Abstract
Recent observations indicate that, in several types of human cancer, only a phenotypic subset of cancer cells within each tumor is capable of initiating tumor growth. This functional subset of cancer cells is operationally defined as the “cancer stem cell” (CSC) subset. Here we developed a CSC model for the study of human colorectal cancer (CRC). Solid CRC tissues, either primary tissues collected from surgical specimens or xenografts established in nonobese diabetic/severe combined immunodeficient (NOD/SCID) mice, were disaggregated into single-cell suspensions and analyzed by flow cytometry. Surface markers that displayed intratumor heterogeneous expression among epithelial cancer cells were selected for cell sorting and tumorigenicity experiments. Individual phenotypic cancer cell subsets were purified, and their tumor-initiating properties were investigated by injection in NOD/SCID mice. Our observations indicate that, in six of six human CRC tested, the ability to engraft in vivo in immunodeficient mice was restricted to a minority subpopulation of epithelial cell adhesion molecule (EpCAM)high/CD44+ epithelial cells. Tumors originated from EpCAMhigh/CD44+ cells maintained a differentiated phenotype and reproduced the full morphologic and phenotypic heterogeneity of their parental lesions. Analysis of the surface molecule repertoire of EpCAMhigh/CD44+ cells led to the identification of CD166 as an additional differentially expressed marker, useful for CSC isolation in three of three CRC tested. These results validate the stem cell working model in human CRC and provide a highly robust surface marker profile for CRC stem cell isolation.
Keywords: CD44, CD166/ALCAM, tumor differentiation, tumor heterogeneity
A growing body of evidence is increasingly lending support to the idea that human cancer can be considered as a stem cell disease (1–3). According to the “cancer stem cell” (CSC) theory, tumors are not to be viewed as simple monoclonal expansions of transformed cells, but rather as complex tissues where abnormal growth is driven by a minority, pathological CSC pool that, on the one hand, has acquired tumor-related features such as uncontrolled growth and the ability to form metastases and, on the other hand, maintains its inherent capacity to self-renew and differentiate into a phenotypically heterogeneous, although aberrant, progeny. This hypothesis is supported by three key experimental observations initially performed on human acute myeloid leukemia (4) and subsequently extended to human solid tumors (5, 6): (i) Only a minority of cancer cells within each tumor is endowed with tumorigenic potential when transplanted into immunodeficient mice, (ii) tumorigenic cancer cells are characterized by a distinctive profile of surface markers and can be differentially and reproducibly isolated from nontumorigenic ones by flow cytometry, and (iii) tumors grown from tumorigenic cells contain mixed populations of both tumorigenic and nontumorigenic cancer cells, thus re-creating the full phenotypic heterogeneity of the parent tumor.
Currently, cancer cell subpopulations selectively endowed with tumorigenic potential are operationally defined as CSCs and have been prospectively identified from selected types of human solid cancer, such as breast (5), brain (6, 7), colon (8, 9), head and neck (10), and pancreatic cancer (11). However, the CSC working model is still being subjected to intense debate (12), and data published on colorectal cancer (CRC) indicate that a subgroup of primary CRC is likely to be negative for the marker currently used for isolation of colorectal CSCs (Co-CSCs) (9). In the present study, we developed an alternative, very robust protocol for the isolation of human Co-CSCs.
Results
Surface Marker Expression Profile of Primary Normal and Malignant Colonic Epithelia.
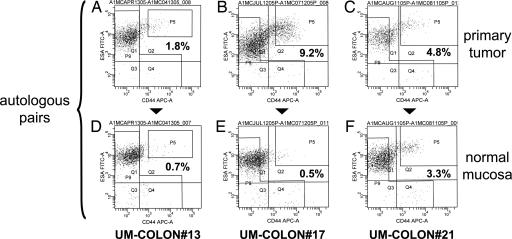
First, we decided to study the in vivo surface marker expression profile of human CRC cells, starting from a panel of fresh primary tumor tissues directly harvested from human patients undergoing surgery [supporting information (SI) Table 2]. In parallel, as a normal control, we also analyzed healthy autologous epithelium removed alongside the tumors as routinely performed during surgical colectomies. We focused our first screening on the expression of two markers that were previously described as useful in the isolation of human breast CSC: CD44 and the epithelial cell adhesion molecule (EpCAM), also known as epithelial-specific antigen (ESA) (5). Primary tissues, including both normal and malignant specimens, were disaggregated into single-cell suspensions and analyzed by flow cytometry (Fig. 1). Based on the above two markers, we were able to discriminate two main populations of epithelial cells: EpCAMhigh/CD44+ and EpCAMlow/CD44−. Both primary CRC tumors and normal colonic epithelium contained an identical profile of cell populations, although several tumors appeared enriched in the percentage of EpCAMhigh/CD44+ cells (Fig. 1). Overall, in normal colorectal mucosa (n = 15), the frequency of EpCAMhigh/CD44+ cells ranged from 0.15% to 5% (mean = 1.6%) of total live tissue cells (DAPI−) and from 0.16% to 10% (mean = 2.6%) of total live epithelial cells (DAPI−, Lin−). In primary CRC tumors (n = 12), the frequency of EpCAMhigh/CD44+ cells ranged from 0.03% to 38% (mean = 5.4%) of total live tumor cells (DAPI−) and from 0.20% to 58% (mean = 11.4%) of total live epithelial cells (DAPI−, Lin−).
Fig. 1.
EpCAM/CD44 expression profiles in primary CRC tumors and normal colonic tissues. Analysis of EpCAM/CD44 expression in primary tissues revealed similar profiles among primary CRC tumors (A–C) and normal colorectal epithelium (D–F). Both normal and malignant tissues contained two main cell subsets: EpCAMhigh/CD44+ and EpCAMlow/CD44−. To minimize experimental variability and contributions of genetic background, primary tumors were compared with autologous normal mucosa and analyzed on the same day. EpCAM expression was analyzed by using the B29.1 anti-ESA monoclonal antibody (Biomeda, Foster City, CA). Percentages reported in flow plots indicate the percentage of cells contained within gate P5.
Differential Tumorigenicity of CRC Phenotypic Subpopulations.
Next, we decided to evaluate whether the previous two phenotypic populations (EpCAMhigh/CD44+ and EpCAMlow/CD44−) were endowed with differential tumorigenic properties. To ensure consistency and reproducibility in the setup of experimental procedures, we decided to address this question first in a panel of human CRC xenograft lines, newly established in our laboratories from fresh primary tumor samples (SI Table 2). All xenograft lines were originally implanted and subsequently passaged as solid tumors and were never cultivated or expanded in vitro. On histological examination, tumor xenografts grew as adenocarcinomas, formed gland-like structures, and scored positive for expression of colon-specific differentiation markers such as cytokeratin-20 (CK20) and neutral epithelial mucins (SI Table 2 and SI Fig. 6). Remarkably, on flow-cytometry analysis, the EpCAM/CD44 expression profile of CRC xenografts mirrored that of primary tumors (Fig. 2), with the highest degree of similarity being observed between each xenograft and the individual autologous primary tumor from which it was originated (SI Fig. 7). Most interestingly, the relative proportion of the two populations varied among different xenograft lines, but was robustly conserved within each line on successive in vivo transplantation (SI Fig. 8). Thus, the relative proportion of the two populations was a unique feature of each xenograft. Overall, in CRC xenografts (n = 8), the frequency of EpCAMhigh/CD44+ cells ranged from 0.8% to 38% (mean = 15.2%) of total live epithelial cancer cells (DAPI−, Lin−).
Fig. 2.
The EpCAM/CD44 expression profile of human CRC xenografts recapitulates that observed in primary CRC tumors. (A–D) Analysis of EpCAM/CD44 expression in human CRC xenografts grown in NOD/SCID mice confirmed the existence of two main cancer cell subsets: EpCAMhigh/CD44+ and EpCAMlow/CD44−. Relative frequencies of the two subpopulations varied among different xenografts. EpCAM expression was analyzed by using the B29.1 anti-ESA monoclonal antibody (Biomeda). Percentages reported in flow plots indicate the percentage of cells contained within gate P5.
In several independent experiments performed on six distinct xenograft lines, the two populations were purified by FACS and injected back into nonobese diabetic/severe combined immunodeficient (NOD/SCID) mice (SI Fig. 9). The results showed a substantial difference in tumorigenic properties. Tumors frequently arose on injection of 200 to 500 EpCAMhigh/CD44+ cells, whereas 104 EpCAMlow/CD44− cells consistently failed to form tumors (Table 1 and SI Table 3). Most important, tumors grown from EpCAMhigh/CD44+ cells maintained a differentiated phenotype and reproduced the morphologic and phenotypic heterogeneity of their parent lesions, including formation of epithelial gland-like structures, production of neutral epithelial mucins, and heterogeneous expression of differentiation markers such as CK20 (SI Fig. 6). Moreover, when analyzed by flow cytometry, they contained both EpCAMhigh/CD44+ and EpCAMlow/CD44− populations in proportions similar to those of their parent lesions (Fig. 3). Taken together, these observations suggested that in human CRC xenografts a population with stem-like properties can be reproducibly and consistently isolated based on EpCAM and CD44.
Table 1.
Comparison of tumorigenic cell doses among distinct CRC phenotypic subpopulations
| Lin− phenotypic subpopulations* | Cell doses and tumor formation† |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 10,000 | 8,000 | 6,000 | 5,000 | 4,000 | 2,000 | 1,000 | 800 | 400 | 200 | 150 | |
| CD44+ | 2/5‡ | — | — | — | — | — | — | — | — | — | — |
| CD44− | 0/5‡ | — | — | — | — | — | — | — | — | — | — |
| ESAhigh/CD44+ | — | — | — | 4/5 | — | 17/19¶ | 13/15 | 2/5 | 16/20 | 21/28 | — |
| All the rest (mainly CD44−/ESAlow) | 0/3§ | 0/5 | 0/5 | — | 0/5 | 1/30 | 0/19 | — | 0/10 | 0/20 | — |
| CD44+/CD166+ | — | — | — | — | — | — | 6/10 | — | — | — | — |
| CD44−/CD166+ | — | — | — | — | — | — | 1/10 | — | — | — | — |
| CD44+/CD166− | — | — | — | — | — | — | 0/7 | — | — | — | — |
| CD44−/CD166− | 0/10 | — | — | — | — | — | — | — | — | — | — |
| ESAhigh/CD166+ | — | — | — | — | 3/5‖ | — | — | — | — | — | — |
| All the rest (mainly ESAlow/CD166−) | — | — | — | — | 0/5‖ | — | — | — | — | — | — |
| ESAhigh/CD44+/CD166+ | — | — | — | — | — | — | — | — | — | — | 1/2‖ |
| ESAhigh/CD44+/CD166− | — | — | — | — | — | — | — | — | — | — | 0/2‖ |
| All the rest (mainly ESAlow/CD44−) | — | — | — | — | — | — | — | — | 0/2‖ | — | — |
*All populations are negative for nonepithelial lineage marker expression (Lin−).
†Cell dose, number of cells per injection; tumor formation, number of tumors formed/number of injections; tumor take was considered unsuccessful when no tumor mass was visible after 5 months follow-up. See also SI Tables 2 and 3.
‡Cell dose was 14,000.
§Cell dose was 12.000.
¶Five injections were performed with 2,500 cells and 14 with 2,000 cells.
‖Cells from primary tumor.
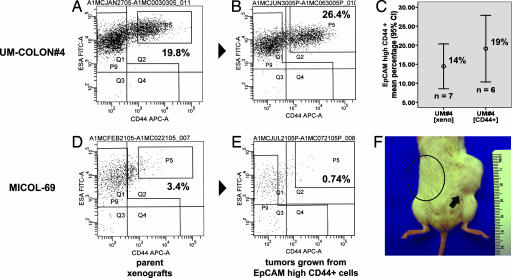
Fig. 3.
Reconstitution of parental EpCAM/CD44 expression profiles in tumors grown from sorted EpCAMhigh/CD44+ cells. Analysis of tumors grown from injection of sorted EpCAMhigh/CD44+ cells (B and E) showed reconstitution of parental expression profiles (A and D), including similar relative frequencies of EpCAMhigh/CD44+ and EpCAMlow/CD44− populations (UM-COLON#4; C). The capacity to form tumors in NOD/SCID mice was restricted to the EpCAMhigh/CD44+ cell population (MICOL-69; F, arrow). No tumor growth was usually observed on injection of EpCAMlow/CD44− cells on the opposite flank of the same animals (circled area). EpCAM expression was analyzed by using the B29.1 anti-ESA monoclonal antibody (Biomeda). Percentages reported in flow plots indicate the percentage of cells contained within gate P5.
Characterization of the Co-CSC Surface Marker Repertoire.
To better characterize the Co-CSC surface marker repertoire and evaluate whether Co-CSC could be further enriched by subfractionation of the EpCAMhigh/CD44+ population, we started a systematic evaluation of markers already described as differentially expressed in other stem cell models, such as CD49f, CD133, and the aldehyde dehydrogenase (ALDH) enzyme (6, 8, 9, 13, 14). Analysis of CD49f expression revealed a consistent and reproducible pattern, similar to that of EpCAM: CD49f was detected on most tumor cells, with higher levels on the CD44+ subpopulation (SI Fig. 10). Expression of CD133 displayed a more variable pattern, with some tumors scoring as homogeneously negative, some as predominantly positive, and others as a mixture of positive and negative cells (SI Fig. 11). A careful study of CD133− tumors, such as UM-COLON#4, revealed that CD133 expression was absent in the primary tumor and remained absent on serial in vivo transplantation in NOD/SCID mice (SI Fig. 12). Combined analysis of CD44 and CD133 expression indicated that when CD133 was expressed CD44+ cells were usually CD133+. In most cases, however, the CD133+ population was larger than the CD44+ one, and CD44+ cells represented a minority subset of the CD133+ population, as clearly visible in tumors that scored as predominantly positive for CD133 (SI Fig. 11). Analysis of ALDH enzymatic activity indicated that EpCAMhigh/CD44+ cells were characterized by higher ALDH levels. ALDH activity, however, was useful for isolation of tumorigenic cells in some, but not all, CRC xenografts (SI Fig. 13).
Identification of CD166 as a Candidate Co-CSC Marker.
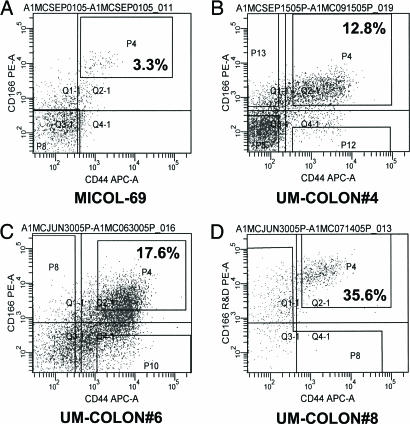
We then decided to focus our investigation on surface markers whose expression had been previously evaluated by immunohistochemistry on primary tumors and was described to be heterogeneous in CRC cells. During this second screening, again performed by flow cytometry, our attention was captured by the CD166 molecule, a mesenchymal stem cell marker that displayed heterogeneous expression patterns in CRC epithelial cells (15, 16) and whose increased expression levels were previously associated with poor clinical outcome in CRC patients (16). We observed that the CD166 molecule was differentially expressed within epithelial cancer cells and that all analyzed tumors contained a distinct EpCAMhigh/CD44+/CD166+ cell subset (Figs. 4 and 5). The differential expression of CD166 was consistently and reproducibly observed in all xenografts, in primary tumors, and in normal colonic epithelium (Figs. 4 and 5 and SI Fig. 14). Overall, in normal colorectal mucosa (n = 7), the frequency of EpCAMhigh/CD166+ cells ranged from 0.5% to 7% (mean = 2.8%) of total live tissue cells (DAPI−) and from 0.5% to 10% (mean = 3.7%) of total live epithelial cells (DAPI−, Lin−). In primary CRC tumors (n = 6), the frequency of EpCAMhigh/CD166+ cells ranged from 1.2% to 16% (mean = 6%) of total live tumor cells (DAPI−) and from 1.5% to 24% (mean = 10%) of total live epithelial cells (DAPI−, Lin−). To evaluate the role of CD166 as a Co-CSC marker, we performed a second round of tumorigenicity experiments on the UM-COLON#4 xenograft line, sorting cancer cells based on the expression of CD44 and CD166 (Fig. 4B). The results showed that, in this tumor, tumorigenic cells are restricted to the CD44+/CD166+ population, indicating that CD166 can be used as an additional Co-CSC marker independent and synergistic with respect to CD44 (Table 1 and SI Table 4).
Fig. 4.
Coexpression of CD44 and CD166 in human CRC xenografts. (A–D) Analysis of CD166 expression in human CRC xenografts revealed that CD166 was differentially expressed within cancer cell populations and that all analyzed tumors contained a distinct CD44+/CD166+ double-positive cell subset.
Fig. 5.
CD44/CD166 and EpCAM/CD166 expression profiles in normal and malignant primary colorectal tissues. Analysis of CD44/CD166 expression profiles in primary tissues confirmed CD44/CD166 coexpression in normal colorectal epithelium (E and G) and primary CRC tumors (C), with a profile similar to that of CRC xenografts (A). As expected, CD166+ cells were predominantly EpCAMhigh (B, D, F, and H). EpCAM expression was analyzed by using the B29.1 anti-ESA monoclonal antibody (Biomeda). Percentages reported in flow plots indicate the percentage of cells contained within gate P9.
Finally, to prove that the EpCAMhigh/CD44+/CD166+ population represented the original reservoir of Co-CSC in vivo, in human patients, and was not enriched in Co-CSC as a consequence of xenografting (e.g., because of modulation of surface marker expression in the mouse s.c. environment), we sorted and injected Co-CSC directly from primary CRC tissues based on expression of EpCAM, CD44, and CD166. Tumorigenicity experiments from primary surgical specimens proved more difficult to standardize because of smaller size and lower cell yields as compared with xenografts. However, we were able to show first that tumorigenic cells are restricted to the EpCAMhigh/CD166+ population and subsequently that their phenotype can be further specified as EpCAMhigh/CD44+/CD166+ using two independent primary tumors (Table 1 and SI Table 4). The reproducibility of these findings across a larger series of cases is currently under investigation.
Discussion
Taken together, our findings extend to CRC the stem cell working model of human neoplasia and provide a robust and reproducible surface marker profile for Co-CSC isolation. Our results suggest that, in several colon tumors, EpCAM and CD44 were more robust as markers of Co-CSC than the recently reported marker CD133 (8, 9) because CD44 appeared to be informative in tumors that do not express CD133 and also allowed for further enrichment of Co-CSC within the CD133+ subset in others. Furthermore, our data indicate that, in several CRC tumors, including both xenografts and primary tumors, CD166 could be used for further enrichment of Co-CSC within the EpCAMhigh/CD44+ population.
Our findings have many potential biological and therapeutic implications (1–3, 17, 18). From a biological point of view, the CSC model introduces an additional conceptual frame and a layer of complexity for the interpretation of intratumor heterogeneity (i.e., heterogeneity among cancer cells within the same tumor lesion), a common feature of both primary and metastatic CRC (19). According to the CSC model, intratumor heterogeneity is not only the result of the coexistence within the same mass of multiple independent tumor subclones originated by the accumulation of divergent genetic mutations, but also of the intermingling of cells that, although genetically monoclonal in origin, differ in their functional state of differentiation. Indeed, one of the markers exploited in this study for Co-CSC isolation (CD44) is a well established, immature differentiation marker in human colonic mucosa (20).
Based on this perspective, the CSC model might help explain unexpected observations in CRC biology, such as the presence of heterogeneous patterns and nonconstitutive mechanisms of telomerase activation within both primary and metastatic CRC lesions (21). Moreover, the CSC model could shed light on the biology of metastases and explain why, despite extensive intratumor heterogeneity, comparison of paired samples of primary tumors and autologous metastases from the same patient frequently reveals high levels of similarity (19, 22–24). This observation is very well established in CRC and spans across diverse parameters, such as tissue morphology (19, 25), repertoire of somatic genetic mutations (26, 27), expression of tumor-suppressor and immunomodulatory proteins (28), and overall transcriptional profile (29). Indeed, if we assume that, in each individual CRC, the differentiation pattern is controlled by the specific repertoire of genetic mutations, we can predict that if two lesions share identical genetic backgrounds and similar genetic abnormalities they will also undergo similar differentiation programs and display similar patterns of intratumor heterogeneity in the expression of differentiation markers. This prediction is in accord with our observation that each CRC xenograft line is individually characterized by a unique and constant ratio of EpCAMhigh/CD44+ versus EpCAMlow/CD44− cell populations, which is maintained in time on serial transplantation.
Finally, the observation that CRC growth is sustained by a minority subpopulation of tumor cells with unique functional properties could assist in the design of new and more effective prognostic tools (30) and antitumor treatments (31). According to the CSC model, therapeutic approaches that are not capable of eradicating the CSC subset are unlikely to be successful because they might be able to kill the majority of tumor cells and induce regression of tumor lesions, but fail to prevent disease relapse and metastatic dissemination (1, 2). Based on this concept, traditional treatments might be reassessed and investigational new therapies developed, focusing on the ability to target CSC and their specific biochemical pathways (2, 3). Similarly, the CSC model might explain why many experimental therapeutic approaches have shown poor clinical results despite extensive preclinical validation in vitro and might provide useful information for their redesign and upgrading, as in the case of immunotherapy against CRC, where target antigen selection might be reevaluated based on expression by Co-CSC (32).
Materials and Methods
Tumor Tissues and Xenograft Lines.
Human CRC tissues used in this study are listed in SI Table 2, together with clinical information related to corresponding patients. All primary tissues were collected under protocols approved by the University of Michigan and Stanford University institutional review boards between 2004 and 2006. Informed consent was obtained from all patients included in the study. All CRC xenograft lines were established by s.c. implantation of solid tissue fragments in 6- to 8-week-old NOD/SCID mice (Charles River Laboratories, Wilmington, MA), with the only exception of MICOL-69, which was established in CD1 “nude” mice (Charles River Laboratories). Briefly, primary CRC tissue specimens were minced with scissors into small (2-mm3) fragments and implanted s.c. by using a 10-gauge Trochar needle through a small incision on the animal's right dorsal flank. Recipient NOD/SCID mice were anesthetized by i.p. injection of a ketamine (75 mg/kg)–xylazine (5 mg/kg) mixture. Once established, solid tumor xenografts were serially passaged by using the same technique. Of the 10 xenograft lines used in this study, one (MICOL-69) was originated at the National Cancer Institute in Milan, Italy, five (UM-COLON#4, #6, #8, #13, #21) were originated at University of Michigan, two (SU-COLON#29, #43) were originated at Stanford University, and two (OMP-C5, C8) were originated at Oncomed Pharmaceuticals, Inc. Xenograft lines established at University of Michigan and Stanford University are part of a collection of 14 independent lines originated from 19 distinct primary CRC specimens, for an estimated engraftment rate of 73% (n = 14 of 19). Histochemical and immunohistochemical analyses of tumor tissues were performed as described in SI Materials and Methods.
Solid Tissue Disaggregation.
Solid tissues, both normal and neoplastic, collected from primary surgical specimens or mouse xenografts were mechanically and enzymatically disaggregated into single-cell suspensions and analyzed by flow cytometry, as described by Al-Hajj et al. (5). Solid tissues were minced with scissors into small (2-mm3) fragments and incubated for 15 min at room temperature in 100 mM phosphate buffer (pH 7.0) with 6.5 mM DTT (Sputolysin Reagent; Calbiochem, La Jolla, CA) to remove mucus contamination. After gentle removal of the DTT solution, tissue fragments were rinsed once with Hank's balanced salt solution (HBSS), resuspended in serum-free RPMI medium 1640 (2 mmol/liter l-glutamine, 120 μg/ml penicillin, 100 μg/ml streptomycin, 50 μg/ml ceftazidime, 0.25 μg/ml amphotericin-B, 20 mmol/liter Hepes) with 200 units/ml Collagenase type III (Worthington, Lakewood, NJ) and 100 units/ml DNase I (Worthington), and incubated for 2 h at 37°C to obtain enzymatic disaggregation. Cells were then resuspended by pipetting and serially filtered by using sterile gauze and 70-μm and 40-μm nylon meshes. Contaminating red blood cells were removed by osmotic lysis (i.e., incubation in ammonium chrloride potassium phosphate hypotonic buffer for 5 min on ice).
Flow Cytometry and Cell-Sorting Experiments.
To minimize experimental variability and loss of cell viability, all experiments were performed on fresh tumor cell suspensions prepared shortly before flow cytometry. Antibody staining was performed in HBSS supplemented with 2% heat-inactivated calf serum and 20 mM Hepes. To minimize unspecific binding of antibodies, cells were first incubated with 0.6% human immunoglobulins (Gammagard Liquid; Baxter, Westlake Village, CA) for 10 min on ice at a cell concentration of 3 to 5 × 105/100 μl. Cells were subsequently washed and stained with antibodies at dilutions determined by titration experiments on each xenograft line. Antibodies used in this study include: anti-human ESA-FITC (clone B29.1; Biomeda), anti-human CD44-APC (clone G44–26; BD Biosciences, San Diego, CA), anti-human CD166-PE (clone 105902; R&D Systems, Minneapolis, MN), anti-human CD49f-PE (clone GoH3; BD Biosciences), and anti-human CD133-PE (clones AC133, 293C3, and AC141; Miltenyi Biotec, Auburn, CA). In all experiments, cells positive for expression of nonepithelial lineage markers (Lin+) were excluded by staining with PE.Cy5-labeled antibodies using two different strategies for primary tissues and mouse xenografts. In experiments on primary human tissues, stromal cells were excluded by simultaneous staining with anti-human CD3-PE.Cy5 (clone UCHT1; BD Biosciences), CD10-PE.Cy5 (clone HI10a; BD Biosciences), CD16-PE.Cy5 (clone 3G8; BD Biosciences), CD18-PE.Cy5 (clone 6.7; BD Biosciences), CD45-PE.Cy5 (clone HI30; BD Biosciences), and CD64-PE.Cy5 (clone 10.1; DakoCytomation, Carpinteria, CA). In experiments on CRC xenografts, mouse cells were excluded by simultaneous staining with anti-mouse CD45-PE.Cy5 (clone 30-F11; BD Biosciences) and anti-mouse H-2Kd-biotin (clone SF1–1.1; BD Biosciences) + streptavidin-PE.Cy5 (BD Biosciences). After 15 min on ice, stained cells were washed of excess unbound antibodies and resuspended in HBSS supplemented with 2% heat-inactivated calf serum, 20 mM Hepes, and 1.1 μM DAPI dilactate (Molecular Probes, Eugene, OR) as viability dye. Analysis of ALDH enzymatic activity was performed by using the Aldefluor system (StemCell Technologies Inc., Vancouver, BC, Canada) following the manufacturer's instructions. Flow-cytometry analysis was performed by using a BD FACSAria cell sorter (Becton Dickinson, San Jose, CA). Forward scatter area versus forward scatter width profiles were used to eliminate cell doublets. Dead cells were eliminated by excluding DAPI+ cells, whereas contaminating human or mouse Lin+ cells were eliminated by excluding PE.Cy5+ cells. In cell-sorting experiments, each cell population underwent two consecutive rounds of purification (double sorting), achieving a final average purity of >95% (SI Fig. 9).
Tumorigenicity Experiments.
Sorted cells were spun down by low-speed centrifugation (850 × g for 5 min) and resuspended in RPMI 1640 supplemented with 10% FBS, 20 mM Hepes, and 2 mM l-glutamine. In all experiments, a small aliquot of cells was set aside to confirm cell counts and viability using conventional techniques (i.e., trypan blue exclusion test). Once cell counts and viability were confirmed, cells were diluted to appropriate injection doses, mixed with BD Matrigel (BD Biosciences) at 1:1 ratio, and injected s.c. in NOD/SCID mice on the ventral side of each flank (SI Fig. 9). To minimize experimental variability due to individual differences in recipient mice, cell populations subjected to comparison were injected on opposite flanks of the same animals (SI Fig. 9). Injected mice were followed for up to 5 months and killed when tumors reached a maximum diameter of 15 mm. Statistical analysis of tumorigenicity experiments was performed as described in SI Materials and Methods. All experiments involving the use of animals were performed in accordance with University of Michigan and Stanford University institutional animal welfare guidelines.
Supplementary Material
Acknowledgments
We thank David J. Adams for extraordinary technical support with flow cytometry and cell-sorting experiments, Dr. Michael W. Becker for precious comments, Nancy McAnsh for help with immunohistochemistry, and Donna Renner-Chuey for help with primary tissue collection. This work was supported by National Institutes of Health Grants CA104987 and CA126524 (to M.F.C.), the Virginia and D. K. Ludwig Foundation (M.F.C.), the Fondazione Italiana per la Ricerca sul Cancro (P.D.), and the California Institute of Regenerative Medicine (P.D.).
Abbreviations
- ALDH
aldehyde dehydrogenase
- CK20
cytokeratin-20
- Co-CSC
colorectal cancer stem cell
- CRC
colorectal cancer
- CSC
cancer stem cell
- EpCAM
epithelial cell adhesion molecule
- ESA
epithelial-specific antigen
- NOD/SCID
nonobese diabetic/severe combined immunodeficient.
Footnotes
Conflict of interest statement: S.J.D., I.-K.P., X.W., T.H., and A.G. are employees of Oncomed Pharmaceuticals, Inc., a biotechnology company that has applied for patents related to this study. M.F.C. is a member of the paid advisory board of Oncomed Pharmaceuticals, Inc., and owns stock options in the company. P.D. and M.F.C are listed as coinventors on patents related to this study.
This article contains supporting information online at www.pnas.org/cgi/content/full/0703478104/DC1.
References
- 1.Reya T, Morrison SJ, Clarke MF, Weissman IL. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 2.Dalerba P, Cho RW, Clarke MF. Annu Rev Med. 2007;58:267–284. doi: 10.1146/annurev.med.58.062105.204854. [DOI] [PubMed] [Google Scholar]
- 3.Jordan CT, Guzman ML, Noble M. N Engl J Med. 2006;355:1253–1261. doi: 10.1056/NEJMra061808. [DOI] [PubMed] [Google Scholar]
- 4.Bonnet D, Dick J. Nature Medicine. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 5.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Proc Natl Acad Sci USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 7.Galli R, Binda E, Orfanelli U, Cipelletti B, Gritti A, De Vitis S, Fiocco R, Foroni C, Dimeco F, Vescovi A. Cancer Res. 2004;64:7011–7021. doi: 10.1158/0008-5472.CAN-04-1364. [DOI] [PubMed] [Google Scholar]
- 8.O'Brien CA, Pollett A, Gallinger S, Dick JE. Nature. 2007;445:106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 9.Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, De Maria R. Nature. 2007;445:111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 10.Prince ME, Sivanandan R, Kaczorowski A, Wolf GT, Kaplan MJ, Dalerba P, Weissman IL, Clarke MF, Ailles LE. Proc Natl Acad Sci USA. 2007;104:973–978. doi: 10.1073/pnas.0610117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, Adsay V, Wicha M, Clarke MF, Simeone DM. Cancer Res. 2007;67:1030–1037. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- 12.Hill RP. Cancer Res. 2006;66:1891–1895. doi: 10.1158/0008-5472.CAN-05-3450. [DOI] [PubMed] [Google Scholar]
- 13.Shackleton M, Vaillant F, Simpson KJ, Stingl J, Smyth GK, Asselin-Labat ML, Wu L, Lindeman GJ, Visvader JE. Nature. 2006;439:84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- 14.Storms RW, Trujillo AP, Springer JB, Shah L, Colvin OM, Ludeman SM, Smith C. Proc Natl Acad Sci USA. 1999;96:9118–9123. doi: 10.1073/pnas.96.16.9118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bruder SP, Ricalton NS, Boynton RE, Connolly TJ, Jaiswal N, Zaia J, Barry FP. J Bone Miner Res. 1998;13:655–663. doi: 10.1359/jbmr.1998.13.4.655. [DOI] [PubMed] [Google Scholar]
- 16.Weichert W, Knosel T, Bellach J, Dietel M, Kristiansen G. J Clin Pathol. 2004;57:1160–1164. doi: 10.1136/jcp.2004.016238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brittan M, Wright NA. Gut. 2004;53:899–910. doi: 10.1136/gut.2003.025478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Radtke F, Clevers H. Science. 2005;307:1904–1909. doi: 10.1126/science.1104815. [DOI] [PubMed] [Google Scholar]
- 19.Brabletz T, Jung A, Spaderna S, Hlubek F, Kirchner T. Nat Rev Cancer. 2005;5:744–749. doi: 10.1038/nrc1694. [DOI] [PubMed] [Google Scholar]
- 20.Wielenga VJ, Smits R, Korinek V, Smit L, Kielman M, Fodde R, Clevers H, Pals ST. Am J Pathol. 1999;154:515–523. doi: 10.1016/S0002-9440(10)65297-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dalerba P, Guiducci C, Poliani PL, Cifola I, Parenza M, Frattini M, Gallino G, Carnevali I, Di Giulio I, Andreola S, et al. Cancer Res. 2005;65:2321–2329. doi: 10.1158/0008-5472.CAN-04-3678. [DOI] [PubMed] [Google Scholar]
- 22.Weigelt B, Peterse JL, van't Veer LJ. Nat Rev Cancer. 2005;5:591–602. doi: 10.1038/nrc1670. [DOI] [PubMed] [Google Scholar]
- 23.Weigelt B, Glas AM, Wessels LF, Witteveen AT, Peterse JL, van't Veer LJ. Proc Natl Acad Sci USA. 2003;100:15901–15905. doi: 10.1073/pnas.2634067100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dalerba P, Ricci A, Russo V, Rigatti D, Nicotra MR, Mottolese M, Bordignon C, Natali PG, Traversari C. Int J Cancer. 1998;77:200–204. doi: 10.1002/(sici)1097-0215(19980717)77:2<200::aid-ijc5>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 25.Brabletz T, Jung A, Reu S, Porzner M, Hlubek F, Kunz-Schughart LA, Knuechel R, Kirchner T. Proc Natl Acad Sci USA. 2001;98:10356–10361. doi: 10.1073/pnas.171610498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Losi L, Benhattar J, Costa J. Eur J Cancer. 1992;28A:1115–1120. doi: 10.1016/0959-8049(92)90468-h. [DOI] [PubMed] [Google Scholar]
- 27.Zauber P, Sabbath-Solitare M, Marotta SP, Bishop DT. Mol Pathol. 2003;56:137–140. doi: 10.1136/mp.56.3.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Menon AG, Tollenaar RA, van de Velde CJ, Putter H, Janssen-van Rhijn CM, Keijzer R, Fleuren GJ, Kuppen PJ. Clin Exp Metastasis. 2004;21:79–85. doi: 10.1023/b:clin.0000017206.08931.42. [DOI] [PubMed] [Google Scholar]
- 29.D'Arrigo A, Belluco C, Ambrosi A, Digito M, Esposito G, Bertola A, Fabris M, Nofrate V, Mammano E, Leon A, Nitti D, Lise M. Int J Cancer. 2005;115:256–262. doi: 10.1002/ijc.20883. [DOI] [PubMed] [Google Scholar]
- 30.Liu R, Wang X, Chen GY, Dalerba P, Gurney A, Hoey T, Sherlock G, Lewicki J, Shedden K, Clarke MF. N Engl J Med. 2007;356:217–226. doi: 10.1056/NEJMoa063994. [DOI] [PubMed] [Google Scholar]
- 31.Guzman ML, Swiderski CF, Howard DS, Grimes BA, Rossi RM, Szilvassy SJ, Jordan CT. Proc Natl Acad Sci USA. 2002;99:16220–16225. doi: 10.1073/pnas.252462599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dalerba P, Maccalli C, Casati C, Castelli C, Parmiani G. Crit Rev Oncol Hematol. 2003;46:33–57. doi: 10.1016/s1040-8428(02)00159-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.