencing and suggests key differences from mouse embryonic X-inactivation.
Keywords: dosage compensation, heterochromatin, histone modification, X-chromosome inactivation, DNA methylation
X-chromosome inactivation occurs early in mammalian development, and most studies of the initiating events have used the mouse because the process can be studied during early development as well as upon differentiation of female ES cells. Differences in the process of inactivation have been noted between ES cells and extraembryonic cells and even among extraembryonic lineages (reviewed in ref. 1), suggesting that there may be different routes to achieving silencing. In the mouse, random X inactivation is observed in the derivatives of the inner cell mass, including ES cells, whereas nonrandom inactivation occurs earlier in the cells that give rise to the extraembryonic tissues. In humans, inactivation is random in both the extraembryonic and embryonic tissues, and thus it is not clear to what extent the events seen in mice model the early events of human X-chromosome inactivation (reviewed in ref. 2). In particular, it has been proposed that Tsix, the antisense regulator of Xist in mice, is an expressed but nonfunctional pseudogene in humans (3). There are also differences in the heterochromatin components of the inactive X, with methylation (m) of lysine (K) 27 of histone H3 (H3K27m3) being more stable on the human inactive X, which also shows an association with the heterochromatin protein HP1, and Barr body staining (4). These differences may reflect both species and cell type differences; however, they underscore the need for a human model system to study initiation of X inactivation. We have taken advantage of one further significant difference between the human and mouse inactivation process to develop such a system in human somatic cells. Although silencing induced by Xist can occur only in a limited developmental window in mice (5), inactivation can be induced in some human somatic cells (6). We now expand these early observations to report an inducible single-copy XIST transgene that can induce silencing of flanking genes.
Inactivation occurs at a time when many events of differentiation are occurring, and, although variations exist among cell types, the following general cascade of events has been described in murine ES cells. The earliest changes observed after Xist coating of the X chromosome are the exclusion of euchromatic histone marks, including histone H3 and H4 acetylation and H3K4m2. As X-linked gene silencing is established, histone modifications such as H3K27m3, H3K9m2, ubiquitinylation of H2A and H4K20m1 are increased, and members of the polycomb repressive complexes (PRC), some of which are responsible for establishment of such histone modifications, are transiently recruited to the inactive X. The association of the histone variant macroH2A and methylation of DNA at CpG islands associated with the 5′ ends of genes are later events (reviewed in ref. 1).
Similar epigenetic changes are observed in autosomal material with transgenic murine Xist (e.g., ref. 7). However, an inducible mouse Xist cDNA construct is only able to induce silencing when induced within 48 h after induction of differentiation (5). In addition, whereas inactivation is reversible upon murine Xist-induced silencing initially, after 72 h of differentiation, silencing becomes irreversible despite loss of Xist expression (5); consistent with results showing that maintenance of human X inactivation does not require XIST in somatic cells (8). We now describe an inducible human XIST cDNA that, in contrast to the mouse, is able to silence flanking genes in a somatic cell line. Furthermore, we show that ongoing silencing, which is accompanied by a subset of chromatin changes, is reversible upon XIST silencing and thus depends on XIST.
Results
Inducible Human XIST Transgenes in Somatic Cells Result in a Localized XIST Focus.
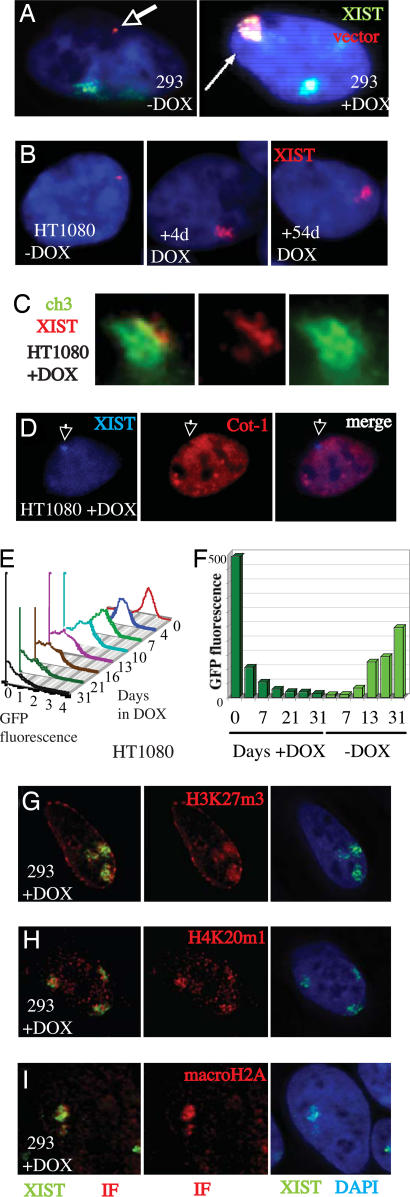
To examine XIST expression from single-copy integrations, we generated an inducible XIST cDNA construct using the Flp-In T-REx system (Invitrogen, Carlsbad, CA). The inducible promoter eliminates selection against sites affected by XIST-mediated silencing and facilitates the analysis of events immediately after the up-regulation of XIST, whereas the FLP-mediated integration into FRT sites permits retargeting to the same chromosomal location to allow comparison between constructs. We introduced a full-length human XIST cDNA transgene into the FRT site of the commercially available Flp-In T-REx 293 (293) cell line (Invitrogen). RNA-FISH showed localization of the XIST cDNA transcript upon induction with doxycycline (DOX; Fig. 1A). Because the 293 cells have two endogenous inactive X chromosomes that confound analysis of the XIST transgene, we generated an FRT integration site in the male HT1080 fibrosarcoma cell line that had previously shown hallmarks of silencing upon exogenous XIST expression (6). In addition, to allow monitoring of silencing in cis, an EGFP reporter gene was cotransfected with the FRT-containing plasmid. As expected for a male cell line, the untransfected HT1080 cells showed no XIST expression (data not shown). Before induction by DOX, a small point signal consistent with a primary transcription focus of XIST was detectable in some cells, suggesting leaky low-level expression (Fig. 1B). Upon induction, the XIST RNA formed a large nuclear focus that did not appear to change in size with increasing days of induction, and the level was similar to that seen in female somatic cells (data not shown). Mapping of the transgene in the HT1080 cells showed integration into chromosome 3q, and the 293 integration is also autosomal because it does not colocalize with X chromosome paint (data not shown). Cohybridization with a chromosome-3 paint probe showed consistent overlap of XIST with the chromosome-3 signal; however, XIST did not coat the entire domain (Fig. 1C). Although previous transgenes of XIST have shown localization (6, 9), as have mouse cDNA constructs (10), human XIST cDNA construct have not previously been used, and their use demonstrates definitively that the human XIST cDNA can localize in cis and that flanking sequences, introns, and splicing are not necessary for cis accumulation.
Fig. 1.
Inducible XIST cDNA construct results in localized focus of XIST and silencing. (A) RNA-FISH analysis of the 293 cell line with the XIST transgene integrated into the FRT site. An ectopic focus of XIST expression (arrow) is observed in addition to the large endogenous signals and increases substantially after induction (+DOX, Right). The transgene signal is distinguished by hybridization with the vector. (B) HT1080 cells containing the XIST cDNA transgene after DOX induction for the times listed. (C) Cohybridization to detect XIST and chromosome 3 DNA show colocalization of the RNA with the chromosome from which it is expressed. Panels show the merged signal as well as only the chromosome paint and the XIST RNA signals. (D) Induction of XIST results in a focus of XIST expression that colocalizes to a reduction in Cot1 staining of the cell. Cells shown were induced for 25 days with DOX. (E) Flow cytometry plots of GFP fluorescence after different days of DOX induction. Multiple plots of fluorescence levels (log scale) from FloJo software for various times of XIST induction are shown. The vertical bars indicate that there is signal that has reached the lower baseline. (F) Bar chart of average fluorescence levels at various time points of DOX induction and then after 31 days of DOX induction, subsequent days without DOX (paler green). As in E plots showed a single peak of fluorescence suggesting that level is due to reduction in amount of GFP rather than percent of cells expressing GFP. (G–I) The 293 cells after 15 days of DOX induction hybridized to detect XIST RNA, H3K27m3 (G), H4K20m1 (H) or macroH2A (I) by immunofluorescence (IF); showing colocalization of the RNA and IF signal.
Inducible Human XIST Expression Results in Silencing That Is Reversible.
To determine whether human XIST expression results in transcriptional silencing, we examined repression in the HT1080 cells by two alternate approaches: silencing of the flanking EGFP reporter and more global transcriptional activity by Cot1 RNA staining. Cot1 DNA has been shown to hybridize to the heteronuclear RNA of the nucleus, with “holes” reflecting XIST-induced X-chromosome silencing as well as silencing of autosomal material (6). As shown in Fig. 1D, a noticeable reduction in Cot1 staining was observed under the XIST signal. We note that the size of the hole was less than is observed for the inactive X chromosome in female somatic cells; however, the Cot1 hole was observed consistently at this ectopic site of XIST expression. Fluorescence-activated cell sorting provided a quantitative assessment of silencing of the downstream EGFP reporter gene. As shown in Fig. 1E, there was a decrease in fluorescence shortly after DOX induction. No such effect was observed with DOX treatment of cells without the XIST integration (data not shown). After 4 days of DOX induction, the expression of GFP was reduced by ≈80%, and the fluorescence levels continued to decrease with increasing days in DOX. Although the peak of fluorescence broadened somewhat over time, there was consistently a single peak, suggesting that reduced levels of fluorescence were due to reduced expression in all cells. The hygromycin gene lies upstream of XIST and was used for selection of FRT-mediated integration, so we did not study its expression in detail because prior selection might have influenced its ability to be silenced; interestingly, however, assessment by RT-PCR and ongoing viability in hygromycin suggest that this gene is not subject to XIST-induced silencing. When DOX was removed from the cells, XIST expression was rapidly silenced returning to close to the low levels observed before DOX induction within 5 days (data not shown). Surprisingly, this rapid silencing of XIST was accompanied by a more gradual reactivation of GFP in the whole cell population (Fig. 1F). Thus, silencing of GFP is XIST-dependent, however the ongoing accumulation, and then gradual loss, of silencing is highly suggestive of the acquisition of chromatin changes.
Epigenetic Changes Accompany XIST Expression and Silencing.
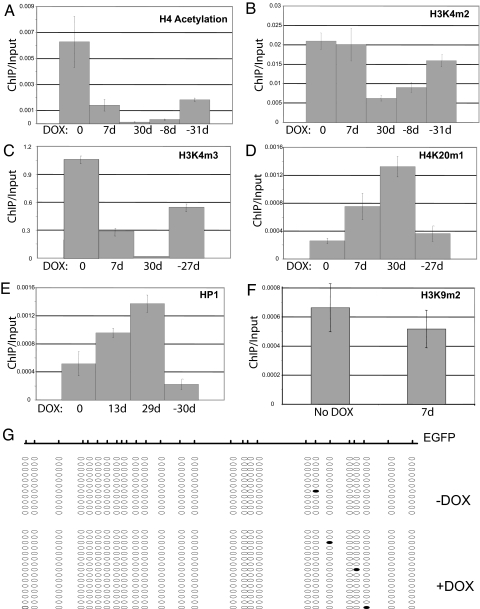
We examined changes in chromatin by immunoflourescence in both the 293 cell line and the HT1080 cell line. The 293 cells showed some accumulation of macroH2A, H3K27m3, and H4K20m1 (Fig. 1 G–I). Interestingly, no detectable accumulation was observed at the XIST focus in the HT1080 cell line, suggesting that there is variability between integration sites in the extent of recruitment of marks of the inactive X. For a higher resolution examination of the role of histone modifications in the establishment of silencing, we examined the EGFP gene promoter in the HT1080 cells by ChIP. Loss of euchromatic marks is an early event in mouse X inactivation, and loss of the active H4 acetylation mark was observed soon after DOX induction of our cells, showing a 4-fold reduction by day 7 (Fig. 2A). H3K4m2, another active chromatin modification, was lost upon long-term DOX induction, however after a week of XIST-induced silencing, the loss of this active mark was not significant, whereas H3K4m3 showed some reduction (Fig. 2 B and C). We observed the acquisition of H4K20m1 and HP1, marks generally associated with inactive chromatin, fairly shortly after silencing is initiated (3- and 2-fold increase in 7 days, respectively, Fig. 2 D and E), with ongoing accumulation. In contrast, several other chromatin modifications associated with an inactive X chromosome were not detected. Although H3K9m2 appears to be present at the promoter of the EGFP reporter gene, this modification was not increased by the induction of XIST expression (Fig. 2F). H3K27m3, H3K9m3, and the polycomb group protein Ezh2 were not detected at the EGFP promoter (data not shown). To examine the level of DNA methylation we performed bisulphite DNA sequencing of the EGFP CMV promoter region. No significant levels of methylation were observed at the promoter before or after DOX induction of XIST (Fig. 2G), consistent with the reversible nature of the silencing observed.
Fig. 2.
Heterochromatic changes associated with XIST expression. (A–F) The amplification of the EGFP promoter in the HT1080 cells was compared by Q-PCR after ChIP relative to the input for the times of DOX induction indicated. The antibodies used for ChIP recognized: histone H4 acetylated at lysines 5, 8, 12, and 16 (A); histone H3 dimethylated at lysine 4 (B); histone H3 trimethylated at lysine 4 (C); histone H4 monomethylated at lysine 20 (D); HP1-γ (E); and histone H3 dimethylated at lysine (F). (G) Schematic showing CpG dinucleotides upstream of the CMV promoter driving EGFP expression. Open circles represent unmethylated Cs detected by bisulphite conversion, and closed circles demarcate methylated Cs resistant to bisulphite conversion. Each line represents an independent clone derived from PCR products of bisulphite modified DNA from uninduced (upper cluster) or 30-day inductions of the HT1080-inducible XIST clone (lower cluster).
The modifications that were observed by ChIP in the HT1080 cells (loss of H4 acetylation and H3K4m2/3; gain of HP1 and H4K20m1) were all XIST-dependent. H4 acetylation and H3K4m3 levels increased when the transgene was silenced upon removal of DOX; however, by 30 days after DOX removal, the H4 acetylation and H3K4m2 levels were only approximately one-third and one-half, respectively, of those observed before XIST expression (Fig. 2B). The other marks appeared to return to pre-XIST levels by 30 days of DOX removal. In summary, we observed a cascade of epigenetic modifications at the EGFP promoter upon XIST-dependent silencing of the gene. The loss of the active mark of H4 acetylation appears to be the first event, followed by gain of H4K20m1 and HP1, loss of H3K4m3, and then loss of H3K4m2. We did not see acquisition of several of the marks generally associated with the inactive X, including DNA methylation, H3K9m2/3, and H3K27m3, which may explain the XIST-dependent nature of the silencing. Upon silencing of XIST and reactivation of EGFP, we observed complete loss of H4K20m1 and HP1 and partial reacquisition of H4 acetylation.
Identification of Regions of XIST Involved in Localization and Silencing.
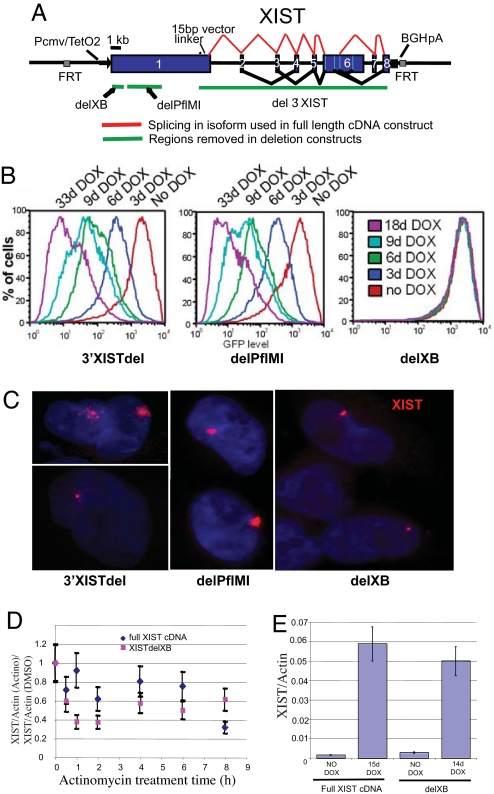
To identify critical regions of the XIST gene involved in localization and/or silencing, we generated a series of deletion constructs and integrated these into the same FRT site adjacent to the EGFP reporter gene in the HT1080 cells. We assayed EGFP silencing upon expression of truncated XIST RNAs and followed XIST localization by FISH (Fig. 3). Analysis of the Xist gene in various eutheria has shown that the internal exons are generally well conserved, as is the 5′ “A” repeat region (11, 12); however deletion experiments have only previously been done with mouse Xist. The murine “C” repeat had been suggested to be important for localization by disruption with PNA oligonucleotides (13). However, extensive deletion analysis has demonstrated that this region is not necessary for Xist function and, furthermore, that redundant regions are involved in mouse Xist localization, with the 5′ A repeats being critical for silencing but not localization (10). Thus, we generated three deletions: one that removed the conserved splice region of the XIST RNA, leaving most of exon 1(del 3′ XIST); a second that removed a 3.8-kb fragment in the central region of exon 1, including the C repeats (delPflMI); and a third deletion of 796 bp, including the 5′ conserved A repeats (delXB). Silencing of the EGFP transgene was unaffected in both the 3′ XIST deletion and the delPflMI (Fig. 3). However, the XIST accumulation in the 3′ deletion was consistently scored by independent observers as being more dispersed than that of the full-length construct. Removal of the 5′ repeats completely eliminated EGFP silencing. In contrast to mouse, where transcripts deleted for the conserved A repeats are able to accumulate and localize to surrounding chromatin but not cause gene silencing, we observed a substantial reduction in the size of the XIST RNA signal by RNA FISH (Fig. 3C). This reduction was also apparent in the 293 cell line, so it is not integration-site- or cell-type-dependent (data not shown). To examine whether the reduced signal size reflected an inability of the transcript to localize or was due to a reduction in transcription or transcript stability, we used quantitative PCR after treatment with actinomycin D, a transcriptional inhibitor, to determine the half-life of the RNA. As shown in Fig. 3D the deletion of the 5′ repeats did not substantially affect the stability of the transcript. In addition, the steady-state levels of the full-length and conserved A repeat deleted transcripts are similar upon DOX induction (Fig. 3E). Therefore, the deletion only affects the transcript's ability to accumulate into a domain. Thus, we conclude that the first 10.5 kb of human XIST are sufficient for silencing, and, that in humans, the conserved A repeat region within this exon is necessary not only for the silencing function of XIST but also for its capacity to accumulate in cis.
Fig. 3.
Deleted XIST constructs show differential silencing and localization. (A) Schematic of the inducible XIST construct shows the regions deleted in each construct. (B) FACS scans of the three XIST deletion constructs induced with DOX for the days listed. (C) Representative images of nuclei after RNA FISH for XIST in the three XIST deletion constructs in HT1080 cells. (D) Quantitative PCR of XIST levels relative to β-actin after actinomycin treatment for the times listed on the x axis. Levels were normalized to paired cultures treated with DMSO as the actinomycin is dissolved in DMSO. No significant difference was observed for the full XIST construct (diamonds) and the 5′ repeat deletion construct (squares). (E) The total level of XIST relative to actin was compared between the full XIST construct (left) and the 5′ repeat deletion (right).
Discussion
XIST is a fascinating functional RNA because it is able to induce the cis-limited silencing of an entire chromosome. However, it is unknown how this RNA localizes in cis and then induces the formation of the facultative heterochromatin of the inactive X. Considerable insights into the process of mammalian X inactivation have been garnered through the use of murine ES cells, but it is not clear to what extent the events in mouse development recapitulate the events occurring during human X chromosome inactivation. Despite one report of induction of X inactivation upon differentiation of human ES cells (14), other studies have generally observed that, even in their undifferentiated state, human ES cells appear to already have an inactive X that expresses XIST (15). The presence of an inactive X in these cells is supported by the presence of X-chromosome methylation in female ES cells (16). Ectopic XIST expression is able to induce some features of silencing in human somatic cells (6), and we have exploited this ability to develop a tractable model system for studying human X inactivation using a human inducible XIST construct. We demonstrate both localization of the XIST RNA and XIST-dependent silencing of cis-linked sequences.
We monitored silencing induced by the XIST transgene by activity of a flanking EGFP reporter gene and Cot1 hybridization. By using the latter approach, XIST-induced transcriptional silencing has been shown to result in a hole in human (6) and mouse (17) cells as well as during meiotic silencing of unpaired DNA (18). It has recently been suggested that the Cot1 hole might reflect an altered nuclear compartment for the inactive X that is more reflective of repetitive element silencing than genic silencing (19, 20). Nonetheless, the acquisition of a Cot1 hole after induction of our XIST transgene suggests that the XIST RNA is acting to silence autosomal human sequences in a manner similar to that observed during normal mammalian inactivation.
We observed reduction of EGFP fluorescence to 20% of preinduction levels in all cells after 4 days of XIST expression. Levels of fluorescence continued to decline with increasing times of induction, and, although some of the gradual decline may be attributed to the stability of EGFP, which has a half-life of > 24 h, the gradual accumulation of chromatin changes is likely contributory. When XIST was silenced, EGFP expression was regained slowly, supporting the idea of progressive chromatin changes and demonstrating that the XIST-dependent silencing observed was reversible. We suggest three alternative explanations for the slow acquisition of silencing and associated chromatin changes, as well as the incomplete acquisition of all changes associated with an inactive X. First, these observations may reflect the expression level of the transgene, which, although approximating that seen in human somatic cells, is severalfold lower than the previously examined multicopy Xic transgenes (6), and the mouse inducible promoter system which uses a transactivator to induce high levels of expression (10).
Second, the flanking sequences of the transgene may influence the ability of XIST to establish silencing, potentially acting cooperatively with the level of XIST expression. Studies of X/autosome translocations have suggested a lower silencing efficiency for autosomal genes (e.g., refs. 21–23), leading to the idea of “way stations” involved in spread of silencing. We propose that sequences flanking XIST may be additional players in the spread of silencing, potentially accounting for differences in the chromatin marks observed by immunohistochemistry for the different integrations into the 293 and HT1080 cell lines and also with the previously described XIST transgene that carried multiple copies of a cosmid retaining flanking sequences (6).
Lastly, differences in developmental stage of the cells, in particular the resistance of somatic chromatin to silencing or the lack of development-specific chromatin modifying pathways, will be a critical effector of silencing competence. Ectopic expression of XIST in HT1080 cells has been shown to result in acquisition of histone hypoacetylation and late replication timing (6), ubiquitination of H2A and recruitment of macroH2A (24), and recruitment of H3K27m3 and H4K20m1 (L.L.H., unpublished work), suggesting that at least some clones of HT1080 are able to recruit extensive silencing marks. Furthermore, we have demonstrated by RT-PCR that our HT1080 cells express the PRC1 genes EZH2 and SUZ12. The inability of murine Xist cDNAs induced after differentiation to induce silencing of flanking genes, whereas human XIST in somatic cells is able to induce silencing (this work and ref. 6) similarly may reflect differences in chromatin responsiveness or presence of active epigenetic pathways rather than a species difference in XIST/Xist function. Indeed, there is a recent report that murine hematopoietic precursor cells reestablish their ability to respond to the Xist transgene (25); thus, the transformed human cells that have been examined (HT1080 is a fibrosarcoma line and 293 cells are adenovirus transformed embryonic kidney cells) may be more similar in their complement of active epigenetic remodeling pathways to these responsive murine cells.
Although the kinetics of inactivation in somatic cells may be distinct from those in ES cells; nonetheless, this somatic-cell-based system is useful to reveal the relationships between steps in the silencing pathway. Indeed, the slower acquisition of changes makes differences between their timing more distinct. As is observed in mouse, a loss of active marks is the earliest event after XIST expression; however, loss of acetylation appears to precede loss of H3K4m2 in our system. This is suggestive that loss of acetylation may be the best candidate as a direct effect of XIST expression, perhaps through XIST-dependent recruitment of histone deacetylases. The next modifications recruited during murine inactivation are methylation of H3 at lysines 9 and 27, which we did not observe in our HT1080 cells, suggesting that these marks are not critical for XIST-dependent silencing, which is supported by the ability of primitive endoderm cells to silence without such marks (26). As we observe reactivation upon silencing of XIST in these cells, it is tempting to speculate that the lack of PRC2 recruitment and DNA methylation prevents the silencing from being permanent. Interestingly, histone methylation is often involved in HP1 recruitment (27); however, we observed recruitment of HP1 despite no increased H3K9 methylation. Thus the recruitment of HP1 may occur through other pathways (28), possibly by interactions with the nuclear membrane (29).
The use of transgenes has allowed the definition of redundant regions of murine Xist that are necessary for localization of Xist and subsequent cis-limited gene silencing (10). In a similar fashion, we have analyzed the requirement for different regions of human XIST for localization and silencing and detected different functionalities than those seen in mouse. Consistent with mouse, only the 5′ 10 kb of XIST is necessary for both localization and silencing, although the terminal third of the gene contributes to formation of the XIST RNA focus. Deletion of the human C repeat region did not affect localization or silencing, also consistent with the murine deletion studies (10). A substantial difference is observed for deletion of the 5′ A repeat region that, in humans, resulted in loss of localization and, in mice, resulted in loss of silencing but ongoing Xist localization (10). A caveat to these comparisons is the differences between the model systems as discussed above. Because the promoter is not affected in this deletion, it is clear that the silencing observed for other constructs is due to XIST function rather than simple transcriptional interference. When the 5′ repeat region was eliminated, XIST was no longer localized within a domain, suggesting that interactions with this region are necessary for localization of the XIST RNA in humans.
The development of a model system that recapitulates the localization and silencing induced by human XIST will allow further exploration not only of the sequences in XIST necessary for function but also of the cis-acting sequences that may influence the spread of XIST and silencing of cis-linked genes. Furthermore, the ability of human XIST to induce silencing in these transformed cells may have important implications for cancer because XIST reactivation through hypomethylation (e.g., ref. 30), or translocation of an autosome to an inactive X might result in transcriptional silencing of cis-linked genes.
Materials and Methods
Construct Generation.
To make the full-length XIST cDNA, exon 1 was amplified from female genomic DNA (Expand Long; Roche, Indianapolis, IN) and then digested with XbaI to obtain nucleotides 105–10747 of the XIST cDNA sequence (accession no. M97168). The fragment was end-filled and cloned into the EcoRV site of the pcDNA5/FRT/TO expression vector (Invitrogen). The 3′ end of XIST was derived from cDNA inserted into the NotI and XhoI sites of pcDNA5/FRT/TO. A 15-bp region of the vector multiple cloning site between the EcoRV and the NotI cloning sites was retained.
Cell Culture and XIST Induction.
HT1080 fibrosarcoma cells were transfected first with pcDNA6/TR to allow DOX induction of the CMV promoter (Invitrogen), followed by cotransfection of pEGFP-N1 (Clontech, Mountain View, CA) and the FRT-containing plasmid pFRT/lacZeo (Invitrogen). Cells were grown in DMEM supplemented with 10% FCS and penicillin/streptomycin and nonessential amino acids at 37°C. The inducible XIST construct was cotransfected with the pOG44 plasmid expressing Flp recombinase for site-specific recombination into the FRT site, followed by hygromycin selection for recombined clones as recommended by Invitrogen.
In Situ Hybridization and Detection.
Cells were fixed and imaged as described (6). Briefly, a monolayer of cells was grown on glass coverslips and permeabilized with triton before fixation in 4% paraformaldehyde. For XIST detection, DNA probes G1A, an ≈10-kb genomic plasmid extending from the fourth intron to the 3′ end of the XIST gene, and HbC1a which spans the conserved A repeat region in exon 1, were nick-translated by using biotin-11dUTP or digoxigenin-16-dUTP (Roche). Distinguishing the endogenous XIST transcripts from the transgene-derived cDNA in the female 293 cells was done by using the labeled empty pcDNA5/FRT/TO vector as a probe (Invitrogen). For Cot1 expression analyses by RNA-FISH, Cot1 DNA (Invitrogen) was labeled and hybridized as above.
ChIP.
We followed the protocol from Upstate Biotechnology (Lake Placid, NY); briefly, the histones were cross-linked to the DNA by treating 107 cells with 1% formaldehyde for 10 min at 37°C and then quenching with 125 mM glycine. The cells were lysed and the DNA sonicated to lengths between 200 and 1,000 bp. The sonicated cell supernatant was precleared with salmon sperm DNA/protein A Agarose-50% and was then incubated overnight with the antibody of interest. Antibodies used were: 5 μl of anti-H3K27m3 (07–449; Upstate Biotechnology); 10 μl of anti-H4K20m1 (07–440; Upstate Biotechnology); 5 μl of anti-H3K9m3 (07–442; Upstate Biotechnology); 5 μl of anti-H3K4m2 (07–030; Upstate Biotechnology); 5 μl of anti-H3K9m2 (07–212; Upstate Biotechnology); 5 μl of anti-H3K4m3 (8580; Abcam, Cambridge, MA); 10 μl of anti-H4 Acetyl (06–866; Upstate Biotechnology); and 10 μl of anti-HP1gamma (05–690; Upstate Biotechnology). The antibody/histone complexes were then collected by using salmon sperm DNA/protein A Agarose, and, after the cross-links were reversed and DNA purified, quantitative PCR was used to determine the level of DNA bound (expressed as a fraction of the input DNA).
Bisulphite Modification and Sequencing.
Five-hundred nanograms of purified, EcoRV-digested DNA from HT1080 cells containing the full XIST cDNA construct without DOX induction and after 36 days of DOX induction was bisulphite treated by using the EZ DNA Methylation-Gold kit (D5005; ZYMO Research, Orange, CA). One microliter of modified DNA was amplified for 35 cycles with CMVbs-d (5′-TTTGYGTTATTTTTTGATTTTGTGGATAAT-3′) and CMVbs-rcorr (5′-CCAACTCRAAAATAAACACCACCCCRA-3′) to generate a 784-bp product that is specific for the EGFP gene. One microliter of the PCR product was used in a 30-cycle nested reaction with CMVbs-d and CMVbs-rC (5′-CAATAAAACRAAATTATTACRACATTTTAA-3′). PCR product was isolated by using the gel purification kit (Qiagen, Valencia, CA), and the insert was cloned by using the pGEM-T Easy cloning system with JM109 cells, followed by sequencing.
Acknowledgments
We thank Laura Carrel (Pennsylvania State University, University Park, PA) for the HT1080 HH1 cell line; Ziny Yen for bisulphite sequence analysis; and Meg Bryon for in situ analysis. This work was supported by Canadian Institute of Health Research (CIHR) operating Grant MOP-13690 (to C.J.B.).
Abbreviations
- H3K27m3
trimethylation of lysine 27 of histone H3
- H3K4m2
dimethylation of lysine 4 of histone H3
- H4K20m1
monomethylation of lysine 20 of histone H4
- H3K9m2
dimethylation of lysine 9 of histone H3
- FRT
Flp recombination target
- DOX
doxycycline.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Heard E. Curr Opin Genet Dev. 2005;15:482–489. doi: 10.1016/j.gde.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 2.Migeon BR. Cytogenet Genome Res. 2002;99:8–16. doi: 10.1159/000071568. [DOI] [PubMed] [Google Scholar]
- 3.Migeon BR. Nat Genet. 2003;33:337. doi: 10.1038/ng0303-337a. [DOI] [PubMed] [Google Scholar]
- 4.Chadwick BP, Willard HF. Sem Cell Dev Biol. 2003;14:359–367. doi: 10.1016/j.semcdb.2003.09.016. [DOI] [PubMed] [Google Scholar]
- 5.Wutz A, Jaenisch R. Mol Cell. 2000;5:695–705. doi: 10.1016/s1097-2765(00)80248-8. [DOI] [PubMed] [Google Scholar]
- 6.Hall LL, Byron M, Sakai K, Carrel L, Willard HF, Lawrence JB. Proc Natl Acad Sci USA. 2002;99:8677–8682. doi: 10.1073/pnas.132468999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee JT, Jaenisch R. Nature. 1997;386:275–279. doi: 10.1038/386275a0. [DOI] [PubMed] [Google Scholar]
- 8.Brown CJ, Willard HF. Nature. 1994;368:154–156. doi: 10.1038/368154a0. [DOI] [PubMed] [Google Scholar]
- 9.Chow JC, Hall LL, Clemson CM, Lawrence JB, Brown CJ. Genomics. 2003;82:309–322. doi: 10.1016/s0888-7543(03)00170-8. [DOI] [PubMed] [Google Scholar]
- 10.Wutz A, Rasmussen TP, Jaenisch R. Nat Genet. 2002;30:167–174. doi: 10.1038/ng820. [DOI] [PubMed] [Google Scholar]
- 11.Chureau C, Prissette M, Bourdet A, Barbe V, Cattolico L, Jones L, Eggen A, Avner P, Duret L. Genome Res. 2002;12:894–908. doi: 10.1101/gr.152902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nesterova TB, Slobodyanyuk SY, Elisaphenko EA, Shevchenko AI, Johnston C, Pavlova ME, Rogozin IB, Kolesnikov NN, Brockdorff N, Zakian SM. Genome Res. 2001;11:833–849. doi: 10.1101/gr.174901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beletskii A, Hong Y-K, Pehrson J, Egholm M, Strauss WM. Proc Natl Acad Sci USA. 2001;98:9215–9220. doi: 10.1073/pnas.161173098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dhara SK, Benvenisty N. Nucleic Acids Res. 2004;32:3995–4002. doi: 10.1093/nar/gkh746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoffman LM, Hall L, Batten JL, Young H, Pardasani D, Baetge EE, Lawrence J, Carpenter MK. Stem Cells. 2005;23:1468–1478. doi: 10.1634/stemcells.2004-0371. [DOI] [PubMed] [Google Scholar]
- 16.Bibikova M, Chudin E, Wu B, Zhou L, Garcia EW, Liu Y, Shin S, Plaia TW, Auerbach JM, Arking DE, et al. Genome Res. 2006;16:1075–1083. doi: 10.1101/gr.5319906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huynh KD, Lee JT. Nature. 2003;426:857–862. doi: 10.1038/nature02222. [DOI] [PubMed] [Google Scholar]
- 18.Turner JM, Mahadevaiah SK, Fernandez-Capetillo O, Nussenzweig A, Xu X, Deng CX, Burgoyne PS. Nat Genet. 2005;37:41–47. doi: 10.1038/ng1484. [DOI] [PubMed] [Google Scholar]
- 19.Clemson CM, Hall LL, Byron M, McNeil J, Lawrence JB. Proc Natl Acad Sci USA. 2006;103:7688–7693. doi: 10.1073/pnas.0601069103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chaumeil J, Le Baccon P, Wutz A, Heard E. Genes Dev. 2006;20:2223–2237. doi: 10.1101/gad.380906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.White WM, Willard HF, Van Dyke DL, Wolff DJ. Am J Hum Genet. 1998;63:20–28. doi: 10.1086/301922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hall LL, Clemson CM, Byron M, Wydner K, Lawrence JB. Hum Mol Genet. 2002;11:3157–3165. doi: 10.1093/hmg/11.25.3157. [DOI] [PubMed] [Google Scholar]
- 23.Sharp AJ, Spotswood HT, Robinson DO, Turner BM, Jacobs PA. Hum Mol Genet. 2002;11:3145–3156. doi: 10.1093/hmg/11.25.3145. [DOI] [PubMed] [Google Scholar]
- 24.Smith KP, Byron M, Clemson CM, Lawrence JB. Chromosoma. 2004;113:324–335. doi: 10.1007/s00412-004-0325-1. [DOI] [PubMed] [Google Scholar]
- 25.Savarese F, Flahndorfer K, Jaenisch R, Busslinger M, Wutz A. Mol Cell Biol. 2006;26:7167–7177. doi: 10.1128/MCB.00810-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kunath T, Arnaud D, Uy GD, Okamoto I, Chureau C, Yamanaka Y, Heard E, Gardner RL, Avner P, Rossant J. Development (Cambridge, UK) 2005;132:1649–1661. doi: 10.1242/dev.01715. [DOI] [PubMed] [Google Scholar]
- 27.Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 28.Singh PB, Georgatos SD. J Struct Biol. 2002;140:10–16. doi: 10.1016/s1047-8477(02)00536-1. [DOI] [PubMed] [Google Scholar]
- 29.Polioudaki H, Kourmouli N, Drosou V, Bakou A, Theodoropoulos PA, Singh PB, Giannakouros T, Georgatos SD. EMBO Rep. 2001;2:920–925. doi: 10.1093/embo-reports/kve199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laner T, Schulz WA, Engers R, Muller M, Florl AR. Oncol Res. 2005;15:257–264. doi: 10.3727/096504005776404607. [DOI] [PubMed] [Google Scholar]