Abstract
Aggregation of proteins and peptides is a widespread and much-studied problem, with serious implications in contexts ranging from biotechnology to human disease. An understanding of the proliferation of such aggregates under specific conditions requires a quantitative knowledge of the kinetics and thermodynamics of their formation; measurements that to date have remained elusive. Here, we show that precise determination of the growth rates of ordered protein aggregates such as amyloid fibrils can be achieved through real–time monitoring, using a quartz crystal oscillator, of the changes in the numbers of molecules in the fibrils from variations in their masses. We show further that this approach allows the effect of other molecular species on fibril growth to be characterized quantitatively. This method is widely applicable, and we illustrate its power by exploring the free-energy landscape associated with the conversion of the protein insulin to its amyloid form and elucidate the role of a chemical chaperone and a small heat shock protein in inhibiting the aggregation reaction.
Keywords: protein aggregation, quartz crystal microbalance, biosensors
It is increasingly clear that peptides and proteins are inherently susceptible to aggregation and amyloid formation (1), and that these processes underlie directly or indirectly pathological events associated with a wide range of medical disorders, including Alzheimer's disease and the prion conditions (2–4). Aggregation is also a major problem in biotechnological and pharmaceutical applications, where it interferes with the characterization and production of therapeutically active polypeptides (5). Although much has been discovered about amyloid formation (6–8), it has proved to be extremely challenging to effect quantitative in vitro measurements of the kinetics and thermodynamics of fibril growth, which are essential for developing a systematic approach to explore strategies that prevent or delay the onset of the formation of protein aggregates and to characterize the fundamental parameters governing the aggregation process (9). A range of techniques has therefore been developed to study the kinetic aspects of these events (10–13), and the prevailing methods include monitoring the increase in light scattering of a growing amyloid suspension, or the change in fluorescence or absorption of dyes that bind to amyloid (10). However, the heterogeneity of the resulting aggregation products and the present lack of detailed knowledge of their structure at the molecular level makes in some cases the quantitative interpretation of the data challenging. For example, light-scattering measurements are highly sensitive to processes other than fibril growth, such as fibril–fibril association, sedimentation of the aggregates, and the rheological properties of the suspension or gel (13). Similarly, understanding the mechanism of binding of dyes remains a topic of current research; the stoichiometry of binding is difficult to measure, and in certain cases, dyes can bind nonspecifically to nonamyloid structures (14), such as amorphous aggregates, or even to soluble native protein (10, 12). Additionally, the presence of dyes that interact with the growing fibrils can influence the kinetics of the reaction to be measured (11, 12). More recently, other methods have been developed that enable the growth of individual fibrils to be directly monitored with atomic force microscopy (AFM) (15–20), total internal reflection fluorescence microscopy (21), or experiments that detect the growth of an ensemble of fibrils by surface plasmon resonance (22) or dynamic light scattering (13). In addition, methods have emerged that concentrate on monitoring, by mass spectrometry (23) or high-performance liquid chromatography (24), the changes in the concentrations of the precursor protein as a measure of its incorporation into growing fibrils.
In this article, we describe an alternative approach to following amyloid formation and show that it is possible to measure accurately and directly the kinetics of amyloid growth by monitoring in real time the increase in the mass of the fibrils and hence establish the change in the number of protein molecules in the aggregated phase, using a method based on quartz crystal microbalance (QCM) sensors (25, 26). We then show that such quantitative measurements enable insights to be gained into the thermodynamical parameters and molecular mechanisms behind the aggregation reaction.
Results
Quantitative Measurements of Fluctuations in Fibril Mass.
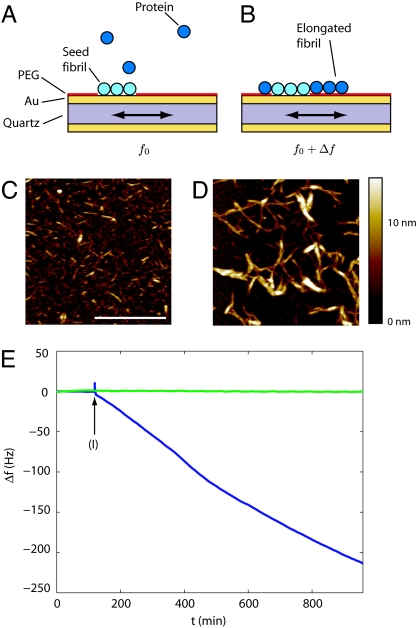
We have used the protein insulin to demonstrate the potential of micro–balance sensor-based kinetic measurements. We first created well defined sites for amyloid growth on the surface of a QCM sensor by adsorbing on to it short fibril segments prepared by sonication of preformed fibrils (Fig. 1 A and C). The rest of the exposed surface was then passivated by using an inert mono–layer (see Materials and Methods), which prevents any nonspecific protein adsorption, an essential requirement for precise measurements. When the sensor is brought into contact with a fresh solution of insulin (Fig. 1B, blue trace), individual insulin molecules bind onto the seed fibrils, resulting in a mass increase. On the other hand, in the absence of seed fibrils, no mass increase was observed even in the presence of soluble insulin (Fig. 1B, green trace), demonstrating that the mass changes directly monitor the growth of the fibrils. To obtain quantitative information on the kinetics of this process we monitored the resonance frequency f of surface shear waves on the sensor: when the mass on the surface increases by Δm the frequency decreases by Δf, as described by the Sauerbrey equation (27): Δm = −C Δfnorm, where C is the mass sensitivity constant, fnorm = fn/n is the normalized frequency, and n = 3, 5, 7 is the overtone number corresponding to the observed higher harmonics. The high sensitivity of such kinetic measurements can be seen to stem at a fundamental level from the fact that the frequency of a signal can essentially be measured with higher precision than any other physical quantity and should therefore preferentially be used, a concept highlighted, for example, in the Nobel lecture by T. W. Hänsch (28).
Fig. 1.
Growth kinetics from quartz crystal oscillator measurements. Seed fibrils deposited onto a gold surface (A) create well defined growth sites, and upon addition of protein solution the fibrils grow (B), resulting in a decrease in the resonance frequency of the quartz crystal oscillator (E, blue line); in the absence of seed fibrils (E, green line) no shift is measured. The length before (C) and after (D) growth on the gold surface from AFM images is in agreement with the frequency shift measured in E as described. The fibrils were exposed to insulin solution at the time indicated by (I). In C and D the scale bar is 1 μm, and the fibril suspension was diluted by a factor of 5 relative to the QCM measurements to facilitate visualization.
A typical measurement of the oscillation frequency shift is shown in Fig. 1E. When the surface is exposed to a control solution containing no protein, the total mass of the fibrils on the surface remains constant, and hence the oscillation frequency does not change (Fig. 1E, 0–119 min). After incubation in the protein solution at 1 mg/ml (I in Fig. 1E), Δf decreases, because of the sequential attachment of protein molecules to the fibril ends (Fig. 1E, 120–960 min). The change in hydrodynamical mass is directly proportional to the number of molecules attaching to the fibrils. In this article, we generally focus only on relative changes in the growth rates in response to changes in intrinsic or extrinsic factors; we can nevertheless estimate the order of magnitude of the absolute growth rates. For example, in Fig. 1E, we observe a frequency shift of 219 ± 1 Hz over 14 h, corresponding to a mass increase of dm/dt = 83 ± 0.2 pg/s or an approximate average growth rate of dN/dt = 8.1 · 109 ± 1.4 · 106 individual insulin molecules per second. We can estimate the surface density σ ∼ 250 ± 100 μm−2 of the seed fibrils from AFM images (Fig. 1C), and therefore our results indicate an approximate elongation rate constant of 9.2 ± 3 × 103 s−1·M−1 for 0.17 mM protein concentration, corresponding to on average one molecule attaching to a fibril every 3.1 ± 1.2 s. Calculations based on the diffusion coefficient of insulin (see Materials and Methods) suggest that approximately one in 105 molecular collisions with the end of a fibril results in attachment. We note that this rate constant is similar in magnitude to the results from previous high-temperature measurements of insulin fibril growth in solution extrapolated to room temperature (17).
To check independently these results by AFM (Fig. 1 C and D) we convert the growth rate to a length increase as a function of time: density arguments (17, 29, 30) together with the dimensions of the fibrils (30) imply that there are three to seven molecules per cross-section, and previous x-ray diffraction studies suggest that there is one molecule every 0.48 nm along the fibril axis in an amyloid fibril (31); therefore, the elongation rate of 0.32 molecules per s per fibril is equivalent to a length increase of 0.03 ± 0.01 nm/s. Based on these assumptions, over 14 h of continuing growth a total length increase of at least 1.4 ± 0.7 μm is expected, a value of similar order of magnitude than the direct observations by AFM in Fig. 1 C and D. The shorter length of fibrils in AFM compared with the estimate from the QCM measurements can be rationalized by the additional contributions to the hydrodynamic mass from water molecules hydrating the proteins and viscoelastic effects. The very slow growth rate at room temperature is still detectable because of the high sensitivity of the sensor; indeed only 5 min of growth are required for the accurate determination of the rate, such that the data in Fig. 1C suggest that a single sensor remains fully functional for the time required for >100 individual measurements.
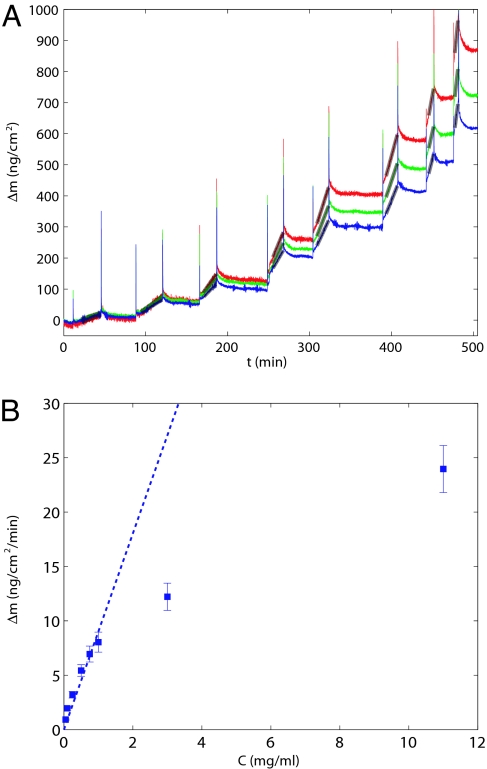
To probe the process by which protein molecules are incorporated into growing amyloid fibrils, we first examined the dependence of the growth rate on the concentration of soluble insulin as shown in Fig. 2. At low concentrations, we observed the reaction rate to increase linearly with the protein concentration, indicative of a bimolecular reaction between the fibril ends and the precursor protein in solution as has been observed in other systems (32). Interestingly, however, at high concentrations, we observed a less than linear relationship between the growth rate and protein concentration. This result is in agreement with previous measurements on the aggregation of insulin (33), and similar behavior has been reported for different amyloid systems such as the NM domain of yeast prions (32), and suggests that at higher concentration a conformational rearrangement occurring after the precursor protein has attached to the fibril becomes the rate-limiting step. Such a two-step “dock and lock” process has also been observed for the aggregation of the Aβ peptide (34, 35) and could be a common feature of the aggregation of proteins and peptides. In addition, high concentrations could promote the formation of oligomers in solution, thereby decreasing the availability of free precursor protein. In this article, we focus on the lower concentration region and probe the physico–chemical aspects associated with the attachment of protein molecules to the fibrils, resulting in amyloid growth.
Fig. 2.
Concentration dependence of fibril elongation. (A) The sensor with the seed fibrils attached was successively exposed to different concentrations of soluble insulin, resulting in different growth rates measured from the linear fits (gray lines). Three different frequency overtones were simultaneously monitored, n = 3 (red), n = 5 (green), n = 7 (blue). (B) Shown is the average (squares) and the standard error (error bars) of the slopes for the different overtone numbers as a function of the protein concentration. The dashed line shows a one parameter fit through zero to the linear portion of the data.
Thermodynamics of Amyloid Growth.
Having established the robustness of this method, we now use it to explore the thermodynamic parameters that characterize fibril elongation, such as the free-energy barrier that a protein has to overcome to transform from its solvated structure into the fibrillar state and the corresponding enthalpic and entropic contributions to this energy. The elongation rate was measured at different temperatures. Initially (Fig. 3A, 0–20 min), at a given stabilized temperature (Fig. 3B), the oscillation frequency of the system was constant. An injection of a solution containing insulin resulted in a frequency decrease with time caused by the mass loading from the addition of molecules to the growing aggregates (Fig. 3A, gray band, 20–50 min). When the protein solution was replaced with water the mass remained constant (Fig. 3A, 50 min), the temperature could then be changed (Fig. 3B), and the solution reintroduced to probe the growth (Fig. 3A, gray band, 70–100 min) of the same fibrils under the different conditions after a new equilibrium was reached (Fig. 3A, 50–70 min) at the higher temperature. Measurements of this type in Fig. 3C show that the rates of fibril growth accurately follow Arrhenius type behavior with an enthalpic activation barrier of ΔH‡ = 24.4 ± 1 kcal/mol in the temperature range tested. This observation does not exclude deviations from Arrhenius behavior over wider temperature ranges, as can be the case for protein folding (36). The value of the activation energy, equivalent to the bond energies of approximately five hydrogen bonds, suggests that significant conformational remodeling is involved in the conversion to the amyloid state. Transition-state theory predicts (13, 36) that the growth of the number of monomers N incorporated into fibrils is exponentially modulated by the Gibbs free energy ΔG‡ of the transition state between the native structure of the protein and its configuration in the fibrillar state: dN/dt = Γexp(−ΔG‡/kBT), where Γ is a kinetic prefactor. In general, in this article we analyze relative changes in the free energy, and therefore the value of Γ does not appear. However, following a procedure described in ref. 13 and outlined in Materials and Methods, an order of magnitude value can be given, enabling us to estimate the absolute value for the Gibbs free energy of activation ΔG = −1/(kBT)ln(dN/dt/Γ) = 6.1 ± 2 kcal/mol. This allows for further deconvolution of the elongation barrier into the enthalpic ΔH‡ = 24.4 ± 1 kcal/mol and entropic TΔS‡ = −16.8 ± 2 kcal/mol contributions. The data reveal that the rate of insulin aggregation is controlled by a competition between two effects of similar orders of magnitude: the process is entropically favorable but enthalpically unfavorable. A similar conclusion has been drawn for a different type of approach with the Aβ peptide (13), suggesting that such a competition could be a general feature of protein aggregation, as is the case for protein folding (36). Interestingly, values for some of the thermodynamical parameters of human insulin aggregation have recently been determined in solution by using light scattering techniques (37), and the values for ΔG, ΔH, and TΔS are within 5–30% of the present values we observed with the QCM technique, suggesting that the growth of amyloid near a surface is very similar to the corresponding process occurring in solution.
Fig. 3.
Temperature dependence of protein aggregation kinetics. (A and B) Growth rates (A) at different temperatures (B) were extracted from the linear fits (gray, A) of the mass loading as a function of time in the growth phases, and three different overtones were simultaneously monitored, n = 3 (red), n = 5 (green), n = 7 (blue). (C) Shown is the average (squares) and the standard error (error bars) of the slopes for the different overtone numbers as a function of the temperature. The frequency shifts during and just after temperature change are not related to mass change, and measurements were only performed after equilibration at the new temperature (gray vertical bands) as described.
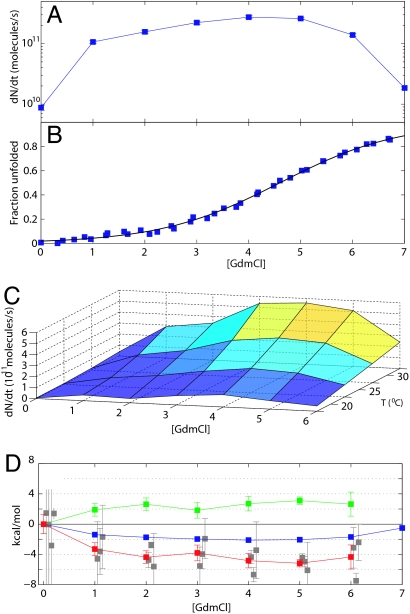
Another fundamental property we can now measure quantitatively is the dependence of the aggregation rate on denaturant concentration, thereby providing a direct analogy to experiments on protein folding (36). Here, we measure the rate of fibril elongation at a fixed temperature while successively introducing an increasing concentration of the denaturing agent guanidinium chloride (GdmCl) into the growth solution. We find that the aggregation rate in Fig. 4A initially increases significantly with denaturant concentration, reaches a maximum, and then starts to decrease. To relate this effect to the changes in the structure of the protein in solution, we determined, using equilibrium unfolding monitored by CD spectroscopy (see Materials and Methods), the fraction of protein molecules that are unfolded under the conditions used for the kinetic measurements, as shown in Fig. 4B. This analysis reveals that the maximum of the growth rate coincides approximately with the midpoint of the unfolding at 4 M GdmCl.
Fig. 4.
Denaturant-dependent acceleration and deceleration of amyloid growth. (A) The growth rate of one set of fibrils on the QCM sensor was probed under increasing concentrations of GmdCl. (B) The maximum in the rate coincides approximately with the midpoint of the unfolding transition as monitored by CD. (C and D) Additionally, varying the temperature results in multidimensional landscapes for aggregation rates (C), from which the evolution of the enthalpic (ΔΔH‡, red) and entropic (TΔΔS‡, green) contributions to change in the free energy barrier (ΔΔG‡, blue) can be computed (D). (D) The activation energies were measured independently three times (gray squares), and the average was taken (red squares). Error bar indicates standard error. Additionally for 0 M GdmCl, the value from Fig. 2C is shown. The growth rates are shown here as nominal numbers of molecules converted from the total change in hydrodynamic mass as described; this conversion does not influence the thermodynamical parameters (D).
The elongation rate increase at low denaturant concentration (0–3 M GdmCl) indicates that the species competent for fibril formation is not the native state under these conditions, but one that is at least partially unfolded (38, 39). In contrast, at higher denaturant concentrations we expect two main factors together to lead to a decrease in the rates: first, the destabilization of the intramolecular interactions caused by the presence of denaturant will also decrease the strength of the intermolecular interactions driving the aggregation process (40), and second, the probability of forming critical contacts for the aggregation reaction will decrease with the increasing radius of gyration (41) of GdmCl-induced unfolding. In accordance with our measurements in Fig. 4A, these competing effects lead to a rate profile that first rises to a maximum and then decreases again, in agreement with previous predictions from considerations based on molecular dynamics simulations (40). These results are also consistent with the observation that the lag phase before fibril formation that is seen for unseeded insulin solutions can be reduced by the presence of GdmCl (42). However, as our measurements are specifically sensitive to the elongation process, we can uniquely separate the relative influences of destabilization on the nucleation vs. elongation processes. Indeed lag times (42), which are mainly sensitive to the nucleation rate, are minimal for less strongly destabilizing conditions (1–2 M GdmCl) than for which we measure a maximal elongation rate (4 M GdmCl). Two factors could underlie these differences: first, the nucleation step is likely to involve a larger number of critical intermolecular contacts than the elongation process and would therefore be preferentially disrupted by the presence of GdmCl, and second, the conformational requirements for nucleation could involve a more native-like precursor (43), as opposed to the elongation of the fibrils, which is more efficient for a more unfolded structure.
The approach described in this article allows the growth of the same ensemble of fibrils to be followed under a variety of conditions simply by exchanging the solution in contact with the fibrils. This is of great significance because of the complexity in reproducing different samples of fibrils that have identical properties. With our approach it is now possible, for example, to map out multidimensional aggregation landscapes, such as that shown in Fig. 4C, by simply repeating the steps illustrated in Fig. 3 for a range of denaturant concentrations and temperatures. From such data, we can then follow how the thermodynamical parameters of the system underlying the aggregation under these conditions are affected by the presence of denaturant in the solution: the Arrhenius plots from the data in Fig. 4C yield the change in the enthalpic barrier ΔΔH‡([GdmCl]), and the values of the rates yield the change in the Gibbs free energy ΔΔG‡([GdmCl]) and the entropic contribution TΔΔS‡([GdmCl]) as shown in Fig. 4D. The change in the Gibbs free energy (2.1 ± 0.1 kcal/mol) is comparable with the total unfolding free-energy barrier ΔGN-U = 2.5 ± 0.5 kcal/mol from Fig. 4B, suggesting that the rate of fibril formation, through ΔG‡ is directly linked to the stability of the folded state of the precursor protein through ΔGN-U.
Effect of Chemical and Small Heat Shock Protein (sHsp) Chaperones on Aggregation Rates.
The power of the QCM approach also allows us to explore perturbations to the kinetics of amyloid growth resulting from the presence of additional components and to study their interactions with the molecular species involved in aggregation reaction. Of particular interest from a biological perspective is the inhibition of the growth of amyloid structures, and we have focused on the effects of a naturally occurring osmolyte trimethylamine N-oxide (TMAO) and a sHsp αB-crystallin.
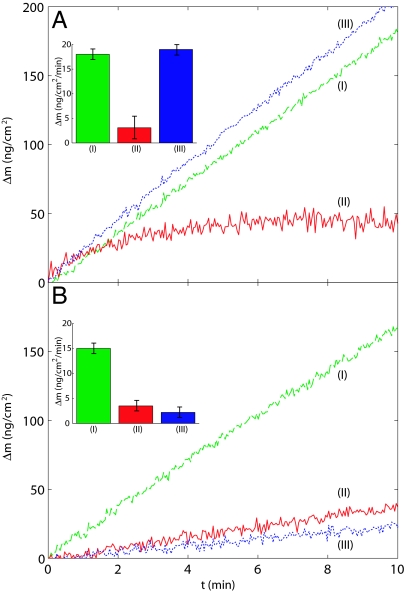
Organic osmotic solute (osmolyte) systems such as TMAO have been shown to have an important role in a variety of organisms (44) in protecting biologically functional forms of proteins, particularly under conditions where severe environmental stress such as high temperature or the presence of urea are involved (44, 45). We set out to investigate the question of how this stabilizing effect of such osmolytes, often called chemical chaperones, influences amyloid formation; to this effect, fibrils were deposited onto the sensor surface and initially exposed to a solution of insulin (I in Fig. 5A). The solution was then exchanged to one containing 1 M TMAO in addition to insulin. The rate of mass increase, (II in Fig. 5A) under these conditions was <25% of that in the absence of the chemical chaperone, suggesting that the presence of TMAO prevented the efficient transition of the soluble insulin into its fibrillar form. A similar trend was observed in parallel growth experiments carried out in solution and monitored with turbidity measurements. Osmolytes are known to enhance the thermodynamical stability of native states through the entropic penalty associated with preferential hydration of partially denatured regions of the protein (44–46), thereby driving the refolding process. Given that aggregation requires partially unfolded states (data in Fig. 4), we can attribute the origin of the inhibition to this fact. Finally, when an insulin solution without TMAO is again brought into contact with the fibrils, the growth resumes at the initial rate (Fig. 5A Inset), showing that the inhibition of the aggregation reaction by TMAO is caused predominantly by interactions with the soluble precursor proteins and that no changes in the ability of the fibrils to seed the elongation reaction persist after the removal of the chemical chaperone.
Fig. 5.
Inhibition of amyloid growth by a chemical chaperone and a sHsp. (A) Insulin fibrils were first exposed to an insulin solution in 50 mM glycine buffer, pH 2.5 and 100 mM NaCl (I, green dashed line). The solution was replaced by a mixture containing the same amount of insulin but in addition 1 M TMAO (II, red solid line), and finally a solution of insulin in buffer alone was injected (III, blue dotted line), allowing normal growth to resume. (Inset) The growth rates observed during the successive stages are shown. (B) The effect of sHsp on fibril growth was probed by exposing the fibrils first to an insulin solution in buffer (I, green dashed line) as in A and then to a mixture of 0.5:1 molar ratio of sHsp to insulin (II, red solid line). Finally, a solution of insulin in glycine buffer was introduced (III, blue dotted line) and the resulting low growth rate demonstrates the inactivation of the growth sites by the sHsp molecules. Kinetic data are shown starting from 120 s after the injection of the solutions.
A different picture presents itself when we probe the effect of αB-crystallin on fibril growth. αB-crystallin is a representative example of a common class of molecular chaperone, the sHsp, and has been shown to bind to exposed hydrophobic regions of partially unfolded proteins, and in certain cases to inhibit the formation of amyloid fibrils (47, 48). Our kinetic data in Fig. 5B clearly demonstrate that the presence of 25 μM of sHsp (a 0.5:1 molar ratio of sHsp to insulin) has an inhibitory effect similar in magnitude to that of 1 M TMAO; however, contrary to the case of TMAO, once the fibrils have been exposed to sHsp, they lose their ability to efficiently act as growth sites for further amyloid deposition. This effect can be visualized in the kinetic trace (III in Fig. 5B), where the growth rates of fibrils previously exposed to sHsp (II in Fig. 5B) still are only <25% of the rates observed for fresh fibrils. This observation suggests that sHsp chaperones can inhibit fibril elongation in deactivating the growth sites by binding to them and thereby reducing their ability to elongate. Interestingly, such a binding effect has been reported under the same conditions for a different fibril system, where the presence of bound αB-crystallin molecules was found to protect the fibrils from depolymerization under conditions that normally result in disaggregation (47). This mechanism of inhibition of the elongation of fibrils is of particular interest, as molecular chaperones are often assumed to act simply by binding to aggregation–prone species present in the solution phase. More generally, the present methodology should allow such phenomena to be investigated in molecular detail and could be a valuable approach for accurate high-throughput screening of agents that are capable of binding to amyloid fibrils and inhibiting or otherwise partaking in their elongation.
Discussion
In conclusion, the experiments discussed here demonstrate that highly quantitative measurements of protein aggregation rates can be made and shed light on the fundamental kinetic and thermodynamical parameters that govern this process. Furthermore, the approach of measuring amyloid growth kinetics with a quartz crystal oscillator overcomes some of the major limitations and crucial problems of traditional methods, in particular, through the elimination of effects associated with the formation of amorphous aggregates and the nonstoichiometric nature of dye binding. Moreover, this technique allows fundamentally new types of experiments to be carried out through which the growth of an identical ensemble of fibrils can be monitored under different conditions, thereby enabling systematic in vitro investigation of factors affecting the growth and potential degradation of amyloid aggregates. Such reactions are of great consequence in, for example, the discovery of the nature of interactions within the cellular environment and the search for potential drugs to block or reverse the formation of amyloid plaques. In the present case, we have used this method to show that the rate of fibril formation is directly linked to the stability of the folded state of the precursor protein, and we have been able to characterize differences in the mechanisms by which sHsps and chemical chaperones can inhibit fibril growth.
Materials and Methods
Sensor Functionalization.
Insulin amyloid fibrils were formed from an acid (pH 2.0) aqueous solution of bovine insulin (Sigma, Dorset, U.K.) at 10 mg/ml heated at 60°C for 24 h and left at room temperature for 7 days. By using a probe sonicator, the length distribution of the fibrils, first diluted 1:500 into water at pH 2.0, was reduced to ≈100 nm as shown in Fig. 1C, and the resulting seed fibril fragments were deposited onto the gold QCM sensor surface. The seed fibrils were left to adsorb onto the surface in a controlled environment of 100% humidity for 60 min. Then, the exposed gold surface not covered by fibrils was passivated with an inert PEG thiol [CH3O(CH2CH2O)6SH; Polypure, Oslo, Norway] monolayer, which was grown by replacing the seed solution by a 0.5% solution of PEG-thiol in water adjusted to pH 2.0 and incubated for 60 min. Finally, the sensor was inserted into the QCM, and the microfluidic system including the sensor was flushed repeatedly with an acid (pH 2.0) aqueous solution and left to equilibrate for 12 h. We used a D300 instrument (Q-Sense, Västra Frölunda, Sweden) to monitor the frequency shifts, the resonance frequency was recorded at several harmonics (5, 15, 25, and 35 MHz) simultaneously, and the sensors used were QSX 301-Standard Gold crystals (Q-Sense) with a frequency/mass sensitivity coefficient of 17.7 ng/cm2 per Hz. The temperature in the 80-μl chamber was stable to within 0.05°C. Growth kinetics were acquired by using insulin dissolved into water adjusted to pH 2.0 with dilute hydrochloric acid, except for the growth inhibition studies where conditions following ref. 47 were used (50 mM glycine buffer at pH 2.5 and 100 mM NaCl). For the temperatures, measurement times, and average lengths of fibrils used in this study, fracturing (17) is not expected to occur.
AFM.
The seed fibrils were deposited onto two identical flat gold substrates, and the exposed areas were passivated as for the sensor preparation steps described above. Then one of the substrates was dried immediately, and the other one was incubated in 1 mg/ml aqueous insulin solution at pH 2.0 for 14 h at room temperature, then dried and imaged in tapping mode using a PicoPlus AFM (Molecular Imaging, Tempe, AZ).
Measurement of the Temperature Dependence of Growth Rates.
The growth rates of amyloid fibrils were measured from linear fits to the mass loading data while the fibrils were exposed to an insulin solution at 1 mg/ml. The insulin solution was replaced by water at pH 2.0 before the temperature was changed to stop the fibril growth, and an equilibration period of 45–120 min was required for the oscillation frequency to stabilize at the new temperature. The measurements of the activation energy ΔH‡ from the Arrhenius plots as described enable the extrapolation to room temperature kRT of previous high-temperature measurements kT of insulin fibril growth: kRT = kT exp(−ΔH‡/kB(1/TRT − 1/T)).
Calculation of the Kinetic Prefactor.
An order of magnitude estimate for the prefactor Γ can be given by following ref. 13: Γ = 103 c NA rreac F D, where c is the protein concentration in the solution, NA is the Avogadro number, rreac is the encounter volume of the order of 10−27 m3, D = 1.0 × 10−10 m2/s the diffusion coefficient of insulin (49), and F is the total number of fibrils per cm2 on the sensor that can be estimated from AFM images (Fig. 1C) to be F = 2.5 ± 1 × 1010. There are two ends per fibril available for the attachment of molecules, and as the fibrils are on the surface the space angle available for the molecules to approach the fibrils is reduced from 4π to 2π.
Measurement of the Denaturant Dependence of the Growth Rates.
Insulin solutions at 1 mg/ml and pH 2.0 were prepared with increasing (0–7 M) amounts of GdmCl and left to equilibrate for 12–24 h at room temperature. Before the insulin/GdmCl mixture was injected into the QCM, a solution of GdmCl at the corresponding concentration was used first to ensure a flat baseline. In this way, all of the concentrations from 0 to 7 M were measured. The temperature was then changed as shown in Fig. 3C, and the measurements series was repeated on the same sensor for three different temperatures. The Arrhenius plots for different temperatures but fixed GdmCl concentration were used to evaluate the enthalpic activation barrier, and the entropic contribution was extracted from the kinetic prefactor, as described. The error bars on the enthalpic barrier were calculated from the R2 values of the fit to the Arrhenius equation. Because of the low growth rates and the resulting low signal-to-noise ratio in 7 M GdmCl, we were not able to determine the activation energy for this concentration.
Analysis of Equilibrium Unfolding.
Insulin at a final concentration of 1 mg/ml was incubated in GdmCl solutions adjusted to pH 2 with HCl for 12–24 h at room temperature to allow for equilibration. CD measurements were acquired on a spectropolarimeter (Jasco, Easton, MD) using a thermostatically controlled 0.2-mm pathlength cuvette at 25°C. Measurements were recorded at 222 nm with the signal averaged over 60 s. The equilibrium unfolding data were fitted to a two-state model, which assumes the following dependence of the observed signal Sobs on denaturant concentration [D]:
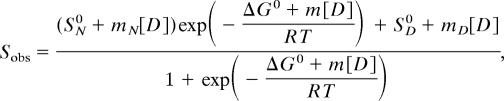 |
where SN0 and SD0 are the intercepts, mn and mD are the slopes of the pretransition and posttransition regimes, T is the absolute temperature in Kelvin, and R is the ideal gas constant.
TMAO and αB-crystallin Induced Inhibition of Amyloid Growth.
Experiments were performed under conditions following ref. 47: 50 mM glycine buffer at pH 2.5 and 100 mM NaCl. The higher ionic strength of the solution resulted in an increase of the growth rates when compared with experiments in 10 mM HCl. Bovine insulin was used at a concentration of 0.3 mg/ml, human recombinant αB-crystallin was used at 1.2 mg/ml (corresponding to a molar ratio of 0.5:1 of αB-crystallin to insulin), and TMAO was used at 1 M. Human recombinant αB-crystallin was expressed and purified as described (50).
Acknowledgments
We thank Scott Buckley, John A. Carver, and Chris Waudby for helpful discussions and Heath Ecroyd for help with the preparation and purification of the αB-crystallin. This work was supported by the Interdisciplinary Research Collaboration in Nanotechnology. W.M.S. is a Cambridge Overseas Trust Scholar. G.L.D. is a C.J. Martin Fellow of the National Health and Medical Research Council, Australia. S.A is supported by the Human Frontier Science Program. C.M.D. is supported, in part, by grants from the Wellcome Trust and the Leverhulme Trust.
Abbreviations
- QCM
quartz crystal microbalance
- ATM
atomic force microscopy
- GdmCl
guanidinium chloride
- TMAO
trimethylamine N-oxide
- sHsp
small heat shock protein.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Dobson CM. Nature. 2003;426:884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- 2.Pepys MB. Philos Trans R Soc London B. 2001;356:203–210. doi: 10.1098/rstb.2000.0766. discussion 210–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohen FE, Kelly JW. Nature. 2003;426:905–909. doi: 10.1038/nature02265. [DOI] [PubMed] [Google Scholar]
- 4.Aguzzi A, Haass C. Science. 2003;302:814–818. doi: 10.1126/science.1087348. [DOI] [PubMed] [Google Scholar]
- 5.Fowler SB, Poon S, Muff R, Chiti F, Dobson CM, Zurdo J. Proc Natl Acad Sci USA. 2005;102:10105–10110. doi: 10.1073/pnas.0501215102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nelson R, Sawaya MR, Balbirnie M, Madsen AØ, Riekel C, Grothe R, Eisenberg D. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krishnan R, Lindquist SL. Nature. 2005;435:765–772. doi: 10.1038/nature03679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanaka M, Collins SR, Toyama BH, Weissman JS. Nature. 2006;442:585–589. doi: 10.1038/nature04922. [DOI] [PubMed] [Google Scholar]
- 9.Dobson CM. Philos Trans R Soc London B. 2001;356:133–145. doi: 10.1098/rstb.2000.0758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nilsson MR. Methods. 2004;34:151–160. doi: 10.1016/j.ymeth.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 11.Demaimay R, Harper J, Gordon H, Weaver D, Chesebro B, Caughey B. J Neurochem. 1998;71:2534–2541. doi: 10.1046/j.1471-4159.1998.71062534.x. [DOI] [PubMed] [Google Scholar]
- 12.Khurana R, Uversky VN, Nielsen L, Fink AL. J Biol Chem. 2001;276:22715–22721. doi: 10.1074/jbc.M011499200. [DOI] [PubMed] [Google Scholar]
- 13.Kusumoto Y, Lomakin A, Teplow DB, Benedek GB. Proc Natl Acad Sci USA. 1998;95:12277–12282. doi: 10.1073/pnas.95.21.12277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Linder E, Lehto VP, Virtanen I. Acta Pathol Microbiol Scand A. 1979;87:299–306. doi: 10.1111/j.1699-0463.1979.tb00056.x. [DOI] [PubMed] [Google Scholar]
- 15.Goldsbury C, Kistler J, Aebi U, Arvinte T, Cooper GJ. J Mol Biol. 1999;285:33–39. doi: 10.1006/jmbi.1998.2299. [DOI] [PubMed] [Google Scholar]
- 16.Goldsbury C, Frey P, Olivieri V, Aebi U, Müller SA. J Mol Biol. 2005;352:282–298. doi: 10.1016/j.jmb.2005.07.029. [DOI] [PubMed] [Google Scholar]
- 17.Smith JF, Knowles TPJ, Dobson CM, Macphee CE, Welland ME. Proc Natl Acad Sci USA. 2006;103:15806–15811. doi: 10.1073/pnas.0604035103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harper JD, Wong SS, Lieber CM, Lansbury PT. Biochemistry. 1999;38:8972–8980. doi: 10.1021/bi9904149. [DOI] [PubMed] [Google Scholar]
- 19.Green JD, Goldsbury C, Kistler J, Cooper GJS, Aebi U. J Biol Chem. 2004;279:12206–12212. doi: 10.1074/jbc.M312452200. [DOI] [PubMed] [Google Scholar]
- 20.Hoyer W, Cherny D, Subramaniam V, Jovin TM. J Mol Biol. 2004;340:127–139. doi: 10.1016/j.jmb.2004.04.051. [DOI] [PubMed] [Google Scholar]
- 21.Ban T, Hoshino M, Takahashi S, Hamada D, Hasegawa K, Naiki H, Goto Y. J Mol Biol. 2004;344:757–767. doi: 10.1016/j.jmb.2004.09.078. [DOI] [PubMed] [Google Scholar]
- 22.Hasegawa K, Ono K, Yamada M, Naiki H. Biochemistry. 2002;41:13489–13498. doi: 10.1021/bi020369w. [DOI] [PubMed] [Google Scholar]
- 23.Larson JL, Ko E, Miranker AD. Protein Sci. 2000;9:427–431. doi: 10.1110/ps.9.2.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Nuallain B, Shivaprasad S, Kheterpal I, Wetzel R. Biochemistry. 2005;44:12709–12718. doi: 10.1021/bi050927h. [DOI] [PubMed] [Google Scholar]
- 25.Dobson CM, Knowles TP, Welland ME. United Kingdom Patent Appl. 2006 0609382.7. [Google Scholar]
- 26.Okuno H, Mori K, Jitsukawa T, Inoue H, Chiba S. Chem Biol Drug Des. 2006;68:273–275. doi: 10.1111/j.1747-0285.2006.00446.x. [DOI] [PubMed] [Google Scholar]
- 27.Sauerbrey G. Z Phys. 1959;155:206–222. [Google Scholar]
- 28.Hänsch TW. Annalen Physik. 2006;15:627–652. [Google Scholar]
- 29.Jiménez JL, Guijarro JI, Orlova E, Zurdo J, Dobson CM, Sunde M, Saibil HR. EMBO J. 1999;18:815–821. doi: 10.1093/emboj/18.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiménez JL, Nettleton EJ, Bouchard M, Robinson CV, Dobson CM, Saibil HR. Proc Natl Acad Sci USA. 2002;99:9196–9201. doi: 10.1073/pnas.142459399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sunde M, Serpell L, Bartlam M, Fraser PE, Pepys MB, Blake C. J Mol Biol. 1997;273:729–739. doi: 10.1006/jmbi.1997.1348. [DOI] [PubMed] [Google Scholar]
- 32.Collins SR, Douglass A, Vale RD, Weissman JS. PLoS Biol. 2004;2:1582–1590. doi: 10.1371/journal.pbio.0020321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rogers SS, Krebs MRH, Bromley EHC, van der Linden E, Donald AM. Biophys J. 2006;90:1043–1054. doi: 10.1529/biophysj.105.072660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nguyen PH, Li MS, Stock G, Straub JE, Thirumalai D. Proc Natl Acad Sci USA. 2007;104:111–116. doi: 10.1073/pnas.0607440104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Esler WP, Stimson ER, Jennings JM, Vinters HV, Ghilardi JR, Lee JP, Mantyh PW, Maggio JE. Biochemistry. 2000;39:6288–6295. doi: 10.1021/bi992933h. [DOI] [PubMed] [Google Scholar]
- 36.Fersht A. Structure and Mechanism in Protein Science. New York: Freeman; 1999. [Google Scholar]
- 37.Mauro M, Craparo EF, Podestà A, Bulone D, Carrotta R, Martorana V, Tiana G, Biagio PLS. J Mol Biol. 2007;366:258–274. doi: 10.1016/j.jmb.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 38.Uversky VN, Fink AL. Biochim Biophys Acta. 2004;1698:131–153. doi: 10.1016/j.bbapap.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 39.Lai Z, Colón W, Kelly JW. Biochemistry. 1996;35:6470–6482. doi: 10.1021/bi952501g. [DOI] [PubMed] [Google Scholar]
- 40.Klimov DK, Straub JE, Thirumalai D. Proc Natl Acad Sci USA. 2004;101:14760–14765. doi: 10.1073/pnas.0404570101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hall D, Hirota N, Dobson CM. J Mol Biol. 2005;351:195–205. doi: 10.1016/j.jmb.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 42.Ahmad A, Millett IS, Doniach S, Uversky VN, Fink AL. Biochemistry. 2003;42:11404–11416. doi: 10.1021/bi034868o. [DOI] [PubMed] [Google Scholar]
- 43.Bouchard M, Zurdo J, Nettleton EJ, Dobson CM, Robinson CV. Protein Sci. 2000;9:1960–1967. doi: 10.1110/ps.9.10.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yancey PH, Clark ME, Hand SC, Bowlus RD, Somero GN. Science. 1982;217:1214–1222. doi: 10.1126/science.7112124. [DOI] [PubMed] [Google Scholar]
- 45.Wang A, Bolen DW. Biochemistry. 1997;36:9101–9108. doi: 10.1021/bi970247h. [DOI] [PubMed] [Google Scholar]
- 46.Devlin GL, Parfrey H, Tew DJ, Lomas DA, Bottomley SP. Am J Respir Cell Mol Biol. 2001;24:727–732. doi: 10.1165/ajrcmb.24.6.4407. [DOI] [PubMed] [Google Scholar]
- 47.Raman B, Ban T, Sakai M, Pasta SY, Ramakrishna T, Naiki H, Goto Y, Rao CM. Biochem J. 2005;392:573–581. doi: 10.1042/BJ20050339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Treweek TM, Morris AM, Carver JA. Aust J Chem. 2003;56:357–367. [Google Scholar]
- 49.Kamholz AE, Schilling EA, Yager P. Biophys J. 2001;80:1967–1972. doi: 10.1016/S0006-3495(01)76166-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Horwitz J, Huang QL, Ding L, Bova MP. Methods Enzymol. 1998;290:365–383. doi: 10.1016/s0076-6879(98)90032-5. [DOI] [PubMed] [Google Scholar]