Abstract
Copy-number variants (CNVs) are an abundant form of genetic variation in humans. However, approaches for determining exact CNV breakpoint sequences (physical deletion or duplication boundaries) across individuals, crucial for associating genotype to phenotype, have been lacking so far, and the vast majority of CNVs have been reported with approximate genomic coordinates only. Here, we report an approach, called BreakPtr, for fine-mapping CNVs (available from http://breakptr.gersteinlab.org). We statistically integrate both sequence characteristics and data from high-resolution comparative genome hybridization experiments in a discrete-valued, bivariate hidden Markov model. Incorporation of nucleotide-sequence information allows us to take into account the fact that recently duplicated sequences (e.g., segmental duplications) often coincide with breakpoints. In anticipation of an upcoming increase in CNV data, we developed an iterative, “active” approach to initially scoring with a preliminary model, performing targeted validations, retraining the model, and then rescoring, and a flexible parameterization system that intuitively collapses from a full model of 2,503 parameters to a core one of only 10. Using our approach, we accurately mapped >400 breakpoints on chromosome 22 and a region of chromosome 11, refining the boundaries of many previously approximately mapped CNVs. Four predicted breakpoints flanked known disease-associated deletions. We validated an additional four predicted CNV breakpoints by sequencing. Overall, our results suggest a predictive resolution of ≈300bp. This level of resolution enables more precise correlations between CNVs and across individuals than previously possible, allowing the study of CNV population frequencies. Further, it enabled us to demonstrate a clear Mendelian pattern of inheritance for one of the CNVs.
Keywords: copy number polymorphism, human genome variation, structural variants
It was recently established that copy-number variants (CNVs), kilobase- to megabase-sized deletions and duplications, are abundant in healthy individuals (1–3) and cause a level of genomic variation similar to that resulting from SNPs (4). CNVs may play a major role in phenotypic variation (1–4). They frequently overlap with genes (1–4) and were shown to be associated with AIDS-susceptibility (5) and immunologically mediated renal disease (6). However, compared with SNPs, knowledge on CNVs is relatively limited: although >3,000 copy-number variable regions are currently described in the Database of Genomic Variants (2, 4), almost all corresponding breakpoint sequences are unknown (7). [At the time of analysis, only for three CNVs (i.e., deletions) were breakpoint coordinates available (8), all of which were based on an analysis of a single individual involving large-scale DNA sequencing (3).] Thus, it is usually unclear whether commonly observed deletions/duplications at a particular locus are due to a single frequently occurring CNV (recurring instances of a CNV with matching breakpoints) or are due to several CNVs with distinct breakpoints that overlap partially [the former, i.e., CNVs with shared breakpoints that occur in >1% of the population are here referred to as copy number polymorphisms (7) or CNPs]. This lack of knowledge, a major obstacle for genotype-phenotype association studies, is largely due to limits of technologies used for CNV detection. Widely applied platforms for CNV identification across individuals are thought to achieve effective resolutions in the tens to hundreds of kilobases [defining effective (or predictive) resolution as the median distance in base pairs between predicted and actual breakpoints], resolutions suitable for detecting the presence of many CNVs (1, 2, 4, 9) but insufficient for precise breakpoint mapping. Thus, genes can be assigned to CNVs only in a general and sometimes provisional manner. Recently, three studies exploited data generated in the course of extensive SNP genotyping efforts (10, 11): clusters of apparent genotyping errors/inconsistencies were detected (12, 13), and haploid source material was hybridized against a microarray platform designed for SNP genotyping (14), enabling identification of many frequently occurring (mostly) smaller deletions (median <10 kb), several of which overlap with previously reported CNVs (1–3). However, duplications were generally not considered, and no breakpoint sequences were reported. Finally, Tuzun et al. (3) used a strategy involving fosmid-paired-end sequencing to fine-map breakpoints in a single individual (with estimated resolutions of 40 kb for deletions and 8–40 kb for duplications). This led to many relevant results, e.g., insertions including sequences not represented in the human reference genome (3), and eventually DNA sequencing is likely to become the method of choice for detecting the boundaries of CNVs, in a similar fashion as a comparison of human genome assemblies has recently yielded numerous candidate CNV breakpoint sequences (15). Nevertheless, microarray-based approaches are more economical and can be readily applied at large scale, enabling the mapping of CNV breakpoints across many individuals and to associate these with phenotypic information. Although the vast majority of CNV boundary coordinates are unknown, their discovery and genome-wide mapping will be valuable for genetics and genomics.
Recently, high-resolution comparative genome hybridization (HighRes-CGH) has been developed (16, 17), a technology that can detect signatures of CNVs at large-scale. Its underlying principle, array comparative genome hybridization [(array)CGH (18)], involves cohybridization of differentially fluorescent-labeled genomic DNA from both an individual and a reference to a microarray and can be applied cost-efficiently across many samples. HighRes-CGH achieves an unprecedented resolution owing to recent advances in high-density tiling microarrays (19–21), allowing for an immense number of distinct oligonucleotide probes per chip. For instance, when using HighRes-CGH with a chromosome 22-wide 85-bp tiling path step size (denoting its theoretical resolution; i.e., the median distance between genomic regions specifically targeted by probes) in conjunction with PCR, we recently managed to identify the breakpoints of a 1.4-Mb disease-associated deletion that had previously been characterized only at standard cytogenetic resolution (16).
However, HighRes-CGH data are hard to interpret in an ad hoc fashion, and, as yet, no systematic computational approach has been developed for mapping human CNVs at the level of base pairs. Above all, the considerable enhancement in theoretical resolution of HighRes-CGH comes with additional costs: the noise level obtained from microarray readouts is high because of the short hybridizing probes and the complexity of genomic DNA, causing cross-hybridization. Novel computational approaches are thus required to benefit from the technology's theoretical resolution. In particular, tiling microarrays open new avenues; their high-resolution data enable us to make use of correlations between array signals and the actual nucleotide sequence. Here, we present an approach for CNV breakpoint identification that integrates signals from HighRes-CGH arrays and nucleotide sequence statistically, facilitating the accurate detection of CNV breakpoints. A thorough analysis of 10 individuals demonstrates its predictive power: >400 breakpoints were mapped and, in eight instances, predicted coordinates are confirmed through DNA sequencing.
Results
An Approach for Systematic CNV Breakpoint Discovery.
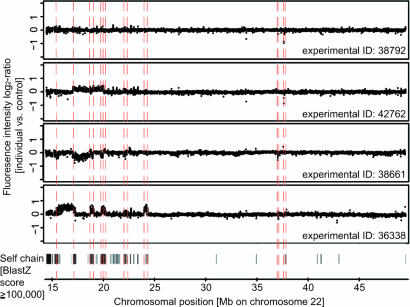
Toward resolving the current situation in which most CNV breakpoints are unknown, we sought to develop an approach for systematic breakpoint fine-mapping. In particular, accumulating evidence suggested a prevalence of CNV breakpoints near/within recently formed segmental duplications (SDs) in the genome (1, 2, 9, 22), which represent potential mediators of CNV formation through nonallelic homologous recombination. Thus, we reasoned that this correlation may, in turn, be used to improve breakpoint prediction. Indeed, when comparing HighRes-CGH data and genomic sequence features, we noted a visible correlation between CNV boundaries and SDs (Fig. 1).
Fig. 1.
Association of breakpoints and SDs. Genomic locations of SDs are indicated by BlastZ (32) self-chain matches to the human reference sequence (black vertical bars). SDs coinciding with deletion/duplication breakpoints are highlighted by a red dashed line. The association of breakpoints and SDs [consistent with earlier observations (1, 2, 9, 16, 22)] indicates that nucleotide sequence signatures can facilitate breakpoint mapping.
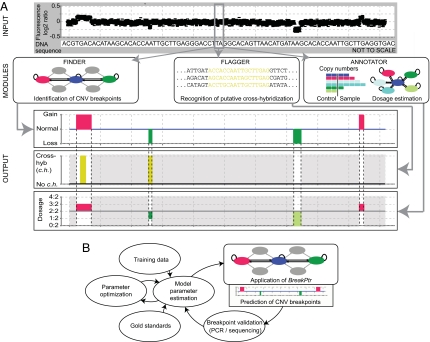
We thus decided to specifically develop an application that allows incorporating aspects of nucleotide sequence. Namely, we developed Break-Pointer (BreakPtr), a hidden Markov model (HMM)-based formalism combining data informative for breakpoint prediction (Fig. 2). Its module Finder predicts CNV breakpoints from HighRes-CGH data and nucleotide sequence, which, by default, are integrated statistically by using a discrete-valued bivariate HMM (dbHMM). The model assigns chromosomal regions to seven distinct states (23), corresponding to “unaffected genomic regions,” “deletions,” and “duplications” as well as four “transition states” (the latter four states directly consider the nucleotide sequence signatures of breakpoints; Fig. 3). In particular, the dbHMM emits discrete symbols for each genomic coordinate targeted by a microarray probe, with each symbol corresponding to a single bin of the dbHMM's emission distribution (see Fig. 3 and Materials and Methods for more details). CNV breakpoints are assigned to state boundaries. Furthermore, the Annotator module of BreakPtr, implemented after Finder (Fig. 2A), identifies actual copy number ratios (i.e., “dosage”) for each CNV. Finally, the Flagger module uses sequence analysis to identify potentially false-positive predictions that may have resulted from cross-hybridization.
Fig. 2.
Overview of BreakPtr and its parameter optimization procedure. (A) Data from HighRes-CGH experiments are statistically integrated with nucleotide sequence signatures. Finder fine-maps CNV breakpoints. The subsequently implemented Annotator provides information in terms of copy number ratios, and Flagger identifies putative cross-hybridization for regions for which Finder has predicted CNVs (i.e., regions colored in light gray are disregarded). (HighRes-CGH signals shown in the figure do not correspond to original data but were generated for visualization purposes.) (B) Parameter optimization. Training data and gold standards are used to estimate initial parameters. Parameters are then optimized by using an EM-based algorithm (25). Finally, CNV breakpoints are predicted, and sequenced. A new round of parameter estimation is initiated subsequently by using further knowledge from validated breakpoints.
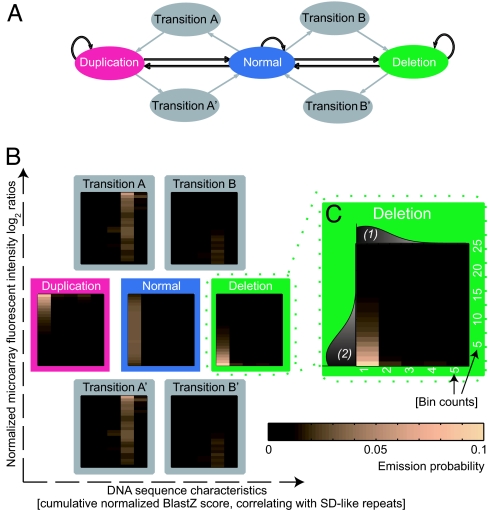
Fig. 3.
Hidden Markov models (HMMs): architecture and parameters. (A) HMMs: arrows indicate transitions used by the dbHMM (gray and black arrows) and by the univariate HMM (black arrows only), e.g., for the core parameterization. (B) Emission distributions for the dbHMM shown as heat maps, here exemplified by a 5 × 25-bin-model (x and y axes refer to each individual heat map). (C) Scheme illustrating the incorporation of discretized signals into bins: (1) scores quantifying DNA sequence characteristics, i.e., SD-like repeats (horizontal axis; schematically depicted distributions (in gray) are drawn for visualization purposes only); (2) normalized microarray fluorescent intensity log2-ratios (vertical axis).
Only a very limited amount of data on CNV breakpoints is currently available; however, a sharp increase in such data is anticipated in the near future. Consequently, we developed BreakPtr in a data-quantity-sensitive fashion. In particular, the approach uses a flexible parameterization enabling breakpoint mapping with variable amounts of training data and gold standards. (We defined training data as HighRes-CGH data containing at least one approximately mapped CNV. Gold standards, according to our definition, are publicly available genomic DNA samples with exactly mapped breakpoints. Two of the latter, i.e., samples with known disease-associated deletions, were available to us at the time of analysis.) Alternative parameterizations of BreakPtr are implemented based on the cubic dependency of available data points and the number of histogram bins (which relates to the number of parameters in the model; Fig. 3) according to Scott (24). This allows the full parameterization, i.e., the seven-state dbHMM requiring ≈2,500 parameters, to gradually collapse to alternative models with fewer parameters in an intuitive fashion [i.e., through reducing numbers of bins and states of the model; see supporting information (SI) Text and SI Fig. 4]. At the extreme end of this collapse is the “core” parameterization, a univariate three-state HMM not considering nucleotide sequence information and requiring 10 parameters.
Integrating experimental validations into the overall analysis of array data with BreakPtr should allow us to leverage a small amount of additional knowledge to incrementally improve the set of gold standards and refine our breakpoint predictions. We thus implemented a concept related to semisupervised machine-learning, allowing iterative optimization of BreakPtr parameters through incremental incorporation of validations (Fig. 2 B and Materials and Methods). In brief, (i) initial parameter estimation was performed by using a set of known or approximately mapped deletions and duplications; (ii) an expectation maximization (EM)-based algorithm (25) was used for parameter optimization; (iii) CNVs and their breakpoints were predicted; (iv) breakpoints were validated by DNA sequencing; (v) this process was iterated, which allowed refinement of parameters and predicted CNVs.
Fine-Mapping CNV Breakpoints.
After developing BreakPtr, we tested the approach in detail, focusing on human chromosome 22 and the β-globin locus (16) (a 100-kb region on chromosome 11). In total, 10 samples were analyzed, including eight subjects with known genetic disorders (16) and two “healthy” individuals (see SI Table 2). Because of the small set of available gold standards, BreakPtr was initially applied by using the core parameterization. Parameter estimation was performed by using a set of experiments targeting approximately mapped chromosomal aberrations that involve the 22q11 chromosomal band (see SI Table 3). Parameters of the state corresponding to unaffected genomic DNA were estimated based on an experimental control (Materials and Methods). In total, 232 putative CNVs were identified by BreakPtr (i.e., 464 breakpoints, flanking 121 duplications and 111 deletions, with median size 15 kb and mean 85 kb; see SI Table 4), many of which may be widespread in humans. In particular, 67 (29%) overlap with the genomic coordinates of previously reported CNVs listed in the Database of Genomic Variants (2). By taking into account estimated mapping resolutions of previous studies, we tentatively assigned refined CNV breakpoint coordinates to 36 of the 108 genomic locations to which breakpoints had previously been approximately mapped. (Note that breakpoint-mapping resolutions of previously carried out surveys, i.e., the expected uncertainties of mapping, were, in several instances, unknown to us and thus estimated by using criteria given in SI Text). Altogether, predicted CNVs intersected with 210 different genes. Because our survey included patients with known chromosomal disorders, not all of these genes may intersect CNVs in healthy individuals. Nevertheless, 91 genes did not overlap with the respective critical regions of the previously diagnosed chromosomal disorders (16) and are thus candidates for genes commonly varying in copy number.
Using BreakPtr (core model), we further reanalyzed the association between CNV breakpoints and SDs (SI Text). Indeed, for >2/3 of the predicted CNVs on chromosome 22, at least one breakpoint intersected with a SD. This represents a >4-fold enrichment over random (i.e., compared with “shuffled” CNVs with randomized genomic locations), consistent with previous estimates at lower resolution (9).
Benchmarking of Predictions.
Agreement with previously mapped breakpoints.
To evaluate our breakpoint predictions critically, we first focused on breakpoints that were previously precisely mapped in the regions analyzed here. Indeed, we identified all four previously sequenced breakpoints (16, 26) in the individuals available to us at nucleotide level (Table 1): both of the physical boundaries of a 619-bp heterozygous deletion causing β-thalassemia (26) and the breakpoints of a 1.4-Mb heterozygous deletion associated previously with 22q11-deletion syndrome (16). Furthermore, the heterozygosity (16, 26) of both deletions was correctly identified by BreakPtr.
Table 1.
Experimental validation of predicted breakpoints
| Subject ID(s) | Coordinates of breakpoints (hg17): | Dosage (agrees with prediction) | Present in healthy individuals | Validation |
|---|---|---|---|---|
| 05–029 | Chromosome 11 5203062 and 5203681 | Heterozygous deletion; 1:2 (yes) | No [deletion involved in disease (16)] | PCR, DNA sequencing (16) |
| 04–018 | Chromosome 22 17977963* and 19359814* | Heterozygous deletion; 1:2 (yes) | No [deletion involved in disease (16)] | PCR, DNA sequencing (16) |
| 04–018 | Chromosome 22 21548126 and 21566356 | Heterozygous deletion; 1:2 (yes) | Yes† | PCR, DNA sequencing |
| 93–171F, 04–018 | Chromosome 22 33969719 and 33970693 | Homozygous deletion; 0:2 (yes) | Yes‡ | PCR, DNA sequencing |
CNVs in normal individuals.
We next assessed whether BreakPtr can be used also for identifying the breakpoints of CNVs in healthy individuals. Initially, we designed primers to sequence the breakpoints of an 18-kb heterozygous deletion predicted to disable the first coding exon of the IGLC1 gene (Ig-λ constant region 1; RefSeq: BC012159). The deletion, which may be involved in normal variation of the immune system, overlaps a genomic region of previously reported copy-number variation (2). Second, we attempted to identify the breakpoints of a BreakPtr-predicted ≈1-kb homozygous deletion located in a region for which, to our knowledge, no CNVs have as yet been reported. The latter deletion intersects with a conserved noncoding element upstream of the HMG2L1 gene (which encodes high-mobility group protein 2-like 1; RefSeq: HMG2L1), and may thus cause variation at the level of gene-regulation. Subsequent to PCR analysis, we sequenced all four breakpoints, leading to the discovery of 18,231 bp and 975 bp deletions (Table 1; and SI Figs. 5 and 6). Furthermore, the observed PCR bands supported the predicted copy number ratios. By comparing genomic coordinates of predicted and validated breakpoints, we determined an effective resolution of BreakPtr of ≈330 bp (taking into account both the four earlier mapped and the four previously uncharacterized breakpoints).
Comparison of core and full parameterization.
We further compared BreakPtr's core parameterization to the full model. In particular, when using the small set of the only four earlier mapped (disease-associated) breakpoints for estimating the parameters for dbHMM transition states (i.e., Transitions B and B′ in Fig. 3A), the full model yielded an improved effective resolution (≈280 bp). Before significance can be established, more breakpoint sequences need to be solved. Nevertheless, when using the full model, the fraction of predicted CNVs overlapping with previously reported CNVs already showed a slight increase (from 29% to 31%), and we expect that with the availability of larger sets of gold standards the full parameterization should cause robust improvements over alternative models.
Use of BreakPtr to refine previously mapped breakpoints.
We believe that the considerable overlap of our predictions with previously reported CNVs indicates that BreakPtr will help in refining many approximately mapped breakpoints. To exemplify this, we analyzed GM15510, a sample derived from a healthy subject previously studied by Tuzun et al. (3) by using fosmid paired-end sequencing: four of six CNVs (67%) previously identified (3) in the chromosomal regions studied here intersected with CNVs predicted by BreakPtr (core parameterization). For these, high-resolution breakpoint assignments are available from SI Table 4. Note that all cases missed by BreakPtr represent insertions detected by fosmid paired-end sequencing (3) and, thus, are not necessarily CNVs, by definition, because they may at least partly contain sequences not present in the human reference genome or products of balanced translocations not affecting gene dosage, which are not detectable by HighRes-CGH. We expect BreakPtr to be suited for refining the coordinates of many previously reported CNVs.
Breakpoint Fine-Mapping Suggests Abundance of CNPs and Mendelian Transmission.
The fine-mapping of CNV breakpoints should enable in-depth analysis of CNV frequency and inheritance across individuals; specifically, correspondences between partially overlapping CNVs cannot be reliably assessed in the absence of precisely mapped breakpoints. For instance, several CNVs reported in our study appear to be common: 11% of the 232 predicted CNVs were observed in at least two unrelated individuals (when applying a margin of ±330 bp for breakpoint identification) and, thus, most likely represent CNPs, i.e., common CNVs. (Because of the relatively small number of individuals analyzed here, the actual fraction of CNPs will presumably be considerably higher; see SI Table 5.) To further exemplify this, we carried out a pilot study examining by PCR the distribution of the previously uncharacterized 975-bp CNV across 19 HapMap individuals (including relatives and unrelated subjects). PCR results suggest Mendelian transmission of the deletion [in agreement with recent observations concerning CNV inheritance (4, 8)] and common occurrence in different populations. Altogether, the CNV was detected in ≈20% of the surveyed chromosomes of HapMap individuals, consistent with our provisional estimate based on BreakPtr predictions in the 10 individuals analyzed by HighRes-CGH (SI Fig. 7 and SI Table 4). This indicates that the predictive resolution of BreakPtr alone enables analyzing CNV frequency and inheritance.
Discussion
We have presented BreakPtr, an approach enabling systematic fine-mapping of CNV breakpoints across individuals. Several algorithms for predicting CNVs from array-CGH and related data [e.g., such as that based on considerably lower-resolution bacterial artificial chromosome-based arrays (2, 9), representational oligonucleotide microarray analysis (1), or SNP genotyping arrays (4)] have already been described (see, e.g., refs. 27 and 28). This includes, for instance, hypothesis-driven approaches such as HMM-based algorithms [see, e.g., refs. 27 and 29 or the CNAT algorithm available from Affymetrix (Santa Clara, CA) for scoring SNP genotyping arrays] or data-driven approaches like the circular binary segmentation algorithm (28) (for a recent comparison of algorithms, see e.g., ref. 30 and references therein). These approaches were developed and so far applied only for detecting more gross changes in copy number, and not for fine-mapping CNV breakpoints by using HighRes-CGH data (for which they may as yet not be practical; see SI Text). Our HMM-based approach has enabled us to exploit DNA sequence information for CNV prediction in a data quantity-sensitive fashion. We expect that in the data-rich near future, this approach may represent a robust improvement over methods that do not consider the association between microarray data and sequence. We further envision that yet additional data types may be incorporated into HMM-based algorithms provided that an association with breakpoints exists. For instance, given the current drop in DNA sequencing costs, CGH analysis and sequencing may soon be integrated computationally, e.g., by combining DNA read counts with array signals.
Finally, to evaluate the prospect of performing breakpoint validations on a large-scale, we studied the design requirements of HighRes-CGH experiments. For instance, when removing half of the probes of the chromosome 22 microarray analyzed here, effective resolutions at 0.5–1 kb were observed (data not shown), resolutions well suited for breakpoint validation. Given the ever-increasing feature density of microarray slides, surveys such as the one described here will soon be performed on a genome-wide scale. For instance, Nimblegen (Madison, WI) has recently begun producing arrays with 2.1 million probes: if by using those arrays with a 170-bp tiling path step size, only nine microarrays per individual may enable genome-wide breakpoint mapping (thereby, BreakPtr analysis is unlikely to be limiting; see SI Text). Eventually, large-scale fine-mapping and sequencing of breakpoints will shed new light on CNV origin, inheritance, population frequency, and associations of CNVs with phenotypes.
Materials and Methods
Microarray Experiments and Data Retrieval.
Microarrays covering chromosome 22 with ≈385,000 different probes at an 85-bp tiling path step size were designed as described (16). Labeled genomic DNA of human subject and reference samples (the latter sample, i.e., the control, comprising a pool of genomic DNA from seven healthy male individuals, from Promega, Madison, WI) were cohybridized to the arrays (16, 17). Fluorescence intensities were obtained for each spot (16) (“probe”). Fluorescence intensity normalization was performed by using the Qspline algorithm (31). We further included and reanalyzed HighRes-CGH data from a previous study (16).
CNV Breakpoint Prediction by Using the Full Parameterization.
HighRes-CGH data were scored by using BreakPtr (source codes available from http://breakptr.gersteinlab.org). Its full model encompasses a seven-state dbHMM operating with two emission channels: i.e., it uses normalized fluorescent intensity log2-ratios and a value quantifying the redundancy of the underlying DNA sequence derived from BlastZ (32) alignments [which can be used for identifying SDs (32, 33)]. Normalized BlastZ-scores (32) from genome-wide human-vs.-human (i.e., BlastZ-self chain) alignments depleted of lineage-specific common (interspersed) repeats were retrieved from the University of California (Santa Clara, CA) Genome Browser (http://genome.ucsc.edu; default parameters according to the Self-Chain Track, i.e., minimum BlastZ raw score = 10,000; normalized BlastZ-score = raw score/no. of bases aligned). We used cumulative scores that were obtained by summing up normalized BlastZ-scores for each BlastZ-hit intersecting with the genomic coordinate of a probe. This measure correlates with the redundancy of the nucleotide sequence, in particular SDs, and we thus considered it for incorporation into the dbHMM. BreakPtr's transition states reflect the propensity of breakpoints to coincide with SDs. Breakpoints are predicted also if not coinciding with SDs, because the model architecture allows transition states to be omitted. The dbHMM emits discrete symbols for each genomic coordinate targeted by a probe, with each symbol corresponding to a bin of the emission distribution associated with particular microarray values and nucleotide sequence composition (Fig. 3). Bins were constructed in the following way: cumulative scores were divided among N1 = 5 bins, with bin sizes selected by using the condition to place approximately equal numbers of data points into each bin. Normalized fluorescence intensity log2 ratios were assigned to N2 = 100 bins according to the following procedure: values between −1 and 1 were assigned to N2 − 2 bins covering equally sized fluorescence intensity log2-ratio intervals. Further, log2 ratios < −1, and log2 ratios >1, were assigned to additional bins. Predictions were robust to bin size selection. Most probable state assignments were found by using the Viterbi algorithm (23). BreakPtr assigns breakpoints to locations of transition to (or from) “deletion” and “duplication” states. In this particular study, given the small amount of available gold standards, emission distributions of the full model were refined by Gaussian smoothing (after parameter estimation) by using parameters that resemble the distribution of emission values of the normal state (i.e., unaffected genomic DNA). This step is unnecessary if the criteria based on Scott (24) are fulfilled (see below).
Alternative Parameterizations.
Alternative parameterizations (core and intermediate models) are implemented according to Scott (24), depending on the amount of available data (see SI Text and SI Fig. 4). For instance, the core model uses a univariate continuous three-state HMM with Gaussian emission and 10 parameters. Focusing on HighRes-CGH data, it is not trained on DNA sequence signatures. Alternative models use the same structure of modules (Finder/Annotator/Flagger; Fig. 2) as the full model.
Training/Optimization of BreakPtr.
BreakPtr was iteratively optimized by using training data and gold standards. For instance, the core model was trained as follows: (i) parameter estimation was based on a labeled training set, i.e., parameters for CNV-states were estimated from approximately mapped breakpoints (see SI Table 3). For the normal state, a control microarray experiment [same DNA applied to both channels (16)] was used. Transition probabilities pb were estimated for each transition between states by dividing the number of previously known aberrations (based on approximately mapped deletions/duplications) in the concatenated training set by the number of probes. Then, transition probabilities pw within states were set:
(ii) Parameter optimization was carried out by using a larger unlabeled training set (in practice, the entire set of arrays is used). (iii) Breakpoints were predicted, and (iv) validated. These four steps were iterated, by using refined breakpoints for each round. Steps i and ii can be viewed as a special case of semisupervised learning (34), because unlabeled data are used to improve a model based on labeled data. Steps iii and iv are closely related to active sampling/learning, because a subset of unlabeled data is selected and validated, and results are subsequently incorporated in the rescoring process. BreakPtr's alternative parameterizations are optimized similarly to the core model: Transition states were trained based on data points corresponding to an interval ±1,000 bp from a mapped breakpoint. Transition-state emission distributions were assumed to be identical for both (the telomeric and centromeric) boundaries of CNVs. BreakPtr allows transition probabilities to be refined by using EM or to be adjusted manually. The latter, e.g., allows the stringency of CNV detection to be adjusted gradually if a CNV reported by using a complementary technology was initially missed.
Dosage Estimation.
Copy number ratios were identified by using a six-state HMM (Annotator). Parameters were estimated from biological samples (if corresponding samples were missing, parameters were manually specified; see SI Table 3 and SI Fig. 8).
Identification of False-Positive Signals Resulting from Cross-Hybridization.
Cross-hybridization of probes may, to a certain extent, affect HighRes-CGH experiments; putative cross-hybridization is identified by using a sequence alignment-based approach (this module, Flagger, is described in detail in SI Text).
Validation of Breakpoints.
Genomic regions surrounding predicted breakpoints were amplified by conventional PCR or vectorette PCR and sequenced (16).
Supplementary Material
Acknowledgments
We thank J. Rozowsky, P. Kim, O. Emanuelsson, T. Gianoulis, A. Tanzer, and G. Fuchs for critical reading; R. Baertsch (University of California, Santa Clara, CA) for help with the BlastZ-chains; and X. Chen and N. Carriero for assistance. We benefited from the Yale Center for High Performance Computation in Biology and Biomedicine and National Institutes of Health (NIH) Grant RR19895-02 funding the instrumentation. J.O.K. was supported by a Marie Curie Fellowship. A.E.U., M.S., S.M.W., and M.B.G. were supported by NIH Grant P50 HG02357-01.
Abbreviations
- (array-)CGH
array comparative genome hybridization
- CNP
copy number polymorphism
- CNV
copy number variant
- EM
expectation maximization
- HMM
Hidden Markov Model
- dbHMM
discrete-valued bivariate HMM
- HighRes-CGH
high resolution CGH
- SD
segmental duplication.
Footnotes
The authors declare no conflict of interest.
Data deposition: Microarray data have been deposited in the Gene Expression Omnibus repository (accession no. GSE6010).
This article contains supporting information online at www.pnas.org/cgi/content/full/0703834104/DC1.
References
- 1.Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, Maner S, Massa H, Walker M, Chi M, et al. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 2.Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 3.Tuzun E, Sharp AJ, Bailey JA, Kaul R, Morrison VA, Pertz LM, Haugen E, Hayden H, Albertson D, Pinkel D, et al. Nat Genet. 2005;37:727–732. doi: 10.1038/ng1562. [DOI] [PubMed] [Google Scholar]
- 4.Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, et al. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gonzalez E, Kulkarni H, Bolivar H, Mangano A, Sanchez R, Catano G, Nibbs RJ, Freedman BI, Quinones MP, Bamshad MJ, et al. Science. 2005;307:1434–1440. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- 6.Aitman TJ, Dong R, Vyse TJ, Norsworthy PJ, Johnson MD, Smith J, Mangion J, Roberton-Lowe C, Marshall AJ, Petretto E, et al. Nature. 2006;439:851–855. doi: 10.1038/nature04489. [DOI] [PubMed] [Google Scholar]
- 7.Feuk L, Carson AR, Scherer SW. Nat Rev Genet. 2006;7:85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- 8.Newman TL, Rieder MJ, Morrison VA, Sharp AJ, Smith JD, Sprague LJ, Kaul R, Carlson CS, Olson MV, Nickerson DA, et al. Hum Mol Genet. 2006;15:1159–1167. doi: 10.1093/hmg/ddl031. [DOI] [PubMed] [Google Scholar]
- 9.Sharp AJ, Locke DP, McGrath SD, Cheng Z, Bailey JA, Vallente RU, Pertz LM, Clark RA, Schwartz S, Segraves R, et al. Am J Hum Genet. 2005;77:78–88. doi: 10.1086/431652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Altshuler D, Brooks LD, Chakravarti A, Collins FS, Daly MJ, Donnelly P, Consortium TIH. Nature. 2005;437:1299–1320. [Google Scholar]
- 11.Hinds DA, Stuve LL, Nilsen GB, Halperin E, Eskin E, Ballinger DG, Frazer KA, Cox DR. Science. 2005;307:1072–1079. doi: 10.1126/science.1105436. [DOI] [PubMed] [Google Scholar]
- 12.Conrad DF, Andrews TD, Carter NP, Hurles ME, Pritchard JK. Nat Genet. 2006;38:75–81. doi: 10.1038/ng1697. [DOI] [PubMed] [Google Scholar]
- 13.McCarroll S, Hadnott T, Perry G, Sabeti P, Zody M, Barrett J, Dallaire S, Gabriel S, Lee C, Daly M, et al. Nat Genet. 2006;38:86–92. doi: 10.1038/ng1696. [DOI] [PubMed] [Google Scholar]
- 14.Hinds DA, Kloek AP, Jen M, Chen X, Frazer KA. Nat Genet. 2006;38:82–85. doi: 10.1038/ng1695. [DOI] [PubMed] [Google Scholar]
- 15.Khaja R, Zhang J, Macdonald JR, He Y, Joseph-George AM, Wei J, Rafiq MA, Qian C, Shago M, Pantano L, et al. Nat Genet. 2006;38:1413–1418. doi: 10.1038/ng1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Urban AE, Korbel JO, Selzer R, Richmond T, Hacker A, Popescu GV, Cubells JF, Green R, Emanuel BS, Gerstein MB, et al. Proc Natl Acad Sci USA. 2006;103:4534–4539. doi: 10.1073/pnas.0511340103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Selzer RR, Richmond TA, Pofahl NJ, Green RD, Eis PS, Nair P, Brothman AR, Stallings RL. Genes Chromosomes Cancer. 2005;44:305–319. doi: 10.1002/gcc.20243. [DOI] [PubMed] [Google Scholar]
- 18.Pinkel D, Segraves R, Sudar D, Clark S, Poole I, Kowbel D, Collins C, Kuo WL, Chen C, Zhai Y, et al. Nat Genet. 1998;20:207–211. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- 19.Kapranov P, Cawley SE, Drenkow J, Bekiranov S, Strausberg RL, Fodor SP, Gingeras TR. Science. 2002;296:916–919. doi: 10.1126/science.1068597. [DOI] [PubMed] [Google Scholar]
- 20.Bertone P, Stolc V, Royce TE, Rozowsky JS, Urban AE, Zhu X, Rinn JL, Tongprasit W, Samanta M, Weissman S, et al. Science. 2004;306:2242–2246. doi: 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Z, Pang AW, Gerstein M. BMC Evol Biol. 2007;7(Suppl 1):S14. doi: 10.1186/1471-2148-7-S1-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lupski JR, Stankiewicz P. PLoS Genet. 2005;1:e49. doi: 10.1371/journal.pgen.0010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rabiner L. Proceedings of the IEEE; New York: IEEE Comput Soc; 1989. pp. 257–285. [Google Scholar]
- 24.Scott DW. Biometrika. 1979;66:605–610. [Google Scholar]
- 25.Dempster A, Laird N, Rubin D. J R Stat Soc Ser B. 1977;39:1–38. [Google Scholar]
- 26.Bhardwaj U, Zhang YH, Lorey F, McCabe LL, McCabe ER. Am J Hematol. 2005;78:249–255. doi: 10.1002/ajh.20269. [DOI] [PubMed] [Google Scholar]
- 27.Fridlyand J, Snijders A, Pinkel A, Albertson D, Jain A. J Multivar Anal. 2004;90:132–153. [Google Scholar]
- 28.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Biostatistics. 2004;5:557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 29.Colella S, Yau C, Taylor JM, Mirza G, Butler H, Clouston P, Bassett AS, Seller A, Holmes CC, Ragoussis J. Nucleic Acids Res. 2007;35:2013–2025. doi: 10.1093/nar/gkm076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Willenbrock H, Fridlyand J. Bioinformatics. 2005;21:4084–4091. doi: 10.1093/bioinformatics/bti677. [DOI] [PubMed] [Google Scholar]
- 31.Workman C, Jensen LJ, Jarmer H, Berka R, Gautier L, Nielser HB, Saxild HH, Nielsen C, Brunak S, Knudsen S. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-9-research0048. research0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwartz S, Kent WJ, Smit A, Zhang Z, Baertsch R, Hardison RC, Haussler D, Miller W. Genome Res. 2003;13:103–107. doi: 10.1101/gr.809403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bailey JA, Gu Z, Clark RA, Reinert K, Samonte RV, Schwartz S, Adams MD, Myers EW, Li PW, Eichler EE. Science. 2002;297:1003–1007. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- 34.Chapelle O, Schölkopf B, Zien A. Semi-Supervised Learning. Cambridge, MA: MIT Press; 2006. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.