Abstract
Fragile X syndrome is caused by the inactivation of the X-linked FMR1 gene, leading to the loss of its encoded protein FMRP. Although macroorchidism and defects in neuronal architecture and function have been associated with lack of FMRP, the exact molecular mechanism underlying this disease remains unclear. We have reported previously that in the brain and testis of mice, FMRP specifically interacts with a distinct mRNA nuclear export factor NXF2 but not with its close relative NXF1, a ubiquitously expressed essential mRNA nuclear export factor. This interaction marked NXF2 as a putative functional partner of FMRP. Here, we demonstrate by immunoprecipitation and quantitative real-time RT-PCR that, in cultured mouse neuronal cells, both FMRP and NXF2 are present in Nxf1 mRNA-containing ribonucleoprotein particles. Further, we show that expression of NXF2 leads to the destabilization of Nxf1 mRNA and that this effect is abolished when Fmr1 expression is reduced by siRNA, arguing that both proteins collaborate to exert this effect. Importantly, these findings correlate well with our observations that in both mouse hippocampal neurons and male germ cells where the expression of FMRP and NXF2 is most prominent, the expression of NXF1 is relatively poorly expressed. Our studies thus identify Nxf1 mRNA as a likely biologically relevant in vivo target of both FMRP and NXF2 and implicate FMRP, in conjunction with NXF2, as a posttranscriptional regulator of a major mRNA export factor. Such regulation may prove important in the normal development and function of neurons as well as of male germ cells.
Keywords: germ cell, neuron, RNA stability
Fragile X syndrome, which affects ≈1/4,000 males and 1/8,000 females, is the leading heritable form of mental retardation and is associated with a variety of learning disorders and behavioral problems (for review, see refs. 1 and 2). The disease also leads to macroorchidism in postpubertal male patients. Fragile X syndrome is almost always caused by an expansion of the CGG repeat in the 5′ untranslated region of the X-linked FMR1 gene, which results in transcriptional silencing and loss of expression of its encoded fragile X mental retardation protein (FMRP) (for review, see refs. 1 and 2).
FMRP expression is widespread but is especially high in the brain and testis (3, 4; for review, see ref. 5). In the brain, FMRP has been implicated in dendritic spine maturation, synapse formation, and synaptic plasticity (for review, see refs. 2, 5, and 6). As a nucleocytoplasmic shuttling RNA-binding protein, FMRP participates in mRNA transport and translational control (for review, see refs. 5–7). At the steady state, FMRP is predominantly cytoplasmic, localized in messenger ribonucleoprotein (mRNP) complexes that associate with polyribosomes (3, 8, 9). A minor fraction of the protein has also been detected in the nucleoplasm and in association with nuclear pores (8), consistent with a role in mRNA nuclear export. In addition, FMRP has been found in large mRNP complexes traveling along dendrites and at the postsynaptic sites of protein synthesis, suggesting its involvement in dendritic mRNA transport and translational control (10, 11). Indeed, studies have variously shown FMRP-mediated enhancement and/or suppression of translation of some transcripts, and such regulation may also be linked to its nucleic acid chaperone activity and to noncoding small RNAs and the RNAi machinery (for review, see refs. 7, 12, and 13 and references therein). It has thus been postulated that the cognitive symptoms of fragile X syndrome may at least in part derive from the dysregulated translation of target mRNAs, leading to abnormal neuronal cell morphology and function (for review, see refs. 2 and 14). However, it remains to be determined whether FMRP affects the stability of associated mRNAs, and if so, whether loss of such regulation might also contribute to the pathogenesis of the disease.
As a multifunctional protein, FMRP appears to be able to interact with a broad range of mRNA targets and protein partners in different cellular and subcellular compartments and in a dynamic fashion. The specificity and functional outcome of these interactions most likely depends on what components and which compartment it is associated with. For the past several years, significant efforts have been made to identify FMRP mRNA targets (15–17). However, there is little overlap among the targets reported by different groups, and only a handful of these targets have been validated in vivo (18). Identifying more biologically relevant in vivo targets is thus critically important. In the case of FMRP-interacting proteins, a growing number of them have been described (19; for review, see ref. 20 and references therein), although in most cases the functional relevance of these interactions has not yet been confirmed. We have shown previously that FMRP specifically interacts with NXF2, a distinct family member of the evolutionarily conserved nuclear export factor proteins, in the mouse brain and testis where both proteins are predominantly expressed (21). In this work, we have further characterized the functional and mechanistic aspects of this interaction. We find that both FMRP and NXF2 are associated with mRNPs containing the mRNA of the major mRNA export factor NXF1 and act to regulate its stability in neuronal cells.
Results
The Expression of FMRP and NXF2 Inversely Correlates with That of NXF1 in Neurons and Male Germ Cells.
We have shown previously that FMRP and NXF2 are highly expressed in the hippocampal neurons of mouse brain (21). In the testis, these proteins are likewise expressed together and are restricted to the primitive sperm-producing cells, the spermatogonia (21). Taken together with the observation that FMRP interacts with NXF2 but not with NXF1 in both the brain and testis, we asked whether there exists a relationship between the expressions of these proteins in these cells. To address this question, we first performed immunofluorescence and immunohistochemistry experiments to examine their expression. Consistent with our previous findings (21), whereas FMRP (Fig. 1 A and D) and NXF2 (Fig. 1 B and E) are highly enriched in the spermatogonia (white arrowheads) that localize at the periphery of the seminiferous tubules, their expression is barely detectable in the more mature germ cells (white arrows). In contrast, NXF1 expression is high in the more mature germ cells (Fig. 1 C and F, white arrows) but relatively low in the spermatogonia (arrowheads).
Fig. 1.
Immunofluorescence of adult mouse testicular cryostatic sections by using antibodies specific for FMRP (A and D), NXF2 (B and E), and NXF1 (C and F). (A–C) High-power images of the selected areas (white boxes in D–F). Each panel is a merged image of DAPI nuclear staining (blue) and the indicated antibody staining (green for FMRP and red for NXF2 and NXF1). The spermatogonia (arrowheads) are the cells residing at the periphery of the seminiferous tubules of the testis. The more mature germ cells including spermatocytes (large arrows) and spermatids (small arrows) are located closer to the lumen of the tubules. (Scale bars, 20 μm.)
Curiously, such differential expression is also seen in the brain. As shown previously (21), hippocampal neurons are sites where FMRP (Fig. 2 A and D) and NXF2 (B and E) are abundantly expressed. In contrast, the expression of NXF1 in hippocampal neurons is relatively weaker (C and F), especially compared with NXF1 staining in cells located outside the hippocampal area (Fig. 2C, white arrows) where the signal is stronger than that in the hippocampal neurons (Fig. 2C, white arrowheads). Therefore, in both the brain and testis in cells where FMRP and NXF2 are highly expressed, NXF1 is present at a relatively low level. This apparent differential expression raised an intriguing possibility that there may exist a regulatory relationship between these genes in these highly specialized cells.
Fig. 2.
Immunohistochemistry of adult mouse brain paraffin sections with antibodies specific for FMRP (A and D), NXF2 (B and E), and NXF1 (C and F). (A–C) High-power images of the selected areas (white boxes in D–F). Arrowheads indicate the cell bodies of hippocampal neurons. Note that the NXF1 staining of the hippocampal neurons is relatively weaker than that of cells located away from the hippocampal area (white arrows). (Scale bars, 20 μm.)
FMRP and NXF2 Associate with Nxf1 mRNA-Containing RNPs in Neuronal Cells.
As a first step toward understanding the possible functional interplay between these genes, we wished to determine whether FMRP and NXF2 are associated with Nxf1 mRNA in vivo. This possible association was based on the notion that both FMRP and NXF2 are RNA-binding proteins and that FMRP has been implicated in regulation of gene expression by binding to its target mRNAs (15–18). We tested this possibility in mouse N2a cells, which express appreciable levels of FMRP and NXF1 but undetectable levels of NXF2. We chose these cells for two reasons: (i) they are well characterized neuronal cells and have been used for FMRP studies (9); and (ii) we could easily test the function of NXF2 in conjunction with FMRP in these cells by transiently transfecting them with a FLAG-tagged NXF2 expression plasmid to raise the NXF2 level to close to that normally seen in spermatogonia. To test whether FMRP and NXF2 associate with Nxf1 mRNA, we carried out immunoprecipitation (IP) with antibodies specific for FMRP or NXF2 to isolate FMRP/NXF2-containing mRNPs from N2a cells transfected with the NXF2 expression vector. In parallel experiments, we performed IP with mouse IgG (a negative control for monoclonal antibody IP) or rabbit preimmune serum (a negative control for polyclonal antibody IP). We used RNA samples extracted from IP complexes to generate cDNAs by reverse transcription followed by real-time PCR analysis with gene-specific primers to identify associated mRNAs. The mRNAs we tested included Nxf1, Nxf2, Fmr1, β-adaptin and Gapdh. The housekeeping Gapdh mRNA was used as a negative control for binding to FMRP and NXF2. β-Adaptin mRNA has been reported both to be present in FMRP-containing mRNPs in primary cultured rat hippocampal neurons and to bind purified FMRP in vitro (17). Similarly, FMRP has been shown to bind Fmr1 mRNA in vitro (17, 22). We initially thought that these two mRNAs would be good positive controls for FMRP binding.
Fig. 3A confirms that the anti-FMRP and anti-NXF2 antibodies could specifically immunoprecipitate FMRP and NXF2, respectively. Fig. 3 B and C shows the relative abundance of each mRNA present in the FMRP- (open bars) or NXF2-containing (filled bars) RNPs. Nxf1 mRNA exhibited the most dramatic enrichment (≈12-fold) in both the FMRP and NXF2 mRNPs, followed by Nxf2 mRNA, which showed a ≈5-fold enrichment. Interestingly, the Fmr1 and β-adaptin mRNAs showed only a ≈2-fold enrichment, which is only marginally different from that of the negative control Gapdh mRNA. A similar enrichment pattern was observed when a monoclonal anti-FLAG antibody specific for the transfected FLAG-tagged NXF2 was used in the IP experiment (Fig. 3C). Given that the Nxf1, Nxf2, Fmr1, and β-adaptin mRNAs were expressed at comparable levels (Fig. 3D), the significant enrichment of Nxf1 mRNA in the FMRP/NXF2 mRNPs strongly suggests that Nxf1 mRNA is an in vivo target for FMRP/NXF2 regulation.
Fig. 3.
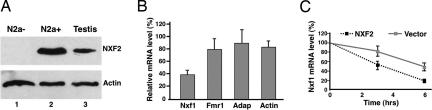
FMRP and NXF2 preferentially associate with Nxf1 mRNA-containing RNPs. FLAG-NXF2 was transfected into N2a cells, and IP was carried out 48 h after transfection. (A) Proteins from purified IP complexes or from 3% of supernatants were resolved on SDS/10% PAGE. The presence of the FMRP and NXF2 proteins in the IP complexes was confirmed by Western blot analyses. (Upper) IP with anti-FMRP (αFMRP) using mouse IgG as a negative control, followed by Western blotting using anti-FMRP. FMRP was in the anti-FMRP IP complexes (lane 4) but not in the IgG IP complexes (lane 3). (Lower) IP with anti-NXF2 (αNXF2) using preimmune serum as a negative control followed by Western blotting using anti-NXF2. NXF2 was in the anti-NXF2 IP complexes (lane 4) but not in the preimmune IP complexes (lane 3). Lanes 1 and 2 indicate that FMRP and NXF2 were present in the cell lysates. (B and C) RNAs were extracted from purified IP complexes followed by qRT-PCR. mRNA levels associated with FMRP (white bars) or NXF2 (gray bars) RNPs are indicated relative to those with negative control IP samples, which were arbitrarily set as 1. (B) A polyclonal anti-NXF2 antibody was used. (C) A monoclonal anti-FLAG antibody was used to immunoprecipitate the transfected FLAG-NXF2. Each bar represents mean ± SEM (B, n = 4; C, n = 2). (D) mRNA expression levels. RNAs were extracted from 10% of IP supernatants, and qRT-PCR was carried out using primers specific for each mRNA. Levels are plotted relative to Gapdh mRNA levels, which were arbitrarily set a value of 100. Each bar represents mean ± SEM (n = 4).
It is important at this point to address the issue of IP with the commercially available anti-FMRP antibody. As stated at the Chemicon website, the anti-FMRP we used does not work in IP. However, based on Fig. 3A and our previous report (21) and that the Eberwine group had also successfully used the same antibody to isolate FMRP·cDNA complexes biochemically (17), we conclude that the ability of this antibody to work well in IP assays may be influenced by different IP conditions used. In addition, there have been reports in the literature that this antibody may cross-react with its closely related family members FXR1P and FXR2P (23). However, we have seen no evidence of cross-reactivity with either protein under the experimental conditions used (21).
Expression of NXF2 Destabilizes Nxf1 mRNA in Neuronal Cells.
To assess the functional relevance of the specific enrichment of FMRP and NXF2 in Nxf1 mRNA-containing RNPs, we took the advantage of the observation that N2a cells do not express detectable levels of NXF2. We first asked whether exogenously expressing NXF2 to a level close to that seen in spermatogonia would affect Nxf1 mRNA expression. Thus, we transfected FLAG-NXF2 or empty vector together with a yellow fluorescence protein (YFP) expression vector into N2a cells and examined the effect of NXF2 expression on steady-state Nxf1 mRNA levels. In these experiments, we generally observed NXF2 levels close (≈2-fold less) to those seen in spermatogonia after normalization against transfection efficiency and the percentage of spermatogonia in a total testicular cell population (Fig. 4A; for calculation details, see Materials and Methods). YFP was used as a marker for fluorescent flow cytometry sorting to enrich transfected cells. Total RNAs from YFP-positive FLAG-NXF2 or empty vector-transfected cells were isolated, and individual mRNA levels were measured by quantitative RT-PCR (qRT-PCR). The expression of NXF2 in YFP-positive cells was confirmed by qRT-PCR (data not shown). Fig. 4B shows mRNA levels from cells transfected with FLAG-NXF2 relative to those transfected with empty vector. Only the level of Nxf1 mRNA was significantly reduced upon NXF2 expression, whereas the levels of three other mRNAs tested (Fmr1, β-adaptin, and β-actin) remained essentially unchanged. This finding correlates well with our earlier observation that the Nxf1 mRNA was highly enriched in the FMRP/NXF2-containing RNPs, whereas Fmr1 and β-adaptin mRNAs were not (Fig. 3B). These results thus suggest that NXF2 may regulate the stability of Nxf1 mRNA. To test this theory, we performed stability assays on Nxf1 mRNA. N2a cells were transfected with FLAG-NXF2 or empty vector, and Nxf1 mRNA levels were measured at the indicated time points after treatment with the transcription inhibitor actinomycin D. The Nxf1 mRNA half-life was reduced by ≈2-fold in response to the expression of NXF2 (Fig. 4C). Considering that the average transfection efficiency of N2a in our hands was ≈53% (ref. 24 and data not shown), were we to normalize for the transfection efficiency, we would see an even more dramatic half-life change. Based on these results, we conclude that NXF2 acts to destabilize Nxf1 mRNA.
Fig. 4.
NXF2 expression results in the destabilization of Nxf1 mRNA. (A) (Upper) Representative Western blots showing NXF2 expression levels in empty vector-transfected (lane 1) or FLAG-NXF2-transfected (lane 2) N2a cells or in the mouse testis (lane 3). (Lower) β-Actin was used as a loading control. (B) N2a cells were transfected with FLAG-NXF2 or empty vector, together with a YFP expression vector. Forty-eight hours after the transfection, cells were harvested and subjected to FACS. YFP-positive cells were collected followed by RNA extraction and qRT-PCR using primers specific for the indicated mRNAs. The levels of mRNAs from FLAG-NXF2-transfected cells are shown relative to those from empty vector-transfected cells, which were arbitrarily set as 100%. Each bar represents mean ± SEM (n = 3≈5). (C) N2a cells were transfected with FLAG-NXF2 or empty vector. Twenty-four hours after transfection, actinomycin D was added to inhibit transcription, and Nxf1 mRNA levels at 0 h, 3 h, and 6 h time points, respectively, were measured by qRT-PCR. The mRNA levels at the 0 h time point were arbitrarily set as 100%. Each time point represents mean ± SEM (n = 4).
Given that both FMRP and NXF2 were enriched in the Nxf1 mRNA-containing RNPs (Fig. 3), we speculated that the observed destabilization might require a contribution of FMRP. To answer this question, we specifically reduced FMRP expression by siRNA and asked whether doing so would eliminate the effect of Nxf1 mRNA destabilization induced by NXF2. First, we wanted to ensure that the siRNA treatment is effective and specific. We transfected a siRNA specific for the mouse Fmr1 or a control siRNA into N2a cells and analyzed mRNA levels 24 h after transfection. The Fmr1 mRNA level was reduced to ≈5% of that in control siRNA-treated cells, whereas the levels of other untargeted mRNAs, including the two closely related Fmr1 family members, Fxr1 and Fxr2, remained statistically unchanged (Fig. 5A). Therefore, Fmr1 mRNA can be effectively and specifically reduced by using siRNA, whereas FMRP protein went down at least by half after 72 h of siRNA transfection (data not shown). Next, we tested the effect of Fmr1 down-regulation on NXF2-induced destabilization of Nxf1 mRNA. We transfected Fmr1-specific siRNA or control siRNA together with FLAG-NXF2 or empty vector into N2a cells. Seventy-two hours later, RNAs were extracted, and levels were analyzed by qRT-PCR. As expected, the expression of NXF2 resulted in the specific reduction of Nxf1 mRNA level (Fig. 5B, open bars). However, when Fmr1 was inhibited by the specific siRNA, NXF2 expression was no longer able to reduce the Nxf1 mRNA level (filled bars). This observation strongly suggests that FMRP is required for the destabilization of Nxf1 mRNA induced by NXF2.
Fig. 5.
FMRP contributes to Nxf1 mRNA destabilization induced by NXF2. (A) The mouse Fmr1 gene is specifically down-regulated by siRNA. An siRNA specific for the Fmr1 gene or a control siRNA was transfected into N2a cells. Twenty-four hours after transfection, RNAs were isolated and analyzed by qRT-PCR using primers specific for the indicated mRNAs. mRNA levels from Fmr1-specific siRNA-treated cells are shown relative to those treated with control siRNA. Each bar indicates mean ± SEM (n = 3≈6). (B) Inhibition of FMRP expression abolishes the Nxf1 mRNA destabilization effect induced by NXF2. Fmr1-specific siRNA or control siRNA, together with FLAG-NXF2 or empty vector, was transfected into N2a cells. Seventy-two hours after transfection, RNAs were extracted, and levels were measured by qRT-PCR. The levels of mRNAs from FLAG-NXF2-transfected cells are shown relative to those from empty vector-transfected cells, which were arbitrarily set as 100%. Each bar represents mean ± SEM (n = 3≈4).
Discussion
In this report, we observed that in both mouse hippocampal neurons and spermatogonial cells, where FMRP and NXF2 are predominantly expressed, the levels of NXF1 are relatively lower than in cells that do not express FMRP and NXF2. We also found that in neuronal cells both FMRP and NXF2 are especially enriched in Nxf1 mRNA-containing mRNPs. Further, by manipulating the level of NXF2 in these cells, we found that expression of NXF2 at a level close to that seen in spermatogonia led to the destabilization of Nxf1 mRNA. Importantly, the destabilization depended on FMRP because reduction of FMRP expression by siRNAs was able to abolish this effect. Based on these findings, we conclude that FMRP and NXF2 are components of Nxf1 mRNA-containing mRNPs and that they function together to regulate Nxf1 mRNA stability in neuronal cells and perhaps as well as in male germ cells.
The results from our IP experiments (Fig. 3 B and C) imply that FMRP and NXF2 are present in Nxf1 mRNA-containing mRNP complexes. Because FMRP and NXF2 interact directly (21) and because NXF2-induced Nxf1 mRNA destabilization depended on FMRP expression (Fig. 5B), we argue that it is quite likely that a fraction of NXF2 and FMRP are present in the same Nxf1 mRNA-containing particles. Because of a similar enrichment of Nxf2 mRNA in the FMRP/NXF2-containing mRNPs, albeit to a lower extent compared with Nxf1 mRNA (Fig. 3 B and C), we speculate that Nxf2 mRNA may also be a target of FMRP/NXF2-mediated regulation, and this observation warrants further investigation. Although β-adaptin and Fmr1 mRNAs were previously implicated as targets for FMRP regulation (17, 22), we failed to detect any specific enrichment of either mRNA in the FMRP/NXF2 complexes. This finding may simply suggest that Nxf1 mRNA has a higher affinity for FMRP/NXF2 than β-adaptin and Fmr1 mRNAs do. Alternatively, this discrepancy may result from the different cell/tissue sources and methodology used in detecting these interactions. Indeed, the Fmr1 mRNA was not among the 432 FMRP-associated mRNAs from mouse brain identified in the IP and microarray studies (16). At present, we do not know whether the FMRP/NXF2–Nxf1 mRNA interaction is direct or indirect. FMRP and NXF2 may each bind Nxf1 mRNA directly and independently. Alternatively, one might bind directly whereas the other is associated only indirectly through protein–protein interactions. It is also possible that both FMRP and NXF2 associate with Nxf1 mRNPs by protein–protein interactions with other yet unknown factors. Finally, FMRP has been reported to recognize some specific sequence motifs such as G-quartets and kissing loops that are present in some target mRNAs (16, 25, 26). If FMRP/NXF2 bind Nxf1 mRNA directly, it would be important to find where they bind and whether the binding site(s) contain specific sequence motifs for a better understanding of the nature of the interactions.
Our work identifies Nxf1 mRNA as a potentially highly significant target for FMRP regulation. Its encoded protein NXF1 is ubiquitously expressed in all tissue cells tested and is believed to be responsible for the nuclear export of bulk cellular mRNAs (for review, see ref. 27). Based on a recent report that Nxf1 mRNA can be alternatively spliced (28), we have chosen a primer pair that only amplifies the full-length NXF1-encoding mRNA in our qRT-PCR analysis to avoid possible complications in data interpretation. Although there have been extensive studies on the function of NXF1, much less is known about the regulation of its expression. However, the Hammarskjöld group (28) has recently demonstrated that there exists at least one alternatively spliced Nxf1 variant that encodes a truncated form of NXF1 and that the expression of this variant is regulated by the full-length NXF1. Our findings represent yet another mechanism of Nxf1 regulation. Moreover, the requirement for the coordinated function of FMRP and NXF2 may indicate neuronal-specific (perhaps also male germ cell-specific) regulation.
There have been reports of mRNA level changes that correlate with changes in FMRP expression (16, 17, 29). However, it is not clear whether the changes reported were at the RNA transcriptional or stability level. Our findings represent a clear example of FMRP regulation at the mRNA stability level. Currently, however, we do not know how this regulation occurs mechanistically. A recent study showed that some FMRP-containing neuronal RNPs are similar in organization and function to the somatic cytoplasmic RNA-processing bodies (P bodies) (30). P bodies contain multiprotein machineries involved in mRNA degradation and translational control (for review, see ref. 31). It is thus possible that some FMRP- and NXF2-containing RNPs may direct Nxf1 mRNA for regulated degradation within P bodies. Although FMRP expression is widespread, NXF2 expression is limited to terminally differentiated neuronal cells and spermatogonial cells (ref. 21 and data not shown). It is therefore not surprising that undifferentiated neuroblastoma N2a cells express FMRP and not NXF2. It is tempting to postulate that FMRP acquires a new function through interaction with NXF2 in highly specialized cells such as hippocampal neurons and spermatogonial cells where a high level NXF1 expression may be detrimental to the function of these cells. Finally, it cannot be excluded at this point that the FMRP/NXF2-mediated regulation of Nxf1 may also occur at the translational level.
It is generally accepted that mRNA nuclear export requires direct binding of export adaptor proteins that recruit NXF1, the principal export receptor, to the mRNP. After binding mRNPs, NXF1 interacts with nuclear pore components to facilitate export (for review, see ref. 27). A number of RNA-binding proteins have been identified as adaptor proteins for NXF1 (32; for review, see ref. 27). Our previous studies suggest that FMRP may function as an adaptor protein for NXF2 to promote the export of a specific subclass of mRNAs in neurons and male germ cells (21). In this work, we revealed another function of the molecular interplay between these two proteins. In summary, we propose that FMRP and NXF2 may function together not only to promote the nuclear export but also regulate the cytoplasmic fate of their target mRNAs, including the Nxf1 mRNA, and that these steps may be mechanistically coupled.
Materials and Methods
Antibodies, Peptides, Plasmids, and siRNAs.
Monoclonal anti-FMRP (AB2160; Chemicon, Temecula, CA), monoclonal anti-FLAG (F3165; Sigma, St. Louis, MO), polyclonal anti-NXF1 (sc-17311; Santa Cruz Biotechnology, Santa Cruz, CA) antibodies, and mouse preimmune IgG (PP54; Chemicon) were purchased. The polyclonal anti-NXF2 antibody and the blocking peptide for the specificity of the antibody have been described previously by us (21). The blocking peptide (sc-17311 P; Santa Cruz Biotechnology) for the specificity of anti-NXF1 was purchased. FLAG-NXF2 was created by cloning the mouse NXF2 ORF into pFLAG-CMV-2 (Sigma). The YFP expression vector (ZsYellow) was purchased from Clontech (Mountain View, CA). Mouse Fmr1-specific siRNA (L-045448-00) and the negative control siRNA (D-001810-01-05) were purchased from Dharmacon (Lafayette, CO).
Animals and Cells.
C57BL/6 mice were maintained in the TAC animal facility at the Yale School of Medicine and treated in accordance with the regulations of the Yale Institutional Animal Care and Use Committee. Mouse brain neuroblast N2a cells (ATCC, CCL-131) were purchased and maintained in DMEM supplemented with 10% FBS and antibiotics.
Cell Transfection and Protein Level Measurement.
N2a cell transfections were carried out essentially as described previously (24). To measure protein levels, cells were harvested by trypsinization, and cell pellets were collected by centrifugation at 800 × g for 2 min in a benchtop microcentrifuge. Cells were then directly lysed in 5 volumes of 2× SDS/sample buffer by heating at 100°C for 5 min with occasional vortexing. Ten to 20 μl of cell lysates were resolved on SDS/10% PAGE followed by Western blot analysis. To check NXF2 levels in the mouse testis, freshly isolated adult mouse testicular tissues were lysed in 5 volumes of 2× SDS/sample buffer by heating at 100°C for 10 min with occasional sonication, and the resulting lysates were analyzed as described above. The relative expression levels of NXF2 in the FLAG-NXF2-transfected N2a cells and in the spermatogonial cells from mouse testis were estimated based on the observation that the transfection efficiency of N2a cells was ≈50% (24) and that the percentage of spermatogonia in a total testicular cell population is ≈3% (33). Thus, if the level of NXF2 in a single transfected N2a cell equals that in a single spermatogonial cell, then the NXF2 level in a sample derived from unsorted N2a cells would be ≈17-fold that from a crude testicular lysate. For example, in Fig. 4A Top, the level of NXF2 in lane 2 is ≈8-fold of that in lane 3. From this finding we estimated that the NXF2 expression in the transfected N2a cells was ≈2-fold less than that in spermatogonia.
Indirect Immunofluorescence.
Cryostatic adult mouse testis sections were fixed for 20 min in 3% paraformaldehyde in TBS (20 mM Tris·HCl, pH 7.4/225 mM NaCl) and washed with TBS for 15 min. Autoimmunofluorescence was inhibited by treatment with 0.1% sodium borohydride in TBS for 40 min. Sections were permeabilized for 5 min in 1% SDS in TBS and washed with TBS for 15 min. Nonspecific sites were blocked by incubation with 10% BSA/10% goat serum in TBS for 1 h. Sections were then incubated with the individual first antibodies diluted in the blocking solution at 4°C overnight. The next day, sections were washed six times for 5 min each with 273 mM NaCl/0.1% BSA in TBS and then stained for 1 h with fluorescence dye-conjugated secondary antibodies (Molecular Probes, Eugene OR) according to the manufacturer's protocol. After washing with TBS for 15 min, sections were counterstained with DAPI, washed with TBS for 15 min, rinsed with ddH2O, and mounted with Vectashield (Vector Laboratories, Burlingame, CA) and visualized with an immunofluorescence microscope (Zeiss, Thornwood, NY). The staining specificity of the antibodies against NXF1 and NXF2 was confirmed by performing immunostaining in the presence of the corresponding blocking peptides during the first antibody incubation. The specificity of the anti-FMRP was confirmed by using Fmr1 knockout brain as a control (4).
Immunohistochemistry.
Adult male mice were perfused with 3% paraformaldehyde, and brains were isolated and postfixed overnight in 3% paraformaldehyde. Brains were embedded in paraffin, and immunohistochemistry experiments using the Vectastain ABC kit (PK-6100; Vector Laboratories) on 5-μm sections were carried out according to the manufacturer's instructions. To reveal nuclear staining of the proteins, nuclear counterstaining with hematoxylin was omitted. The specificity of the antibodies were verified as described above.
Immunoprecipitation, RNA Extraction, and Protein Analysis.
To prepare antibodies, 30 μl of a mixture of protein A– and G–Sepharose beads (A:G = 1:1) was incubated with 10 μl of anti-FMRP, 20 μl of anti-NXF2, 10 μl of anti-FLAG M2 antibody, 3 μl of mouse IgG or 20 μl of rabbit preimmune serum, in 500 μl of buffer H/glycerol [0.5% Triton X-100/150 mM NaCl/2.5 mM MgCl2/10 mM Tris·HCl, pH 7.4/20% (vol/vol) glycerol] at 4°C overnight. Each incubation contained ≈45 μg of IgGs. The next day, beads were washed three times with buffer H and kept on ice until use. To prepare cell lysates, N2a cells transfected with FLAG-NXF2 or empty vector were harvested by manual scraping, and cell pellets were resuspended at 3 × 107 cells per ml in freshly prepared lysis buffer [1% Triton X-100/150 mM NaCl/50 mM Tris·HCl, pH 8.0/0.5 mM PMSF/1 mM DTT/1× protease inhibitor mixture (Calbiochem, San Diego, CA)/400 units/ml RNase inhibitor]. The suspensions were incubated on ice for 20 min with occasional mixing by inversion. Lysates were cleared by centrifugation at 15,000 × g for 15 min to remove insoluble materials, and yeast tRNA was added to the cleared lysates at a final concentration of 40 μg/ml. Five hundred microliters of the lysate (equivalent to 1.5 × 107 cells) was transferred to each tube containing antibody-coated beads, and IP was carried out by rotating the tubes at 4°C for 3 h. After IP, beads were washed three times with buffer H containing 10 μg/ml yeast tRNA by adding 1 ml of the buffer and rotating the tube at 4°C for 2 min each time. RNA extraction from the beads with RNeasy mini kit from Qiagen (Valencia, CA) was carried out according to the manufacturer's instructions except that 30 μl of 55°C RNase-free water was used to elute RNA from each column and that the elute was added back to the column to elute again to increase RNA yield. Reverse transcription was performed by using an iScript cDNA synthesis kit (Bio-Rad, Hercules, CA), and cDNAs were purified with the Qiagen QIAquick PCR purification kit. To detect the FMRP and NXF2 proteins, purified IP complexes or 3% of supernatants were mixed with 2× SDS/sample buffer, and proteins were resolved by SDS/10% PAGE followed by Western blot analyses.
Real-Time PCR.
Real-time PCR was carried out on cDNA (5–50 ng) by using iQ SYBR Green (Bio-Rad) with an iCycler (Bio-Rad). All reactions were done in a 25-μl volume. Primers for the individual mouse genes were as follows: Gapdh forward, 5′-TTAGCACCCCTGGCCAAGG-3′; Gapdh reverse, 5′-CTTACTCCTTGGAGGCCATG-3′; β-actin forward, 5′-GTGGGCCGCTCTAGGCACCAA-3′; β-actin reverse, 5′-CTCTTTGATGTCACGCACGATTTC-3′; Fmr1 forward, 5′-GCCTTGCTGTTGGTGGTTAGC-3′; Fmr1 reverse, 5′-CACAACTCCTGACTTGTCCACGAT-3′; β-adaptin forward, 5′-GCATGTATACCAGGCCTACGAGAC-3′; β-adaptin reverse, 5′-TGGGCTGGCAGAAGGCT-3′; Fxr1 forward, 5′-CGTCGTAGGCGGTCTCGTAG-3′; Fxr1 reverse, 5′-ACCATTCAGGACTGCTGCTT-3′; Fxr2 forward, 5′-CGACTT-CGGCCAGTCAATTCC-3′; Fxr2 reverse, 5′-GCTTCGAGCCTGTCTGCATGC-3′; Nxf1 forward, 5′-GCAACCAACAAGCACTTGA-3′; Nxf1 reverse, 5′-CAAGAAGCGCAGGACCA-3′; Nxf2 forward, 5′-GCCTGCTTCTCCTTGTCA-3′; Nxf2 reverse, 5′-GCCCTCATTCTGGAGAGC-3′. PCR was performed by initial denaturation at 95°C for 5 min followed by 40 cycles of 30 s at 95°C, 30 s at 60°C, and 30 s at 72°C. PCR with water instead of template was used as a negative control. Specificity was verified by melting curve analysis and agarose gel electrophoresis. The threshold cycle (Ct) values of each sample were used in the post-PCR data analysis. In Fig. 3 B and C, the difference in Ct values (ΔCt) between antibody IP (Ctab) and negative control IP (Ctneg) was expressed as ΔCt = Ctab − Ctneg, and the relative fold of binding was calculated as 2−ΔCt. In Fig. 3D, the endogenous housekeeping Gapdh mRNA was used as an internal control for mRNA level normalization. ΔCt between an mRNA of interest and Gapdh was calculated, and the results are expressed as percentage of expression relative to Gapdh, which was arbitrarily assigned a value of 100. In Fig. 5B, mRNA levels were normalized against Gapdh, and the levels of mRNAs from Fmr1 siRNA-treated cells were expressed as the percentage of expression relative to those from control siRNA-treated cells. In Figs. 4B and 5B, mRNA levels from cells transfected with FLAG-NXF2 or with empty vector were first normalized against Gapdh. Then, the normalized mRNA levels from FLAG-NXF2-transfected cells were plotted as the percentage of expression relative to those from empty vector-transfected cells.
RNA Stability Assays.
Cells were transfected with FLAG-NXF2 or empty vector in a 96-well plate. Twenty-four hours later, actinomycin D was added to the cells at a final concentration of 5 μg/ml. Total RNAs were extracted, and levels were measured by qRT-PCR. Nxf1 mRNA levels were plotted after normalization against Gapdh (Fig. 4C).
Acknowledgments
We thank Kexiong Zhang for help with computer analysis and Jianwu Zhao for technical assistance. This work was supported by funding from the Department of Obstetrics, Gynecology, and Reproductive Sciences at the Yale University School of Medicine (to Y.H.).
Abbreviations
- FMRP
fragile X mental retardation protein
- IP
immunoprecipitation
- mRNP
messenger ribonucleoprotein particle
- qRT-PCR
qualitative RT-PCR
- YFP
yellow fluorescent protein.
Note Added in Proof.
Since we submitted this manuscript, another report has appeared in which it was shown that FMRP can regulate the turnover of the PSD-95 mRNA (34).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Bardoni B, Davidovic L, Bensaid M, Khandjian EW. Expert Rev Mol Med. 2006;8:1–16. doi: 10.1017/S1462399406010751. [DOI] [PubMed] [Google Scholar]
- 2.Koukoui SD, Chaudhuri A. Brain Res Rev. 2006;53:27–38. doi: 10.1016/j.brainresrev.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 3.Devys D, Lutz Y, Rouyer N, Bellocq JP, Mandel JL. Nat Genet. 1993;4:335–340. doi: 10.1038/ng0893-335. [DOI] [PubMed] [Google Scholar]
- 4.Bakker CE, Otero Y, Bontekoe C, Raghoe P, Luteijn T, Hoogeveen AT, Oostra BA, Willemsen R. Exp Cell Res. 2000;258:162–170. doi: 10.1006/excr.2000.4932. [DOI] [PubMed] [Google Scholar]
- 5.Willemsen R, Oostra BA, Bassell GJ, Dictenberg J. Ment Retard Dev Disabil. 2004;10:60–67. doi: 10.1002/mrdd.20010. [DOI] [PubMed] [Google Scholar]
- 6.Vanderklish PW, Edelman GM. Genes Brain Behav. 2005;4:360–384. doi: 10.1111/j.1601-183X.2005.00134.x. [DOI] [PubMed] [Google Scholar]
- 7.Miyashiro K, Eberwine J. Proc Natl Acad Sci USA. 2004;101:17329–17330. doi: 10.1073/pnas.0408034101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feng Y, Gutekunst C-A, Eberhart DE, Yi H, Warren ST, Hersch SM. J Neurosci. 1997;17:1539–1547. doi: 10.1523/JNEUROSCI.17-05-01539.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stefani G, Fraser CE, Darnell JC, Darnell RB. J Neurosci. 2004;24:9272–9276. doi: 10.1523/JNEUROSCI.2306-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Antar LN, Afroz R, Dictenberg JB, Carroll RC, Bassell GJ. J Neurosci. 2004;24:2648–2655. doi: 10.1523/JNEUROSCI.0099-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Antar LN, Dictenberg JB, Plociniak M, Afroz R, Bassell GJ. Genes Brain Behav. 2005;4:350–359. doi: 10.1111/j.1601-183X.2005.00128.x. [DOI] [PubMed] [Google Scholar]
- 12.Siomi H, Ishizuka A, Siomi MC. Ment Retard Dev Disabil. 2004;10:68–74. doi: 10.1002/mrdd.20011. [DOI] [PubMed] [Google Scholar]
- 13.Plante I, Provost P. J Biomed Biotechnol. 2006;2006:1–7. doi: 10.1155/JBB/2006/16806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bear MF. Genes Brain Behav. 2005;4:393–398. doi: 10.1111/j.1601-183X.2005.00135.x. [DOI] [PubMed] [Google Scholar]
- 15.Darnell JC, Jensen KB, Jin P, Brown V, Warren ST, Darnell RB. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- 16.Brown V, Jin P, Ceman S, Darnell JC, O'Donnell WT, Tenenbaum SA, Jin X, Feng Y, Wilkinson KD, Keene JD, et al. Cell. 2001;107:477–487. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- 17.Miyashiro KY, Beckel-Mitchener A, Purk TP, Becker KG, Barret T, Liu L, Carbonetto S, Weiler IJ, Greenough WT, Eberwine J. Neuron. 2003;37:417–431. doi: 10.1016/s0896-6273(03)00034-5. [DOI] [PubMed] [Google Scholar]
- 18.Darnell JC, Mostovetsky O, Darnell RB. Gene Brain Behav. 2005;4:341–349. doi: 10.1111/j.1601-183X.2005.00144.x. [DOI] [PubMed] [Google Scholar]
- 19.Davidovic L, Bechara E, Gravel M, Jaglin XH, Tremblay S, Sik A, Bardoni B, Khandjian EW. Hum Mol Genet. 2006;15:1525–1538. doi: 10.1093/hmg/ddl074. [DOI] [PubMed] [Google Scholar]
- 20.Bardoni B, Mandel JL. Curr Opin Gene Dev. 2002;12:284–293. doi: 10.1016/s0959-437x(02)00300-3. [DOI] [PubMed] [Google Scholar]
- 21.Lai D, Sakkas D, Huang Y. RNA. 2006;12:1–4. doi: 10.1261/rna.94306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown V, Small K, Lakkis L, Feng Y, Gunter C, Wilkinson KD, Warren ST. J Biol Chem. 1998;273:15521–15527. doi: 10.1074/jbc.273.25.15521. [DOI] [PubMed] [Google Scholar]
- 23.Kirkpatrick LL, Mcllwain KA, Nelson DL. Genomics. 2001;78:169–177. doi: 10.1006/geno.2001.6667. [DOI] [PubMed] [Google Scholar]
- 24.Zhang M, Guller S, Huang Y. Placenta. 2007 doi: 10.1016/j.placenta-2007-01-012. [DOI] [PubMed] [Google Scholar]
- 25.Darnell JC, Fraser CE, Mostovetsky O, Stefani G, Jones TA, Eddy SR, Darnell RB. Genes Dev. 2005;19:903–918. doi: 10.1101/gad.1276805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Darnell JC, Warren ST, Darnell RB. Ment Retard Dev Disabil Res Rev. 2004;10:49–52. doi: 10.1002/mrdd.20008. [DOI] [PubMed] [Google Scholar]
- 27.Dimaano C, Ullman KS. Mol Cell Biol. 2004;24:3069–3076. doi: 10.1128/MCB.24.8.3069-3076.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Y, Bor Y-C, Misawa Y, Xue Y, Rekosh D, Hammarskjöld M-L. Nature. 2006;443:234–237. doi: 10.1038/nature05107. [DOI] [PubMed] [Google Scholar]
- 29.Xu K, Bogert BA, Li W, Su K, Lee A, Gao FB. Curr Biol. 2004;14:1025–1034. doi: 10.1016/j.cub.2004.05.055. [DOI] [PubMed] [Google Scholar]
- 30.Barbee S, Estes P, Cziko A, Hillebrand J, Luedeman R, Coller J, Howlett I, Geng C, Ueda R. Neuron. 2006;52:997–1009. doi: 10.1016/j.neuron.2006.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kiebler MA, Bassell GJ. Neuron. 2006;51:685–690. doi: 10.1016/j.neuron.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 32.Huang Y, Gattoni R, Stevenin J, Steitz JA. Mol Cell. 2003;11:837–843. doi: 10.1016/s1097-2765(03)00089-3. [DOI] [PubMed] [Google Scholar]
- 33.Mays-Hoopes LL, Bolen J, Singer-Sam J. Biol Reprod. 1995;53:1003–1011. doi: 10.1095/biolreprod53.5.1003. [DOI] [PubMed] [Google Scholar]
- 34.Zalfa F, Eleuteri B, Dickson KS, Mercaldo V, De Rubeis S, di Penta A, Tabolacci E, Chiurazzi P, Neri G, Grant SG, Bagni C. Nat Neurosci. 2007;10:578–587. doi: 10.1038/nn1893. [DOI] [PMC free article] [PubMed] [Google Scholar]