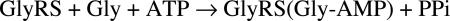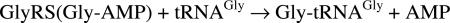Abstract
Functional expansion of specific tRNA synthetases in higher organisms is well documented. These additional functions may explain why dominant mutations in glycyl-tRNA synthetase (GlyRS) and tyrosyl-tRNA synthetase cause Charcot–Marie–Tooth (CMT) disease, the most common heritable disease of the peripheral nervous system. At least 10 disease-causing mutant alleles of GlyRS have been annotated. These mutations scatter broadly across the primary sequence and have no apparent unifying connection. Here we report the structure of wild type and a CMT-causing mutant (G526R) of homodimeric human GlyRS. The mutation is at the site for synthesis of glycyl-adenylate, but the rest of the two structures are closely similar. Significantly, the mutant form diffracts to a higher resolution and has a greater dimer interface. The extra dimer interactions are located ≈30 Å away from the G526R mutation. Direct experiments confirm the tighter dimer interaction of the G526R protein. The results suggest the possible importance of subtle, long-range structural effects of CMT-causing mutations at the dimer interface. From analysis of a third crystal, an appended motif, found in higher eukaryote GlyRSs, seems not to have a role in these long-range effects.
Keywords: crystal structure, structure-function analysis, dimer interface, inherited peripheral neuropathy, aminoacyl tRNA synthetase
Charcot–Marie–Tooth (CMT) diseases are the most common heritable disorder of the peripheral nervous system, having a frequency of occurrence of ≈1 in 2,500 (1). Although different genetic loci associated with the disease have been identified, some of them have provided no obvious explanation for the neuropathological and electrophysiological phenotypes that have been observed (2). This lack of understanding may mean that proteins thought previously to be associated only with non-neuron-specific activities are part of a broader systems biology where connections are made between pathways and activities usually regarded as discrete and isolated. For example, CMT-causing dominant mutations occur in two of the genes coding for human aminoacyl tRNA synthetases: YARS [gene for tyrosyl-tRNA synthetase (TyrRS)] (3) and GARS [gene for glycyl-tRNA synthetase (GlyRS)] (4–9). These enzymes are representatives of the aminoacyl tRNA synthetase family of proteins, which catalyze the first step of protein synthesis, i.e., aminoacylation of tRNAs with their cognate amino acids. They are the only two of the 20 tRNA synthetases presently known to be causally linked to CMT disease.
Although GlyRS is an uncommon example of a human tRNA synthetase, having a single gene encoding both cytoplasmic and mitochondrial forms, the two forms of TyrRS are encoded by separate genes. CMT-causing mutations are found only in the gene for cytoplasmic TyrRS (3). Thus, even though neuropathies associated with mitochondrial disorders are known, it seems unlikely that the CMT disease connection to these two tRNA synthetases is through effects on mitochondrial protein synthesis. Alternatively, because several human aminoacyl tRNA synthetases (including human TyrRS) have expanded functions that enable them to participate in cell signaling networks (10), these expanded functions have raised the possibility of tRNA synthetase connections with the nervous system through pathways beyond aminoacylation.
Considering that at least 10 different mutations in GlyRS have been linked to CMT disease (4–9), and because the structure of this enzyme was not known, we were motivated to determine its structure and see whether that structure would offer clues into the etiology of the disease. We focused on obtaining crystals of wild-type and a CMT-causing mutant of cytoplasmic GlyRS, because none of the CMT mutations fall in the mitochondria-specific N-terminal extension. In addition, the cytoplasmic version is easier to express and isolate than the more extended mitochondrial form. GlyRS is one of the 10 members of the class II tRNA synthetases and, like most members of that class, are α2 dimers. Although the structure of a bacterial ortholog (Thermus thermophilus) of human GlyRS is known (11, 12), the latter enzyme has limited sequence identity with that ortholog and, in addition, has N- and C-terminal extensions and three insertions not present in the bacterial GlyRS structure. The distribution of the 10 CMT disease mutations on the sequence of human GlyRS, and rough placement of these mutations on the structure of T. thermophilus GlyRS, showed that the mutations were widely dispersed across the sequence and structure and, therefore, had no unifying theme to connect them together. Thus, in obtaining the crystal structure of human GlyRS, we were especially motivated to look for characteristics in the structure of a mutant form that could, in principle, suggest some sort of unifying connection between at least some of the CMT disease mutations.
Results
Structures Determined for Wild-Type and the CMT-Causing Mutant G526R GlyRS.
Crystals of the wild-type human GlyRS were obtained earlier (13). These crystals had a modest diffraction quality, and the collected data did not render a successful structure solution by molecular replacement, using the structure of the orthologous T. thermophilus GlyRS as the search model. Efforts were then devoted to improving the crystal quality of the wild-type enzyme and to crystallizing a specific CMT-causing mutant protein. Interestingly, the G526R mutant, under slightly different crystallization conditions compared with that used for the wild-type enzyme, yielded crystals that diffracted to 2.85 Å and with good data quality (Table 1). These crystals, and those formed with the wild-type enzyme, had the same space group, P43212, with each asymmetric unit containing one subunit of the dimeric enzyme. Using molecular replacement with the structure of T. thermophilus GlyRS, this data set allowed us to solve the structure of the G526R mutant enzyme. Subsequently, the structure of wild-type GlyRS was solved at 2.9 Å using, in this instance, molecular replacement with the newly obtained structure of the G526R mutant enzyme.
Table 1.
Data collection and refinement statistics
| Wild type | G526R | |
|---|---|---|
| Space group | P43212 | P43212 |
| Molecules in asymmetric unit | 1 | 1 |
| Data collection | ||
| Resolution, Å | 50.0–2.90 (3.0–2.9) | 50.0–2.85 (2.95–2.85) |
| a, Å | 91.74 | 91.41 |
| c, Å | 247.18 | 246.81 |
| Wavelength, Å | 0.97945 | |
| Rsym,* % | 9.9 (52.4) | 6.6 (37.0) |
| I/σ(I) | 17.6 (3.1) | 36.6 (7.7) |
| Redundancy | 8.9 (7.8) | 13.8 (14.0) |
| No. of unique reflections | 18,765 | 25,080 |
| Completeness, % | 77.2 (80.2) | 98.7 (98.5) |
| Refinement | ||
| Resolution, Å | 36.8–2.90 | 29.2–2.85 |
| No. of reflections used for refinement | 17,766 | 23,727 |
| No. of reflections used for test set | 961 | 1,238 |
| Final R, % | 23.5 | 23.1 |
| Final Rfree,† % | 27.0 | 27.1 |
| rmsd bonds, Å | 0.003 | 0.003 |
| rmsd angles, Å | 0.756 | 0.847 |
| Mean B value, Å2 | 55.0 | 58.2 |
Values in parentheses refer to the highest-resolution shell.
*Rmerge = [ΣhΣi∣Ii(h) − 〈I(h)〉 / ΣhΣiIi(h)] × 100.
†Calculated from 5% of the reflections that were excluded before refinement.
Overall Structural Organization of Human GlyRS.
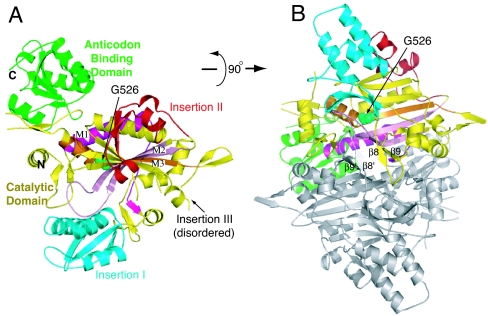
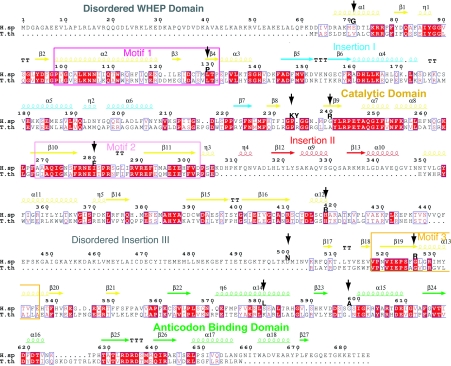
The cytoplasmic human GlyRS has 685 residues composed of an N-terminal WHEP-TRS domain (M1–D62) that is unique for GlyRS from Metazoans, a catalytic domain (D63–V556), and a C-terminal anticodon binding domain (V557–E685). The catalytic domain has the typical core (for class II enzymes) of an extended β-sheet and the three conserved motifs, consisting of a helix–strand (motif 1), strand–loop–strand (motif 2), and strand–helix (motif 3), which are characteristic of all class II tRNA synthetases. In addition, three insertions split the catalytic domain. In comparison to other class II tRNA synthetases, insertion I (F144–N225, between motifs 1 and 2) is unique to and shared by GlyRSs through evolution (14). Insertion II (D307–N348, after motif 2) and insertion III (V440–V504, before motif 3) are specific to eukaryotic GlyRSs (15). Except for the disordering of residues E382–Y386 and A421–V504, which cover the entire insertion III, and of residues D545–E546, our structure of wild-type GlyRS resolved most of the catalytic domain including insertions I and II (Fig. 1A). The N-terminal WHEP-TRS domain is completely disordered, whereas the C-terminal anticodon binding domain is well resolved except for 11 residues at the C terminus (G675–E685).
Fig. 1.
Overall structure of human GlyRS. (A) “Front” view of the asymmetric unit that contains one subunit of the dimeric enzyme. Different domains and insertions of the subunit are colored as indicated. The three conserved sequence motifs (M1, M2, and M3) in the catalytic domain are colored magenta, pink, and orange, respectively. (B) A 90° rotation along the x axis to view the interface between two subunits of the GlyRS dimer. The second subunit is generated by symmetry operation and is not colored. The β8–β9 hairpins are labeled on both subunits to show their interaction on top of the dimer interface.
Interestingly, whereas insertion I is well resolved in human GlyRS, it was not resolved in the structure of T. thermophilus GlyRS (11, 12). Apart from the absence of insertions II and III in T. thermophilus GlyRS, the overall structure of human GlyRS is similar to that of its bacterial ortholog. The rmsd between the two structures is 1.9 Å for 400 Cαs with a similarity Z-score of 36.9 (16).
Fig. 2 gives the structure-based sequence alignment of the human and T. thermophilus GlyRSs. Insertion I of the catalytic domain contains a compact three-helix bundle (α4–α5–α6) that is flanked at one end by a three-strand antiparallel β-sheet (β5–β6–β7). This insertion forms a domain-like entity that is virtually separated from the rest of the catalytic domain (Fig. 1A) and protrudes itself away from the dimer interface (Fig. 1B). Insertion I is likely to be involved in acceptor stem recognition (W.X., unpublished data).
Fig. 2.
Structural alignment of human and T. thermophilus GlyRS with the secondary structure elements placed on top. Ten CMT-causing mutations are marked with vertical arrows.
The eukaryote-specific insertion II associates closely with the catalytic core by extending the central seven-strand β-sheet, by adding β12 and β13, to give a nine-strand β-sheet. The spatial location of this insertion suggests that it contributes to neither substrate binding nor dimerization (Fig. 1B). However, the almost completely disordered insertion III could be involved with tRNA binding across the dimer interface and make contact with the anticodon stem or variable or D-loops (W.X., unpublished data). In that connection, mammalian GlyRS is more discriminating than the T. thermophilus enzyme in that T. thermophilus GlyRS charges tRNAGly from both bacteria and eukaryotes, whereas mammalian GlyRS can charge only eukaryotic tRNAGly (17). Thus, insertion III could provide additional tRNA recognition that is eukaryote-specific.
Compared with T. thermophilus GlyRS, human GlyRS has 27 additional residues at the C terminus (Fig. 2). Most of these are resolved, with the first part folding into helix α18 and a short strand β27. The final 11 residues are disordered, with more than half of them being either positively or negatively charged. In our tRNA complex model (W.X., unpublished data), the extra C-terminal sequences bring the charged C terminus into a space that it could interact with tRNA at the anticodon and thus provide additional tRNA binding and recognition capacity to the anticodon-binding domain.
The disordered N-terminal WHEP domain is predicted to have a helix–turn–helix structure, as shown for human TrpRS (18) and for human Glu-ProRS (19). The WHEP domain is present in several higher eukaryotic tRNA synthetases in different configurations, for example, as an N-terminal appended domain (as in GlyRS or TrpRS), as part of the linker that fuses together the two synthetases of the novel Glu-ProRS, and as a C-terminal appended domain (as in MetRS) (20). The domain usually is flexibly linked to the core enzyme and, probably for that reason, tends to be disordered in crystal structures, if not provided with crystal lattice interactions. We found that the WHEP domain of human GlyRS could be clipped off by limited protease digestion (data not shown). The resulting truncated protein was crystallized. Interestingly, these crystals have the same lattice and space group parameters as those of the crystals for full-length GlyRS. This observation suggested that the absence of the WHEP domain does not likely cause a conformational change in the rest of the enzyme. [A similar observation was made with human TrpRS. And like what was observed for TrpRS (21), deletion of the WHEP domain in GlyRS does not affect aminoacylation activity (data not shown).] In addition, because exactly the same crystals were obtained with both full-length and WHEP-truncated GlyRS, the WHEP domain itself must make little or no contribution to crystal lattice formation. These observations are consistent with this extra domain being completely disordered in the structure of wild-type GlyRS shown here.
Structure of G526R Mutant Predicts a Disrupted Enzyme Activity.
Except for the mutation site (Fig. 3 A and C), the overall structure of the G526R mutant protein is almost identical to that of the wild-type enzyme, with an rmsd = 0.65 Å for the 519 Cαs that are commonly resolved in both structures. (The disordered regions in the G526R mutant protein are almost the same as those seen in wild-type GlyRS.) All specific sites that are associated with conformational discrepancy between the two structures are on loop regions, which are generally flexible. Those regions include loop F231–M238, which contains one of the CMT-causing mutation P234KY (P278KY in the mouse) (7).
Fig. 3.
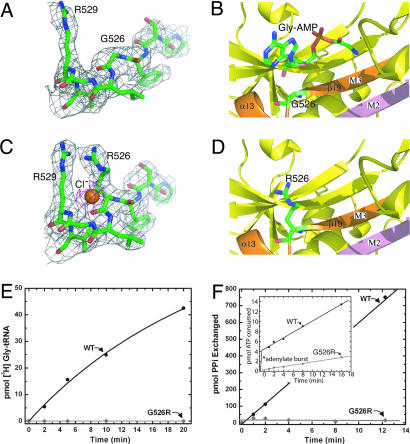
G526R mutation blocks the AMP binding site and thereby inactivates the enzyme. (A) The electron density of the region of G526 in wild-type GlyRS. (B) Gly-AMP modeled in the active site of wild-type human GlyRS with potential interactions between G526 and the AMP moiety. (C) The G526R mutation site is clearly shown by the electron density in the mutant structure. A chloride ion was fitted into the extra observed electron density. (D) G526R mutation in the active site blocks the AMP binding site. (E) Loss of overall aminoacylation activity by G526R mutation. (F) Inactivation of the glycine-dependent ATP–PPi exchange activity by G526R mutation. (Inset) Active-site titration of the G526R mutant form of GlyRS demonstrates a significant defect in the formation of glycyl-adenylate as compared with the wild-type enzyme.
G526 is a strictly conserved residue in the middle of motif 3 and connects strand β19 with the immediately following helix α13 (Fig. 3B). In T. thermophilus GlyRS, the corresponding residue of G526, using its backbone nitrogen, makes an H-bond with O2′ of the ribose of AMP (Fig. 3B). This interaction is conserved for binding to both ATP and Gly-AMP (11). Although the G526R mutation does not disturb other residues in the active site, R526 itself directly blocks the active site (Fig. 3D). The guanadino side chain of R526 stacks with the same side chain guanadino group of R529 and thereby preoccupies the binding site for the AMP moiety (Fig. 3C). Interestingly, an extra electron density was observed in the omit map, and this extra density fits perfectly with a chloride ion (Fig. 3C). Presumably, the negatively charged Cl− is used to shield the mutual repulsion from the two positively charged guanadino groups.
Experimental Demonstration That G526R GlyRS Has an Inactivated Adenylate Synthesis Site.
The overall aminoacylation of tRNAGly occurs in two steps (Schemes 1 and 2). This overall reaction can be measured by monitoring the incorporation of glycine into tRNAGly to form Gly-tRNAGly. Use of this assay showed clearly that, whereas the wild-type enzyme was active as expected, G526R GlyRS was inactive (Fig. 3E).
Scheme 1.
Scheme 2.
Either the first or second reaction (Scheme 1 or Scheme 2) can be responsible for the defect in aminoacylation of G526R GlyRS. For example, a failure to bind tRNAGly, or a failure to transfer the glycyl moiety from Gly-AMP to tRNAGly, would cause inactivation of aminoacylation. Alternatively, disruption of the first reaction, formation of the adenylate, would be another way to disrupt aminoacylation.
We focused on monitoring the first step in two different ways. The most common assay is measurement of glycine-dependent ATP–PPi exchange in the presence of GlyRS. We found that, whereas the wild-type enzyme was fully active in this assay, G526R GlyRS was inactive (Fig. 3F). This inactivity could be due to a failure to synthesize Gly-AMP or, additionally or alternatively, an extremely slow back-reaction of PPi with the bound Gly-AMP. These possibilities can be distinguished by a second assay that monitors the “adenylate burst” (22). In this experiment, GlyRS is mixed with ATP and Gly to form bound Gly-AMP in an initial burst. As the adenylate is slowly released from the enzyme, additional rounds of synthesis can occur. These additional rounds are at a slower rate than the initial burst. We found that, whereas wild-type GlyRS gave the expected burst of synthesis of Gly-AMP (as measured by consumption of ATP), no such burst was observed with the mutant enzyme (Fig. 3F Inset). Thus, the defect in ATP–PPi exchange is caused by an inability to synthesize Gly-AMP, presumably because of the blockage of the AMP binding site by R526.
Structures Suggest That G526R Mutation Enhances Dimer Interaction by Action at a Distance.
Like most class II tRNA synthetases, human GlyRS is reported to be a dimer. Independently, we confirmed that the enzyme runs as a dimer by gel filtration chromatography (data not shown). In the crystal, GlyRS is also a dimer with a 2-fold axis coincident with the crystallographic 2-fold axis of the P43212 unit cell. The extensive dimer interface in GlyRS is conserved between the bacterial T. thermophilus and human enzymes and is mainly composed of three patches (Fig. 4A). The first patch is comprised of residues F78–T137 that contains the entire motif 1 and flanking sequences on both ends. This patch, featured by the long helix α2 in the middle, interacts symmetrically with the same region of the other subunit. The second patch contains residues F224–L242, which fold into a β8–β9 hairpin that is interacting across the dimer interface with the same hairpin motif from the other subunit (β8′-β9′). These hairpins, instead of being buried, are placed on top of the dimer interface (Fig. 1B). The third patch is around residues L252–E291, which includes part of the motif 2 (Fig. 4A), and also has symmetrical interactions with its counterpart in the other subunit.
Fig. 4.
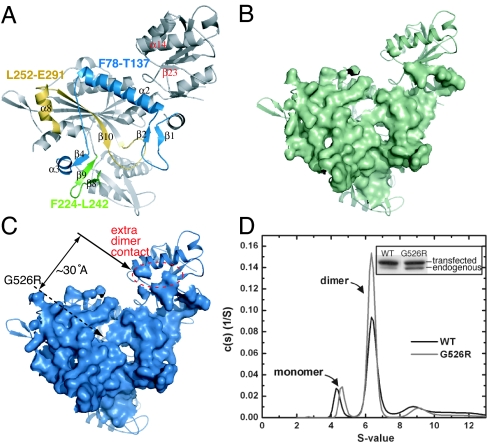
G526R mutation strengthens dimer interaction. (A) “Back” view of the GlyRS subunit that shows the dimerization interface. This view is related to the “front” view in Fig. 1A by a 180° rotation along the y axis. The three patches that give dimer interactions are colored: patch 1 (F78–T137) in cyan, patch 2 (F224–L242) in green, and patch 3 (L252–E291) in gold. (B) Loose dimerization interface of wild-type GlyRS generated by mapping the surface area of one subunit that is within 7 Å of the other. (C) The same dimerization interface generated for G526R mutant. The extra dimer interface, absent in the wild-type enzyme, lies in the anticodon recognition domain and is ≈30 Å away from the mutation site. (D) Analytical ultracentrifugation experiment showing that more dimers are formed by G526R mutant than by wild-type GlyRS. (Inset) Immunoprecipitation experiment showing that G526R mutant GlyRS pulled down more endogenous GlyRS than did the wild-type GlyRS, presumably by forming dimers.
Interestingly, the G526R mutation changes the dimer interface in a subtle but demonstrable way, even though G526 is ≈15 Å away from the nearest part of the dimer interface. Superposition of the dimers of the wild-type and G526R proteins gives an rmsd of 0.71 Å, slightly larger than the earlier number (0.65 Å) for superimposing only the two monomers. This difference suggests a subtle conformational change at the dimer interface caused by the mutation. When the surface area of one subunit that is within 7 Å of the other is then considered, apparent differences between G526R and wild-type GlyRS are more specifically demonstrated (Fig. 4 B and C). This “loose” (7-Å band either side of interface) dimer contact area for the G526R mutant protein is 7% larger than the same area calculated for the wild-type enzyme. This observation suggested that G526R GlyRS formed a more stable dimer than does the wild-type enzyme and, for that reason, the G526R mutant enzyme formed a slightly more compact crystal lattice that diffracted to a higher resolution than did crystals of wild-type GlyRS (Table 1). Some of the extra dimer contact area for G526R GlyRS is from the anticodon binding domain, ≈30 Å away from the G526R mutation site (Fig. 4C). The extra contact from the anticodon binding domain is contributed from residues on helix α14 and the following strand, β23 (Fig. 4A), including S581, whose mutation (S581L) is associated with CMT disease. Thus, this long-range structural effect, caused by the active site mutation, has manifested itself through an apparent strengthened interaction with the other subunit.
Two Independent Measurements Demonstrate Tighter Dimer Formation for G526R Enzyme.
To see whether G526R GlyRS formed a tighter dimer than did the wild-type enzyme, analytical ultracentrifugation was first carried out to evaluate the quaternary structures of the two GlyRSs (Fig. 4D). By sedimentation velocity analysis, two peaks were observed for both wild-type and G526R mutant GlyRS, with the smaller peak (S values of ≈4.30 for the wild type and ≈4.66 for G526R GlyRS) corresponding to the monomers of GlyRS and the larger peak (S values of ≈6.23 for wild type and ≈6.35 for the G526R GlyRS) being at the expected position for the dimer. Remarkably, the G526R mutant protein had a larger peak for the dimer, showing that, under the experimental conditions, it formed more dimers than did the wild-type protein (Fig. 4D).
A second assay was developed to give a picture more representative of those that occur in vivo. Genes encoding wild-type and G526R mutant human GlyRS were transfected into mouse neuroblastoma N2a cells that also expressed endogenous mouse GlyRS. The transfected GlyRSs were labeled with a V5-epitope tag that was placed at the N terminus. (This tag and the linker add a total of 41 aa to the N terminus and, therefore, give a small increase to the size of GlyRS.) Twenty-four hours after the cells were transfected, they were lysed and immunoprecipitated with anti-V5 antibodies to pull down the GlyRSs that were expressed from the recombinant, transfected genes. The precipitates were then redissolved and run on SDS/PAGE. The resolved proteins on the gel were then visualized by Western blot analysis using anti-GlyRS (not anti-V5-tagged) polyclonal antibodies.
For both wild-type and G526R GlyRS gene transfections, the starting lysate (before immunoprecipitation) had a small amount of transfected GlyRS relative to the endogenous GlyRS (≈5%). Therefore, the chance for the transfected GlyRS to form a homodimer with itself was low. After immunoprecipitation, two bands that differ slightly in molecular weight were seen using the anti-GlyRS antibodies (Fig. 4D Inset). In addition to the V5-tagged GlyRS expressed from the transfected DNA, a smaller band was observed. This band corresponded to endogenous GlyRS that was coimmunoprecipitated with transfected GlyRS through formation of a heterodimer. For transfections with the wild-type GlyRS gene, the amount of endogenous GlyRS pulled down was ≈14% of the transfected GlyRS. This number is ≈35% for the transfections with the G526R GlyRS gene (Fig. 4D Inset). By comparing heterodimer formation in these experiments, the results support strongly the idea that the G526R mutation strengthens the dimer interface.
Discussion
The structures presented here raise the possibility that the dispersion of CMT-causing mutations across the sequence of GlyRS could be understood in terms of effects on the dimer interface. Remarkably, the G526R mutation at the active site causes a long-range structural effect that perpetrates into distal areas to strengthen the dimer interface. That the wild-type subunit interacts more strongly with the G526R mutant protein provides a rationale for a dominant phenotype, that is, “subunit poisoning” by heterodimer formation. In studies of a CMT-causing mutant TyrRS (normally an α2 dimer), heterodimers were seen as well and were suggested to be involved in some way in the disease phenotype (3). Not clear, however, is whether it is a change in the interface per se, or the formation of heterodimers, or both, that are relevant. Thus, our results point to a need for further investigations of subunits' interactions and their relationship, if any, to CMT disease.
At the start of our investigations we were interested in the WHEP domain at the N terminus of GlyRS. Our thought was that this domain could bring in another neuron-specific function. However, no CMT-causing mutations have been mapped to the WHEP domain. Thus, if the WHEP domain has a role in CMT disease, then it should in some way communicate with the rest of the enzyme where the mutations are located. However, we obtained crystals of GlyRS with the WHEP domain clipped off. Although we did not solve the structure, the crystals had almost the same crystal lattice parameters as those for wild-type GlyRS, slightly bigger than those for G526R GlyRS (data not shown). This observation suggests that the structure of the non-WHEP-containing main body of GlyRS is independent of the WHEP domain and, therefore, that this domain most likely has little role in CMT disease.
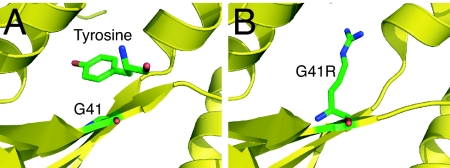
Even though we lacked the appropriate cocrystal, we were able to deduce that that G526R mutation occurs at the site for synthesis of Gly-AMP. This conclusion was strongly supported by direct biochemical assays that demonstrated an inability of the G526R enzyme to synthesize Gly-AMP. Interestingly, one of the CMT-causing mutations in TyrRS is G41R. Like G526 in GlyRS, G41 is located in the active site (Fig. 5A). Using our 1.6-Å cocrystal structure of human TyrRS with tyrosinol (Protein Data Bank ID code 1Q11), it is obvious that the arginine substitution (to give G41R TyrRS) could block tyrosine binding (Fig. 5B), just as the arginine substitution of G526 in GlyRS blocks AMP binding (Fig. 3D). This block of tyrosine binding in G41R TyrRS could explain why the yeast version of the G41R mutant did not complement a yeast strain bearing an inactivating mutation in TYS1 (yeast gene for TyrRS) (3).
Fig. 5.
G41R mutation in TyrRS resembles G526R mutation in GlyRS. (A) Active site of human TyrRS bound with substrate analog tyrosinol. (B) CMT-causing mutation G41R would block tyrosine binding in a similar way as G526R in GlyRS blocks binding of the AMP moiety.
Still, the question of the relationship between enzyme activity and CMT disease is unclear. In the mouse model of CMT disease, a P278KY substitution (corresponding to P234KY in cytoplasmic human GlyRS) is responsible for the dominant disease phenotype (7). However, P234KY human GlyRS is fully active for aminoacylation. Moreover, by using a gene trap insertion that disrupted one allele of GARS, haploinsufficient mice were created. These animals had the expected reduction in activity for GlyRS but showed none of the symptoms for CMT disease or for any other neurological disorder. This experiment raises doubts about the relationship of activity to CMT disease. Moreover, with respect to the haploinsufficient mice, only the wild-type, homodimeric GlyRS is present. Thus, haploinsufficient mice for GARS leave open the question of the relevance of intersubunit interactions to CMT disease and further raise the motivation to investigate the significance of the dimer interface.
Materials and Methods
Crystallization and Data Collection.
Wild-type GlyRS was cloned, expressed, and crystallized as described (13). The G526R mutation was introduced by standard site-directed mutagenesis. Purified G526R GlyRS protein was crystallized in 13–15% PEG 6000, 0.1 M sodium citrate buffer (pH 5.5), and 0.1 M NaCl. Crystals were flash frozen at 100 K, with 20% ethylene glycol added to the reservoir solution as a cryoprotectant. Both wild-type and G526R GlyRS data were collected on beamline 11-1 at the Stanford Synchrotron Radiation Laboratory (Menlo Park, CA).
Structure Determination.
The diffraction data were processed and scaled with HKL2000. The crystals belong to space group P43212, with one molecule of GlyRS per asymmetric unit and a solvent content of 69.2%. The structure of G526R GlyRS was determined by molecular replacement with Phaser (23) using the T. thermophilus GlyRS structure (Protein Data Bank ID code 1ATI) as a search model. Density modification with solvent flattening was done by using DM (24). Iterative model building and refinement were performed by using Coot (25) and Refmac5 (26), respectively. The refined G526R GlyRS structure was used as the starting model to solve and refine the wild-type GlyRS structure. Data collection and refinement statistics are given in Table 1.
Aminoacylation Assays.
A mixture containing 100 mM Hepes (pH 7.5), 20 mM KCl, 10 mM MgCl2, 2 mM DTT, 2 mM ATP, 20 μM l-glycine, and 0.7 μM l-[3H]glycine was added to ≈200 μM total bovine tRNA (≈10 μM tRNAGly) plus 100 nM purified GlyRS (to initiate the reaction), and the sample was incubated at 37°C. Aliquots were removed at specified time points, spotted onto trichloroacetic acid-soaked filter pads, washed with cold 5% trichloroacetic acid, and measured by scintillation counting (27).
Pyrophosphate Exchange Assays.
Glycyl adenylate synthesis was measured by using the glycine-dependent ATP-pyrophosphate (PPi) exchange assay (28). A mixture containing 100 mM Hepes (pH 7.5), 20 mM KCl, 2 mM ATP, 1 mM NaPPi, 2 mM DTT, 1 mM l-glycine, 10 mM MgCl2, and ≈0.01 mCi/ml Na[32P]PPi was added to 0.1–1 mM purified GlyRS. The reaction was incubated at room temperature, and aliquots were removed at specified times and quenched in a mixture containing 40 mM NaPPi, 1.4% HClO4, 0.4% HCl, and 8% (wt/vol) activated charcoal. The charcoal mixture was filtered and washed with a solution of 7% HClO4 and 200 mM NaPPi using Spin-X Centrifuge Filters (Corning, Corning, NY) containing 0.45-μm pore-size cellulose acetate filters. Radioactivity from the centrifuge filters containing the remaining charcoal mixture was measured by scintillation counting.
Active-Site Titration Assays.
A mixture containing 100 mM Hepes (pH 7.5), 20 mM KCl, 10 mM MgCl2, 2 mM DTT, 5 μM ATP, 5 mM l-glycine, and [γ-32P]ATP was added to 5 μM purified GlyRS (to initiate the reaction). The reaction was incubated at 37°C, and aliquots were removed at specified times and quenched in PDVF MultiScreen filter plates (Millipore, Billerica, MA) containing a mixture of 10 mM NaPPi, 7% HClO4, 0.5% HCl, and 10% (wt/vol) activated charcoal. Quenched reactions were mixed well and centrifuged through the filter plate into a collection plate, and 200 μl was transferred to scintillation vials for counting.
Analytical Ultracentrifugation.
Sedimentation velocity and equilibrium analytical ultracentrifugation of wild-type and G526R human GlyRS were performed by using a temperature-controlled Beckman XL-I analytical ultracentrifuge equipped with an An60Ti rotor and photoelectric scanner (Beckman Coulter, Fullerton, CA) Samples (≈400 μl) were loaded by using a Hamilton syringe into a double-sector cell equipped with a 12-mm Epon centerpiece and sapphire windows.
Immunoprecipitation for Dimer Detection.
Mammalian cell expression vector pcDNA3.1/nV5-DEST (Invitrogen, Carlsbad, CA) was used to clone and express wild-type GlyRS and G526R GlyRS with an N-terminal V5 tag. Wild-type or mutant GlyRS was transfected into mouse neuroblastoma cell line N2a, which was cultured in minimum essential medium (Eagle) containing 2 mM l-glutamine, Earle's BSS, 1.5 g/liter sodium bicarbonate, 0.1 mM nonessential amino acids, 1.0 mM sodium pyruvate, and 10% FCS. Twenty-four hours after transfection, cell lysates were prepared in mammalian protein extraction reagent (M-PER; Pierce, Rockford, IL) containing mixture protease inhibitors (Roche, Indianapolis, IN). The cell lysates were concentrated, precleared, and then added into the protein G agarose beads, which were preincubated overnight with anti-V5 antibody (Invitrogen). After 1 h of incubation, beads were washed four times by PBS containing 1% Nonidet P-40, boiled in 2′ SDS loading buffer, and then analyzed in Western blots using polyclonal anti-GlyRS as the primary antibody to detect both transfected and endogenous GlyRS.
Acknowledgments
We thank Dr. Robert Burgess (The Jackson Laboratory, Bar Harbor, ME) and Prof. Alexander Rich (Massachusetts Institute of Technology, Cambridge, MA) for helpful comments on the manuscript. This work was supported by National Cancer Institute Grant CA92577, National Institutes of Health Grant GM15539, and a grant from the National Foundation for Cancer Research.
Abbreviations
- CMT
Charcot–Marie–Tooth
- GlyRS
glycyl-tRNA synthetase
- TyrRS
tyrosyl-tRNA synthetase.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 2PME (for wild-type GlyRS) and 2PMF (for G526R GlyRS)].
References
- 1.Skre H. Clin Genet. 1974;6:98–118. doi: 10.1111/j.1399-0004.1974.tb00638.x. [DOI] [PubMed] [Google Scholar]
- 2.Shy ME. Curr Opin Neurol. 2004;17:579–585. doi: 10.1097/00019052-200410000-00008. [DOI] [PubMed] [Google Scholar]
- 3.Jordanova A, Irobi J, Thomas FP, Van Dijck P, Meerschaert K, Dewil M, Dierick I, Jacobs A, De Vriendt E, Guergueltcheva V, et al. Nat Genet. 2006;38:197–202. doi: 10.1038/ng1727. [DOI] [PubMed] [Google Scholar]
- 4.Antonellis A, Ellsworth RE, Sambuughin N, Puls I, Abel A, Lee-Lin SQ, Jordanova A, Kremensky I, Christodoulou K, Middleton LT, et al. Am J Hum Genet. 2003;72:1293–1299. doi: 10.1086/375039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sivakumar K, Kyriakides T, Puls I, Nicholson GA, Funalot B, Antonellis A, Sambuughin N, Christodoulou K, Beggs JL, Zamba-Papanicolaou E, et al. Brain. 2005;128:2304–2314. doi: 10.1093/brain/awh590. [DOI] [PubMed] [Google Scholar]
- 6.Del Bo R, Locatelli F, Corti S, Scarlato M, Ghezzi S, Prelle A, Fagiolari G, Moggio M, Carpo M, Bresolin N, Comi GP. Neurology. 2006;66:752–754. doi: 10.1212/01.wnl.0000201275.18875.ac. [DOI] [PubMed] [Google Scholar]
- 7.Seburn KL, Nangle LA, Cox GA, Schimmel P, Burgess RW. Neuron. 2006;51:715–726. doi: 10.1016/j.neuron.2006.08.027. [DOI] [PubMed] [Google Scholar]
- 8.Antonellis A, Lee-Lin SQ, Wasterlain A, Leo P, Quezado M, Goldfarb LG, Myung K, Burgess S, Fischbeck KH, Green ED. J Neurosci. 2006;26:10397–10406. doi: 10.1523/JNEUROSCI.1671-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.James PA, Cader MZ, Muntoni F, Childs AM, Crow YJ, Talbot K. Neurology. 2006;67:1710–1712. doi: 10.1212/01.wnl.0000242619.52335.bc. [DOI] [PubMed] [Google Scholar]
- 10.Park SG, Ewalt KL, Kim S. Trends Biochem Sci. 2005;30:569–574. doi: 10.1016/j.tibs.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 11.Arnez JG, Dock-Bregeon AC, Moras D. J Mol Biol. 1999;286:1449–1459. doi: 10.1006/jmbi.1999.2562. [DOI] [PubMed] [Google Scholar]
- 12.Logan DT, Mazauric MH, Kern D, Moras D. EMBO J. 1995;14:4156–4167. doi: 10.1002/j.1460-2075.1995.tb00089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xie W, Schimmel P, Yang X-L. Acta Crystallogr F. 2006;62:1243–1246. doi: 10.1107/S1744309106046434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cusack S. Nat Struct Biol. 1995;2:824–831. doi: 10.1038/nsb1095-824. [DOI] [PubMed] [Google Scholar]
- 15.Freist W, Logan DT, Gauss DH. Biol Chem Hoppe Seyler. 1996;377:343–356. [PubMed] [Google Scholar]
- 16.Holm L, Sander C. J Mol Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 17.Mazauric MH, Keith G, Logan D, Kreutzer R, Giege R, Kern D. Eur J Biochem. 1998;251:744–757. doi: 10.1046/j.1432-1327.1998.2510744.x. [DOI] [PubMed] [Google Scholar]
- 18.Yang X-L, Otero FJ, Skene RJ, McRee DE, Schimmel P, Ribas de Pouplana L. Proc Natl Acad Sci USA. 2003;100:15376–15380. doi: 10.1073/pnas.2136794100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeong EJ, Hwang GS, Kim KH, Kim MJ, Kim S, Kim KS. Biochemistry. 2000;39:15775–15782. doi: 10.1021/bi001393h. [DOI] [PubMed] [Google Scholar]
- 20.Lee SW, Cho BH, Park SG, Kim S. J Cell Sci. 2004;117:3725–3734. doi: 10.1242/jcs.01342. [DOI] [PubMed] [Google Scholar]
- 21.Yang X-L, Otero FJ, Ewalt KL, Liu J, Swairjo MA, Kohrer C, RajBhandary UL, Skene RJ, McRee DE, Schimmel P. EMBO J. 2006;25:2919–2929. doi: 10.1038/sj.emboj.7601154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fersht AR, Ashford JS, Bruton CJ, Jakes R, Koch GL, Hartley BS. Biochemistry. 1975;14:1–4. doi: 10.1021/bi00672a001. [DOI] [PubMed] [Google Scholar]
- 23.McCoy AJ. Acta Crystallogr D. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cowtan K, Main P. Acta Crystallogr D. 1998;54:487–493. doi: 10.1107/s0907444997011980. [DOI] [PubMed] [Google Scholar]
- 25.Emsley P, Cowtan K. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 26.Murshudov GN, Vagin AA, Dodson EJ. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 27.Schreier AA, Schimmel PR. Biochemistry. 1972;11:1582–1589. doi: 10.1021/bi00759a006. [DOI] [PubMed] [Google Scholar]
- 28.Calendar R, Berg P. Biochemistry. 1966;5:1690–1695. doi: 10.1021/bi00869a034. [DOI] [PubMed] [Google Scholar]