Abstract
Robustness to environmental or genetic perturbation, like any other trait, is affected by evolutionary change. However, direct studies on the interplay of robustness and evolvability are limited and require experimental microevolutionary studies of developmental processes. One system in which such microevolutionary studies can be performed is vulva development in the nematode Pristionchus pacificus. Three vulval precursor cells respond to redundant cell–cell interactions, including signals from the gonad and the epidermal cell P8.p. Interestingly, P. pacificus P8.p is involved in cell fate specification of the future vulva cells by lateral inhibition but is incompetent to respond to the inductive signal from the gonad itself. These functional properties of P8.p are unknown from other nematodes, such as Caenorhabditis elegans. We began an experimental and genetic analysis of the microevolution of P8.p function. We show that vulva misspecification events differ between Pristionchus strains and species. Similarly, lateral inhibition and developmental competence of P8.p evolved within the genus Pristionchus and between natural isolates of P. pacificus. Surprisingly, in some recombinant inbred lines of two distinct P. pacificus isolates, P8.p gained competence to form vulva tissue, a trait that was never observed in P. pacificus isolates. Our results suggest differences in developmental stability between natural isolates, and we hypothesize that the remarkable evolvability of redundant cell–cell interactions allows for adaptive evolution of robustness to developmental noise.
Keywords: evolvability, developmental stability
Living organisms are remarkably stable in the face of environmental and genetic perturbation. This property, often called robustness (1) or developmental stability, reduces the amount of phenotypic variation that is visible to selection. However, organisms do evolve, and the accumulation of mutations results in changes of modular components, i.e., molecules, networks, and cells, eventually resulting in developmental and morphological novelty. Robustness itself can evolve as well. This is especially evident in the case of robustness to developmental noise: mutations that confer higher robustness in a given environment might increase in frequency by standard evolutionary mechanisms (1). For a full understanding of the evolution of robustness, research activities in evolutionary genetics and evolutionary developmental biology have to be combined. More precisely, direct studies on the interplay of robustness and evolvability, i.e., the capacity to generate heritable selectable phenotypic variation (2), require experimental microevolutionary studies of developmental processes (3).
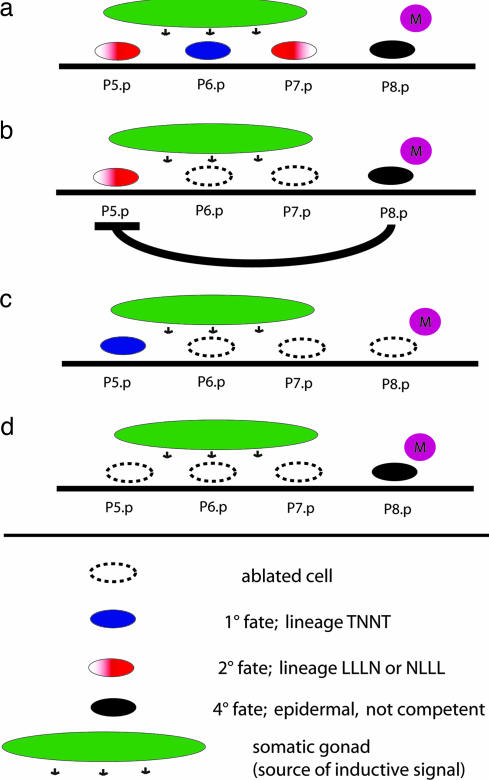
One developmental system in which the robustness of signaling systems can be studied with single-cell resolution and by genetic and molecular analyses is the nematode vulva. In the model organism Caenorhabditis elegans, various cell–cell interactions are involved in vulval cell fate specification and require activities of EGF, Wnt, and lin-12/Notch signaling pathways (4).The diplogastrid nematode Pristionchus pacificus shares many experimental advantages with C. elegans but differs in several aspects of vulva development (5). Although redundant cell–cell interactions and the activity of signaling systems are also crucial for P. pacificus vulva formation, the exact identity of cellular and molecular components differs strongly between the two species (5). In short, the nematode vulva is a derivative of the ventral epidermis, which consists of 12 blast cells (P1.p–P12.p) [supporting information (SI) Fig. 4]. In P. pacificus, four of these 12 blast cells (P5.p–P8.p) are selected to form a vulva equivalence group. Later in development, three of the four vulva precursor cells (VPCs), P(5–7).p, are induced by a signal from the gonad and adopt one of two alternative vulval fates (Fig. 1). P5.p and P7.p have the 2° fate and generate seven progeny each that build the outer part of the vulva. P6.p has the 1° fate and generates six progeny that form the inner part of the vulva. The only surviving blast cell not directly involved in vulva formation is the posterior cell P8.p (Fig. 1a). P8.p remains epidermal and is assigned a 4° fate, because it does not contribute to vulva tissue and, contrary to the homologous cell in C. elegans, is not competent to respond to inductive signal in the absence of other VPCs (6). Thus, the vulva equivalence group of P. pacificus is asymmetric in respect to the gonadal anchor cell, with a fate pattern 2°-1°-2°-4° (Fig. 1a). Interestingly, P8.p is involved in a cell–cell interaction that regulates vulva cell fate specification in a redundant manner. P8.p, together with the mesoblast M, prevents P5.p and P7.p from adopting the 1° fate in the absence of P6.p (Fig. 1b) (6). For example, if P(6,7).p are ablated, P5.p will adopt a 2° fate in the majority of cases (88%), whereas it will adopt a 1° fate if P(6,7).p are ablated together with P8.p (Fig. 1 b and c). This interaction has been designated as “lateral inhibition” (LI) and is unknown from C. elegans and other nematodes. Because of the asymmetric configuration of the Pristionchus vulva equivalence group, we refer to the P8.p effect on P7.p and P5.p as short- and long-range LI, respectively. Interestingly, ablation of P8.p alone does not alter the VPCs' lineages, indicating that LI represents a redundant signaling system that affects 2° cell fate specification of P(5,7).p, together with the inductive signal from the somatic gonad. Thus, P8.p-mediated signaling might contribute to robustness of the developmental module of the vulva.
Fig. 1.
P8.p is the source of an inhibitory signal. (a) P(5–7).p are induced to give rise to vulva tissue by the somatic gonad; P8.p remains epidermal. P5.p and P7.p have the 2° fate (seven progeny) and form the outer part of the vulva. P6.p has the 1° fate (six progeny) and forms the central part of the vulva. (b) P5.p adopts the 2° fate after ablation of P(6,7).p. (c) If P8.p is ablated together with P(6,7).p, P5.p adopts the 1° fate, indicating that P8.p is the source of an inhibitory signal. The same logic applies to ablation of P(5,6).p and P(5,6,8).p, respectively. (d) P8.p itself cannot respond to gonadal signaling, even in the absence of all other VPCs. L, N, and T refer to cell division patterns in the last round of VPC division: L, longitudinal; N, nondividing; T, transversal.
Here we study robustness and evolvability of the developmental program of Pristionchus vulva development by analyzing developmental misspecification events and microevolutionary differences in P8.p-mediated signaling. In recombinant inbred lines (RILs), we investigate the evolvability of this signaling network and observe the emergence of previously undescribed characteristics in redundant aspects of cell–cell signaling. We hypothesize that this remarkable evolvability of redundant cell–cell interactions allows for adaptive evolution of robustness to developmental noise.
Results
Pristionchus Vulva Cell Fate Specification Is Robust.
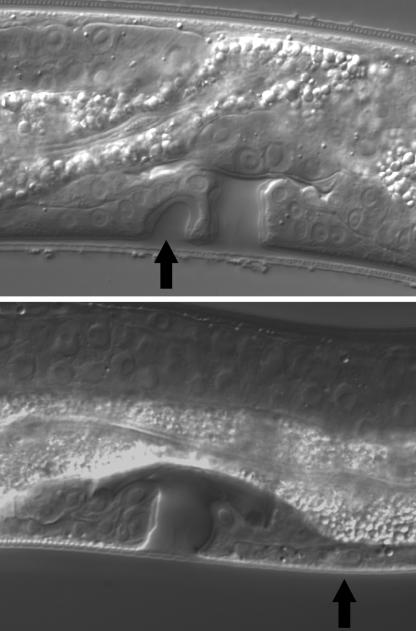
To study to what extent Pristionchus vulva development is affected by developmental noise and to investigate what kind of developmental errors can occur in different genetic backgrounds, we analyzed VPC specification patterns in >4,000 worms belonging to two strains of P. pacificus and two strains of Pristionchus entomophagus (Table 1). In all strains, the developmental program is executed with high precision; only between 0.2% and 1.1% of all examined animals showed VPC misspecifications. In P. pacificus PS1843, vulva-centering mistakes are prevalent: P7.p instead of P6.p had the 1° fate in six of 1,039 PS1843 animals. Very rarely, we also observed that P7.p adopted a 3° fate or was not present. This is in stark contrast to what we find in P. entomophagus: in strain RS145, by far the most common error is a misspecification of P5.p or P7.p, which normally have the 2° fate. In 10 of 1,134 animals (0.9%), P5.p or P7.p formed an independent invagination (“D” fate in Table 1; Fig. 2).
Table 1.
Developmental noise in four Pristionchus strains
| (P4.p) | P5.p | P6.p | P7.p | P8.p | RS106, n = 1,049 | PS1843, n = 1,039 | RS145, n = 1,147 | RS144, n = 1,010 | F2 PxW, n = 503 | |
|---|---|---|---|---|---|---|---|---|---|---|
| Wild type | 2° | 1° | 2° | 4° | 1,047 | 1,030 | 1,134 | 1,006 | 499 | |
| 2° specification | D | 1° | 2° | 4 | 0 | 0 | 9 | 1 | 0 | |
| 2° | 1° | D | 4° | 0 | 0 | 1 | 0 | 0 | ||
| Centering | 3° | D | 1° | 4° | 0 | 2 | 0 | 0 | 0 | |
| 3° | 2° | 1° | 4° | 0 | 3 | 0 | 1 | 0 | ||
| 3° | 2° | 1° | 2° | 0 | 1 | 0 | 0 | 0 | ||
| 1° | 2° | 3° | 4° | 0 | 0 | 1 | 0 | 0 | ||
| 2° competence | 2° | 1° | 3° | 4° | 0 | 2 | 0 | 0 | 3 | |
| 3° | 1° | 2° | 4° | 0 | 0 | 0 | 0 | 1 | ||
| 2.5° | 1° | 2° | 4° | 1 | 0 | 0 | 0 | 0 | ||
| 2° | 1° | 2.5° | 4° | 0 | 0 | 0 | 1 | 0 | ||
| Cell survival | 2° | 1° | X | 4° | 0 | 1 | 0 | 0 | 0 | |
| X | 1° | 2° | 4° | 1 | 0 | 1 | 0 | 0 | ||
| X | 1° | 3° | 4° | 0 | 0 | 0 | 1 | 0 | ||
| D | 2° | 1° | 2° | 4° | 0 | 0 | 1 | 0 | 0 |
RS106 and PS1843 are strains of P. pacificus, and RS144 and RS145 are strains of P. entomophagus. The last column shows data from randomly picked F2 animals of a cross between RS106 from Poland and PS1843 from Washington. See ref. 6 for cell fate definitions.
Fig. 2.
Developmental noise in P. entomophagus and P. pacificus. (Upper) In P. entomophagus RS145, the most common developmental error is a misspecification of P5.p. Instead of adopting the 2° fate, P5.p forms an independent invagination (arrow) that does not connect to the uterus. (Lower) In P. pacificus, very rarely P7.p, instead of adopting the 2° fate, remains undivided (3° fate, arrow).
Interestingly, there are strain-specific differences in the frequency at which any type of misspecification is observed. P. entomophagus RS144 and P. pacificus RS106 both show few “errors” compared with their conspecific strains RS145 and PS1843, respectively (Table 1). To test whether P. pacificus vulva development is affected by genetic perturbation, we determined the frequency of vulva misspecifications in a hybrid genetic background. We analyzed VPC specification patterns in 503 F2 animals from a cross between RS106 and PS1843 and found only four misspecification events (0.8%), which is in the same range as in PS1843 animals (Table 1). Taken together, the Pristionchus vulva is a developmental module that is robust to developmental noise, and misspecification events are in the promille range only. However, the observed misspecification events indicate differences in the exact types of errors in different strains and species. This result might indicate differences in cell–cell signaling pathways that contribute to developmental stability in the respective strains and species.
Cell–Cell Signaling Systems Vary Between Pristionchus Strains and Species.
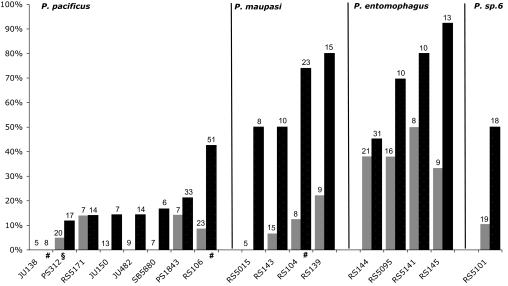
To determine whether the cell–cell signaling systems involved in Pristionchus vulva development undergo microevolutionary changes, we analyzed short- and long-range LI in 489 ablated worms belonging to eight strains of P. pacificus from North America, Asia, and Europe and nine strains of the three closely related hermaphroditic species Pristionchus maupasi, P. entomophagus, and Pristionchus sp. 6. The vulva cell fates of unablated animals in all examined strains and species show the typical 2°-1°-2°-4° pattern as described for the P. pacificus laboratory strain PS312 from California (data not shown). In P. pacificus PS312, the presence of P8.p strongly inhibits P5.p from adopting the 1° fate after P(6,7).p ablation (6) (Fig. 1). To test whether there is natural variation for this trait, we carried out P(6,7).p ablation experiments in 17 Pristionchus strains (Fig. 3, black bars; SI Table 4). Compared with P. pacificus, P5.p has the 1° fate more often in P. maupasi (63% of the animals, averaged over the strains; P ≪ 0.0001), P. entomophagus (74%; P ≪ 0.0001) and Pristionchus sp. 6 (50%; P < 0.05) (Fig. 3; SI Table 4). At the intraspecific level, seven of the eight P. pacificus strains show LI that is not significantly different from PS312 (Fig. 3; SI Table 4). However, in P. pacificus RS106 from Poland P5.p had the 1° fate in 43% of ablated animals (P < 0.05 for the pairwise comparison with PS312). Thus, the influence of long-range LI is reduced in this strain.
Fig. 3.
Natural variation of short- and long-range LI. Percentage of P5.p adopting the 1° fate after P(6,7).p ablation (long-range LI, black bars) and P7.p adopting the 1° fate after P(5,6).p ablation (short-range LI, gray bars). Numbers above bars represent total number of ablated animals in that strain. See SI Table 4 for detailed lineage analysis. §, data from ref. 6; #, data from ref. 7.
Next, we assessed the influence of short-range LI by ablating P(5,6).p and following the cell fate of P7.p (Fig. 3, gray bars). In 16 of 17 strains, P7.p has the 1° fate less often than P5.p in the equivalent ablation experiment, indicating either a stronger influence of P8.p on its neighboring VPC or intrinsic P8.p-independent differences between P5.p and P7.p. On average, P7.p had the 1° fate in 40% of P(5,6).p ablated animals in P. entomophagus, but only in 5–11% of the animals in P. pacificus (P ≪ 0.0001), P. maupasi (P < 0.01), and P. sp. 6 (P < 0.05) (Fig. 3; SI Table 4). Thus, the influence of short-range LI is significantly reduced in P. entomophagus. Taken together, although the VPC fate pattern in unperturbed animals is identical in all four Pristionchus species, cell ablation experiments reveal differences in cell–cell signaling between species and strains. In particular, long- and short-range LI can be uncoupled over microevolutionary time: in seven of eight P. pacificus strains, both, P5.p and P7.p have the 1° fate only rarely in the respective ablation experiments. In contrast, the fate patterns of these two cells differ strongly if the same experiments are performed in P. maupasi, P. sp. 6, and also in the strain RS106 of P. pacificus.
Long-Range LI Is Lost in Some P. pacificus RILs.
To determine the genetic contribution to the difference in LI between P. pacificus strains, we analyzed F1 and F2 animals from crosses between RS106 from Poland and PS1843 from Washington, two strains that differ in the strength of LI (P < 0.05) and are genetically highly polymorphic (7) (Table 2, C and D). When we crossed RS106 (weak LI) and PS1843 (strong LI) and ablated P(6,7).p in the resulting F1 generation, P5.p had the 1° fate in only three of 27 animals, indicating that LI is strong in heterozygous animals (P < 0.01 for the pairwise comparison with RS106). Furthermore, in randomly chosen F2 animals, five of 16 P5.p cells adopted the 1° fate (31%), which is intermediate between the values found in the parental strains but not significantly different from either of them (P > 0.5 for both pairwise comparisons).
Table 2.
| Strain | P5.p 1° (n) | Percent P5.p 1° | 90% CI, % |
|---|---|---|---|
| A, PS1843 (Wash) | 7 (33) | 21 | 10–33 |
| B, RS106 (Pol) | 22 (51) | 43 | 32–55 |
| C, Pol × Wash F1 | 3 (27) | 11 | 1–21 |
| D, Pol × Wash F2 | 5 (16) | 31 | 12–50 |
| E, RIL 12B | 0 (12) | 0 | – |
| F, RIL 14B | 0 (6) | 0 | – |
| G, RIL 10A | 2 (19) | 11 | 0–22 |
| H, RIL 18C | 2 (11) | 18 | 0–37 |
| I, RIL 9A | 3 (13) | 23 | 4–42 |
| J, RIL 2A | 4 (13) | 31 | 10–52 |
| K, RIL 16C | 2 (6) | 33 | 2–65 |
| L, RIL 3A | 5 (14) | 36 | 15–57 |
| M, RIL 13B | 5 (14) | 36 | 15–57 |
| N, RIL 25B | 5 (12) | 42 | 18–65 |
| O, RIL 1A | 5 (11) | 45 | 21–70 |
| P, RIL 11B | 5 (11) | 45 | 21–70 |
| Q, RIL 17C | 5 (10) | 50 | 24–76 |
| R, RIL 4A | 4 (8) | 50 | 21–79 |
| S, RIL 20C | 7 (11) | 64 | 40–87 |
| T, RIL 19C | 13 (14) | 93 | 82–100 |
| Average (16 RILs) | 4.2 (11.6) | 36 | |
| U, RIL C22 | 12 (13) | 92 | 80–100 |
| V, RIL A8 | 16 (18) | 89 | 77–100 |
| W, RIL A8BC7 | 19 (23) | 83 | 70–96 |
| X, RIL A8BC10 | 15 (17) | 88 | 75–100 |
| Y, RIL A8BC18 | 11 (12) | 92 | 79–100 |
P(6,7).p were ablated 0–1 h after hatching, and the cell lineage of P5.p was analyzed as described in ref. 6. A, B, homozygous and isogenic laboratory strains. C, F1 animals. D, segregating F2 population. E–V, homozygous recombinant inbred lines. W−Y, Second-generation RILs. See text for details. Wash, Washington; Pol, Poland. CI, confidence interval.
To study the strength of LI in hybrid genetic backgrounds, we generated RILs of the strains RS106 and PS1843 (SI Fig. 5). We generated 16 RILs that were inbred until F13 and processed them for P(6,7).p ablation. As expected, most RILs showed a pattern of P5.p fate specification that is intermediate to (38% of the RILs) or reminiscent to the parental strains (56%) (Table 2, E–T).
Surprisingly, one of the 16 RILs showed strong transgression, i.e., the occurrence of a more extreme phenotype than in either parental line (8). Specifically, P5.p had a 1° fate in >90% of the animals after ablation of P(6,7).p in RIL19C (Table 2, T; P < 0.001 for the pairwise comparison with RS106; at P < 0.05, significant after Bonferroni correction for 16 pairwise comparisons). Such a pattern is not known from the two parental lines, F1 hybrids thereof, or any other wild isolate of P. pacificus. We therefore screened another set of 58 RILs (inbred until F9) specifically for the absence of LI (SI Table 5). We found two more lines in which P5.p had the 1° fate in ≈90% of P(6,7).p-ablated animals (Table 2, U and V; P < 0.01 and P < 0.001 for the respective pairwise comparisons with RS106; in the case of RILA8 significant at P < 0.05 after Bonferroni correction).
We selected the RILA8 as the parental strain for raising a second set of RILs (SI Fig 5). RILA8 was crossed to RS106, and the resulting second RILs were inbred until the F9 generation. We expected to find RILs that lack LI at a higher ratio than in the first RILs, because RS106-derived factors that are required for the transgression phenomenon are fixed throughout the experiment. Indeed, three of 15 (20%) second RILs lacked LI, in contrast to 5% in the first RILs (Table 2, W, X, and Y; SI Table 6). The proportion of RILs that lack LI is probably an underestimate because of the possibility of false negatives in the screening procedure. We conclude that the recombination of genetic variation in RILs results in a change of cell–cell interaction patterns.
Some RILs Gained Competence of P8.p.
To ask whether other properties of P8.p are also affected in these RILs, we further characterized the RILs that showed transgression with respect to LI. One of the intriguing properties of P8.p is that, unlike the homologous cell in C. elegans, it is not competent to respond to inductive signaling in the absence of P(5–7).p. By ablating P(5–7).p, we found that in RILA8 and derivatives thereof (Table 3, I–L): P8.p is competent to differentiate and form vulva tissue, which is in contrast to all tested wild isolates of P. pacificus (Table 3, A–F). Specifically, P8.p had a vulva fate in 50% (n = 14) and 42% (n = 12) of P(5–7).p ablated animals in RILA8 and RILA8BC7, respectively (Table 3, I and J).
Table 3.
P8.p competence in laboratory strains and RILs
| Strain | P8.p D (n) | Percent P8.p D |
|---|---|---|
| A, PS1843 | 0 (21) | 0 |
| B, RS106 | 0 (12) | 0 |
| C, PS312* | 0 (22) | 0 |
| D, JU482 | 0 (21) | 0 |
| E, SB5880 | 0 (7) | 0 |
| F, JU150 | 0 (6) | 0 |
| G, RIL 19C | 0 | |
| H, RIL C22 | 0 (10) | 0 |
| I, RIL A8 | 7 (14) | 52 |
| J, RIL A8BC7 | 5 (12) | 42 |
| K, RIL A8BC10 | 2 (10) | 20 |
| L, RIL A8BC18 | 3 (4) | 75 |
| M, RIL | 9 (14) | 64 |
| A8BC7BC30 | ||
| N, RIL | 4 (8) | 50 |
| A8BC7BC77 | ||
| O, RIL | 7 (16) | 44 |
| A8BC7BC59 | ||
| P, RIL | 2 (7) | 29 |
| A8BC7BC56 | ||
| Q, RIL | 3 (13) | 23 |
| A8BC7BC10 | ||
| R, RIL | 13 | |
| A8BC7BC47 | ||
| S, RIL | 1 (2) | 50 |
| A8BC7BC20 |
A–F, homozygous and isogenic laboratory strains; G–S, recombinant inbred lines; G−I, J−L, and M−S are first, second, and third generations, respectively. See text for details.
*, data for PS312 from ref. 6.
To study the genetics of this trait, we generated third-generation RILs with RILA8BC7 and RS106 as parental lines. We found seven of 49 third RILs (14%) with P8.p having a vulva fate after P(5–7).p ablation (Table 3, M–S; SI Table 7). Again, this is a lower-bound estimate, given the small number of tested animals per RIL. Thus, our results suggest that random fixation of genotypic combinations in P. pacificus can result in evolutionary innovations. P8.p gained competence to form vulva tissue, a trait that was never observed in P. pacificus isolates but is occasionally observed in other species in the genus Pristionchus (7). We conclude that evolutionary innovations in redundant aspects of cell–cell signaling may arise by recombining preexisting genetic variation.
We genotyped RILA8BC7 with 114 single-strand conformational polymorphism (SSCP) markers, covering all six chromosomes of the P. pacificus genome (SI Fig. 6). Almost all markers showed an RS106 pattern; however, on Chromosome I, a large region of PS1843 genotypes is retained. We found that six of seven third RILs with a P8.p-differentiation phenotype show a PS1843 pattern in the middle of Chromosome I, whereas only eight of 21 randomly chosen “sister” third RILs had a PS1843 genotype at the same region (SI Fig. 7). We assume that several loci in the P. pacificus genome are involved in the regulation of P8.p competence.
Aspects of Redundant Signaling Can Be Uncoupled in RILs and in Nature.
Although two traits, P8.p competence and loss of long-range LI, cooccur in a number of RILs (compare Table 2, V–Y, and Table 3, I–L), they can apparently be uncoupled. For example, RILC22 lacks long-range LI, but P8.p remains always epidermal (Table 2, U, and Table 3, H). Vice versa, RILA8–7-30 has a competent P8.p, but P5.p had a 2° fate in four of six animals after ablation of P(6,7).p, indicating that long-range LI is present in this RIL (Table 3, M, and SI Table 8, A).
Similarly, the competence of P5.p and P7.p to adopt the 1° fate can be changed independently in Pristionchus isolates (Fig. 3 and SI Table 4) and in at least one P. pacificus RIL (SI Table 8, B). This might indicate that long- and short-range LI are controlled by two distinct signals. Alternatively, the signal originating from P8.p might be graded and thus P5.p, but not P7.p, could escape the influence of P8.p if the signaling levels are low. In summary, we demonstrate the emergence of evolutionary innovations within the redundant signaling network of P. pacificus vulva development at several levels. Different aspects of redundant cell–cell signaling can be uncoupled in nature as well as in laboratory-generated RILs.
Discussion
Robustness to genetic or environmental perturbation was recognized a long time ago as a significant phenomenon (9, 10) and continues to be an area of lively discussion (1, 2, 11, 12). Ultimately, analyses of robustness and evolvability need to take into account both evolutionary dynamics within populations as well as developmental dynamics within individual organisms. Taking advantage of the fixed cell lineage of the P. pacificus vulva and the availability of cell ablation technique for experimental manipulation, we investigated the evolution of redundant signaling during cell fate specification.
Invariability of the Vulva Phenotype Despite Natural Variation in Cell–Cell Signaling.
The 2°-1°-2° pattern of the vulva and most other aspects of morphology are strongly conserved within the genus Pristionchus. Indeed, the 18 species of the genus can be classified according to reproductive mode (hermaphrodites or gonochorists), but a reliable species identification based on morphology alone is close to impossible (13, 14). This stability of vulva morphology and cell lineage over evolutionary time, a general phenomenon in many nematode genera, can be attributed to robustness of developmental modules to mutation (1). Thus, constancy of the phenotype at a wide phylogenetic range contrasts with genetic variation of redundant aspects of cell–cell signaling at a microevolutionary scale.
Correlation of LI and Robustness to Developmental Noise.
Is the natural variation in P8.p-mediated LI the outcome of neutral evolution, or is there any conceivable selection pressure that could have favored the emergence of the observed differences between closely related Pristionchus species? We think that changes in P8.p-mediated signaling might influence the robustness of vulva development to developmental noise. Under standard laboratory conditions, misspecifications that affect specifically the 2° fate are of opposite nature in PS1843 and RS145: whereas in PS1843 the only 2°-specific defect is a rare noninduction, in RS145, the most common defect is an ectopic differentiation. Thus, the type of P5.p/P7.p-related misspecification events correlates with the strength of LI in these strains. Although it is not yet clear whether there is a direct link between LI and robustness, our comparison of misspecification events in Pristionchus isolates might be interpreted as the first indication that differences in redundant cell–cell signaling affect developmental stability of Pristionchus vulva development. These differences might reflect adaptations that assure a robust VPC specification in the respective species-specific habitats. Future studies on the exact natural habitats of these nematodes might indicate whether developmental noise changes in different environments.
Evolvability at the Level of Redundant Cell–Cell Signaling.
In our experiments with RILs, we exemplified how redundant VPC specification signals can evolve: although vulva development is robust to genetic perturbation, at the level of redundant signaling, we observe not only transgression (loss of LI) but also novel properties (competence of P8.p). Interestingly, the three aspects of cell–cell signaling studied here in depth, long- and short-range LI and P8.p competence, can be uncoupled experimentally and also can have been uncoupled in nature. Thus, a mechanism exists in Pristionchus that would allow a diversification of redundant signaling events without affecting the phenotypic output (the 2°-1°-2°-4° fate pattern) in most of the animals.
Species-Specific Characteristics of Developmental Noise.
It is illuminating to compare developmental stability in different nematode genera. The most prominent influence of developmental noise in the most-used laboratory strain of C. elegans, N2, concerns the fate of P3.p: in ≈50% of the animals; this cell divides once and adopts the 3° fate (4). Alternatively, it remains undivided and fuses with the epidermis during the L2 stage. The frequency of P3.p divisions varies in different strains of C. elegans from 15% to 59% (15). Moreover, P3.p is part of the vulva equivalence group in C. elegans but not in C. briggsae. Similar strain-specific differences in the division patterns of nonvulval cells were observed in the case of P4.p and P8.p in nematodes of the genus Oscheius (15). However, the 3° cell lineages do not contribute to vulva formation, and differences are not likely to be strongly selected against (15). In contrast, developmental errors in P(5–7).p, the cells that form the vulva in Oscheius, Caenorhabditis, and Pristionchus are rare in all three genera. In Oscheius, the most frequent developmental error is the absence of some Pn.p cells (≈2%), which is in contrast to the results we obtained for Pristionchus strains, where missing Pn.p cells are very rarely found (only 4 of 4,245 animals; Table 1). Therefore, the distinct configurations of the vulva equivalence groups in distantly related nematode species provide a platform for evolutionary comparisons of developmental stability at the cellular level.
Materials and Methods
Strains.
Worms are grown on Escherichia coli OP50 as described previously. Collection procedures and origins of Pristionchus isolates are described elsewhere (13, 14, 16). Because of long inbreeding in the laboratory and the hermaphroditic mode of reproduction, the wild-type strains used in this study are likely to be homozygous at nearly all loci.
Cell Ablation Experiments and Lineage Analysis.
Cell ablation experiments were carried out by using standard techniques described for C. elegans (17), with a Laser Science (Franklin, MA) dye laser (18). Animals were picked into M9 buffer placed on a pad of 5% agar in water containing 5 mM sodium azide as anesthetic. All ablation experiments were carried out 0–1 h after hatching of the larvae (20°C).
For analysis of developmental noise, worms were grown under standard laboratory conditions (20°C) with plenty of E. coli OP50 and analyzed by Nomarski microscopy between late J3 and early J4 stage. In some cases, worms that displayed misspecification events were rescued and the offspring analyzed to rule out the possibility that the misspecification was caused by spontaneous mutations. Cell lineage characters and cell fate terminology have been described in detail (6).
RIL Construction.
RILs were set up by crossing approximately three to five males and hermaphrodites of RS106 and PS1843, respectively. Out-crossed F1 progeny were isolated and, in the F2, single animals were transferred to a new plate and allowed to self-fertilize. Randomly chosen single offspring larvae were transferred to new plates in every generation until at least the F9. During the inbreeding period, worms were kept at 25°C; then, strains were kept by chunking and at 20°C. We raised the following set of RILs: (i) RS106 male × PS1843 [dumpy (dpy)] hermaphrodite; (ii) RS106 male × PS1843(dpy) hermaphrodite, RS106 male × PS1843 [uncoordinated movement (unc)] hermaphrodite, and PS1843male × RS106(dpy) hermaphrodite; (iii) RILA8 male × RS106(wt) hermaphrodite (heterozygosity of single F1 animals from iii was confirmed after egg laying by testing the SSCP marker S28); and (iv) RILA8BC7 male × RS106(dpy).
Sets i and ii were frozen, and lines of sets iii and iv were kept only if interesting phenotypes arose. Screening for loss of LI and P8.p competence in sets ii–iv was carried out by initially ablating one to three animals and increasing numbers only if the phenotype of interest (1° P5.p or differentiating P8.p) was observed.
SSCP Genotyping.
SSCP markers were taken from the P. pacificus genetic linkage map (ref. 19 and www.pristionchus.org). PCR and fragment detection were performed as described (19).
Statistical Analysis.
If not indicated otherwise, P values refer to pairwise χ2 tests. For the analysis of between-species differences in LI (Fig. 3, SI Table 4), data of strains belonging to the same species have been pooled.
Supplementary Material
Acknowledgments
We thank G. Bento and B. Schlager for critically reading the manuscript and the Sommer laboratory for discussions. H.Z. was supported by a predoctoral fellowship of the Boehringer Ingelheim Fonds.
Abbreviations
- VPC
vulva precursor cell
- LI
lateral inhibition
- RIL
recombinant inbred line
- SSCP
single-strand conformational polymorphism.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. S.C. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0610799104/DC1.
References
- 1.Wagner A. Robustness and Evolvability in Living Systems. Princeton: Princeton Univ Press; 2005. [Google Scholar]
- 2.Kirschner M, Gerhart J. Proc Natl Acad Sci USA. 1998;95:8420–8427. doi: 10.1073/pnas.95.15.8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dichtel-Danjoy ML, Felix MA. Trends Genet. 2004;20:268–276. doi: 10.1016/j.tig.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 4.Sternberg P. WormBook. 2005 doi: 10.1895/wormbook.1.6.1. The C. elegans Research Community, www.wormbook.org. [DOI] [PMC free article] [PubMed]
- 5.Hong RL, Sommer RJ. BioEssays. 2006;28:651–659. doi: 10.1002/bies.20404. [DOI] [PubMed] [Google Scholar]
- 6.Jungblut B, Sommer RJ. Development (Cambridge, UK) 2000;127:3295–3303. doi: 10.1242/dev.127.15.3295. [DOI] [PubMed] [Google Scholar]
- 7.Srinivasan J, Pires-daSilva A, Gutierrez A, Zheng M, Jungblut B, Witte H, Schlak I, Sommer RJ. Evol Dev. 2001;3:229–240. doi: 10.1046/j.1525-142x.2001.003004229.x. [DOI] [PubMed] [Google Scholar]
- 8.Lynch M, Walsh B. Genetics and Analysis of Quantitative Traits. Sunderland, MA: Sinauer; 1998. [Google Scholar]
- 9.Schmalhausen II. Factors of Evolution: The Theory of Stabilizing Selection. Chicago: University of Chicago Press; 1949. [Google Scholar]
- 10.Waddington CH. The Strategy of the Genes: A Discussion of Some Aspects of Theoretical Biology. New York: Macmillan; 1957. [Google Scholar]
- 11.Kitano H. Nat Rev Genet. 2004;5:826–837. doi: 10.1038/nrg1471. [DOI] [PubMed] [Google Scholar]
- 12.Siegal ML, Bergman A. Proc Natl Acad Sci USA. 2002;99:10528–10532. doi: 10.1073/pnas.102303999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herrmann M, Mayer WE, Sommer RJ. Front Zool. 2006;3:14. doi: 10.1186/1742-9994-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herrmann M, Mayer WE, Sommer RJ. Zoology (Jena) 2006;109:96–108. doi: 10.1016/j.zool.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 15.Delattre M, Felix MA. Curr Biol. 2001;11:631–643. doi: 10.1016/s0960-9822(01)00202-0. [DOI] [PubMed] [Google Scholar]
- 16.Zauner H, Mayer WE, Herrmann M, Weller A, Erwig M, Sommer RJ. Mol Ecol. 2007;16:1267–1280. doi: 10.1111/j.1365-294X.2006.03222.x. [DOI] [PubMed] [Google Scholar]
- 17.Epstein HS, Shakes DC. Caenorhabditis elegans: Modern Biological Analysis of an Organism. San Diego: Academic; 1995. [Google Scholar]
- 18.Avery L, Horvitz HR. Cell. 1987;51:1071–1078. doi: 10.1016/0092-8674(87)90593-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Srinivasan J, Sinz W, Lanz C, Brand A, Nandakumar R, Raddatz G, Witte H, Keller H, Kipping I, Pires-daSilva, et al. Genetics. 2002;162:129–134. doi: 10.1093/genetics/162.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.