Abstract
There is extensive evidence that some species of ecological generalists, which use a wide diversity of resources, are in fact heterogeneous collections of relatively specialized individuals. This within-population variation, or “individual specialization,” is a key requirement for frequency-dependent interactions that may drive a variety of types of evolutionary diversification and may influence the population dynamics and ecological interactions of species. Consequently, it is important to understand when individual specialization is likely to be strong or weak. The niche variation hypothesis (NVH) suggests that populations tend to become more generalized when they are released from interspecific competition. This niche expansion was proposed to arise via increased variation among individuals rather than increased individual niche breadth. Consequently, we expect ecological generalists to exhibit stronger individual specialization, but this correlation has been repeatedly rejected by empiricists. The drawback with previous empirical tests of the NVH is that they use morphological variation as a proxy for niche variation, ignoring the role of behavior and complex phenotype–function relationships. Here, we used diet data to directly estimate niche variation among individuals. Consistent with the NVH, we show that more generalized populations also exhibit more niche variation. This trend is quite general, appearing in all five case studies examined: three-spine stickleback, Eurasian perch, Anolis lizards, intertidal gastropods, and a community of neotropical frogs. Our results suggest that generalist populations may tend to be more ecologically variable. Whether this translates into greater genetic variation, evolvability, or ecological stability remains to be determined.
Keywords: frequency dependence, individual specialization, niche expansion
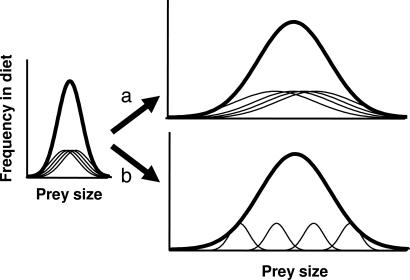
Variation is the raw material for evolution by natural selection. Consequently, evolutionary biologists have been very interested in describing the forces that create and maintain phenotypic variation within natural populations. In 1965, Van Valen (1) proposed the “niche variation hypothesis” (NVH), which suggests that “populations with wider niches are more variable than populations with narrower niches” (2). This hypothesis sought to explain the observation that bird populations inhabiting oceanic islands tend to be more morphologically variable than their mainland counterparts (1). Van Valen suggested that island birds evolve to use a wider diversity of resources when they are released from stabilizing selection imposed by interspecific competitors. This increased niche breadth (“ecological release”) could, in principle, be achieved in two ways. First, all individuals might shift to use the full set of available resources (Fig. 1a). Alternatively, each individual might continue to use a narrow range of resources but diverge from its conspecific competitors to minimize resource use overlap and competition. Increased population diet breadth is thus achieved by greater between-individual variation (Fig. 1b). Van Valen argued that the latter scenario might be “a major cause of variation in at least higher animals and plants. In other words, much variation is probably adaptive in itself and is not part of the genetic or phenotypic load” (1).
Fig. 1.
Illustration of two alternative patterns of population niche expansion. A population that uses a narrow range of prey sizes can increase its population niche breadth (bold lines) in two ways. (a) All individual niche breadths (thin lines) can expand resulting in no increased niche variation among individuals. (b) Individual niche breadths can remain limited, whereas individuals diverge from each other to increase among-individual variation.
Although the NVH is intuitively appealing and has some empirical support (1, 3–7), it also has been heavily criticized on both empirical and theoretical grounds. A reanalysis of some of the data that first inspired the NVH found that island populations of common chaffinches (Fringilla coelebs) were in fact not more variable than their mainland counterparts (8). Other studies have failed to find any relationship between population diet breadth and phenotypic variance [in 7 passerine species (9); 6 species of central African birds (2); 9 species of grasshoppers (10); 39 species of carnivorous mammals (11); as well as blue tits (12, 13), chaffinches (14), hummingbirds (15), and mongooses (16)]. In Anolis lizards, initially supportive evidence that jaw size variance increased with mean jaw size (thought to impart greater niche breadth) (17) was subsequently reinterpreted as a simple consequence of growth trajectories (18). The NVH was further undermined by theoretical models of niche width evolution that found that ecological release should occur by increased within- rather than between-individual niche diversity (19–21). This is because generalist individuals have access to a wider range of resources and therefore have higher fitness than more specialized individuals. The exception occurs when functional tradeoffs limit the abilities of individuals to efficiently use a wide range of prey types; in which case, individual niche breadth might remain limited even as the population diversifies (19, 22).
One of the drawbacks of the previous tests of the NVH is their emphasis on morphological variation. In fact, the logic underlying the NVH does not require that niche expansion lead to increased morphological variation per se. Rather, any phenotypic trait, whether morphological or behavioral, might be favored by selection if it allows an individual to use a novel set of resources and thus mitigate intraspecific competition (23). For instance, the Cocos Island finches (Pinaroloxias inornata) are one of only four resident bird species on Cocos Island, which is covered by tropical wet forest. As expected by the NVH, the finches use a broad array of resources and exhibit substantial niche variation (24). However, the among-individual variation in foraging appears to be entirely behavioral, being uncorrelated with any measured morphological traits. A more appropriate test of the NVH, therefore, would be to examine whether among-individual niche variation itself increases with population niche breadth. We tested for a correlation between niche variation and niche breadth within five disparate taxa (Table 1), by using actual patterns of resource use rather than a morphological proxy.
Table 1.
Information about case studies used
| Common name | Latin name | Type of data | Groups sampled, n | Description of groups used to calculate niche variation | Refs. |
|---|---|---|---|---|---|
| Three-spine stickleback | Gasterosteus aculeatus | Prey taxon counts in stomach contents | 11 | Fish held within each of 10 experimental enclosures plus a wild-caught control group; enclosures were kept at different densities and in distinct microhabitats | 27 |
| Eurasian perch | Perca fluviatilis | Prey taxon counts in stomach contents | 18 | Large and small age classes within a single lake, sampled each year from 1992 to 2000 | 26 |
| Whelk | Nucella spp. | Repeated prey-capture observations | 5 | Two geographic populations each of N. emarginata and N. melones; one population of N. melones was sampled in 2 years | 28, 29 |
| Brazilian savannah frogs | Adenomera sp., Eleutherodactylus sp., Leptodactylus fuscus, Proceratophryssp. | Prey taxon counts in stomach contents | 8 | One geographic population per species, divided into wet- and dry-season groups | 30, 31 |
| Anolis lizards | Anolis sagrei | Prey size distribution in stomach contents | 5 | Different island populations | 25 |
Results
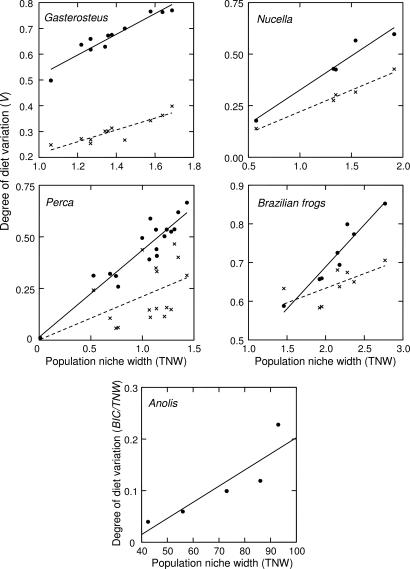
Within-population diet variation increased with population niche breadth in all five taxa examined (Fig. 2). Linear regression confirmed a significant positive slope in each case (Table 2). Diet variation in Nucella snails was established by longitudinal observation of multiple prey capture events over time. In stickleback, perch, and the frogs, diet variation was inferred from variance in stomach contents of individuals in a cross-sectional sample. One concern with such cross-sectional samples was whether the current diet of an individual is representative of its long-term behavior. We addressed this concern in two ways. First, correlations between stomach contents and morphology or stable isotope signatures confirmed that the present diet was representative of long-term behavior. Diet variation in stickleback and perch has been confirmed by both morphological correlations and isotopes (D.I.B., unpublished results; R.S., unpublished results). In frogs, diet variation has been confirmed by stable isotope analyses (25).
Fig. 2.
Correlation between diet variation among individuals (V) and the TNW of the population (see Materials and Methods for details). The empirical results are shown with filled circles. Crosses (and the dotted regression line) indicate the expected trend under a null model in which diet variation arises solely by individuals randomly sampling a limited set of prey from a shared prey distribution. Diet variation for Anolis lizards was measured based on variances of prey sizes (BIC/TNW) rather than the Shannon–Weaver diversity index. Hence, the scale for both diet variation and niche breadth are different from the other taxa.
Table 2.
Results of regression analyses relating diet variation to population TNW
| Taxon | Empirical results |
Null model results |
Test whether empirical ≠ null |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Slope | SE | t | P | r2 | Slope | SE | F | df | P | |
| Three-spine stickleback | 0.401 | 0.043 | 9.32 | <0.001 | 0.906 | 0.229 | 0.037 | 9.25 | 1,18 | 0.007 |
| Eurasian perch | 0.424 | 0.034 | 12.55 | <0.001 | 0.908 | 0.212 | 0.070 | 7.42 | 1,32 | 0.010 |
| Whelk | 0.331 | 0.042 | 7.84 | 0.004 | 0.953 | 0.209 | 0.017 | 7.13 | 1,6 | 0.037 |
| Brazilian savannah frogs | 0.217 | 0.027 | 8.05 | <0.001 | 0.915 | 0.076 | 0.035 | 10.22 | 1,12 | 0.008 |
| Anolis lizards | 0.003 | 0.001 | 3.56 | 0.038 | 0.809 | n/a* | n/a | n/a | n/a | n/a |
The slope (with SE) is given for both the actual regression relationship and the null relationship based on sampling effects. The t, P, and r2 values are for the empirical regression alone; the F and P values are given for the TNW source interaction term in a general linear model comparing the slopes of the empirical and null relationships (″source″). A significant interaction term indicates that the empirical and null slopes differ.
*n/a, not applicable. Note that the Anolis lizard relationship uses a different metric for diet variation, so it is not comparable with the other cases. In addition, raw data are not available for the Anolis comparison, so resampling was not possible.
Second, we tested whether the degree of diet variation could be accounted for by individuals sampling stochastically from a common diet distribution. Individuals can appear to be heterogeneous if there is limited diet information for each individual in a population. Small stomach size or resource competition may constrain the number of prey recorded per individual to just a couple of prey items; in which case, gut contents may underestimate the diversity of prey actually eaten. By extension, cross-sectional diet studies will tend to overestimate the variation among individuals. This artifact becomes more severe as the actual diet breadth increases, because samples of diets of individuals are proportionally smaller and hence more likely to underestimate individual niche breadth. It is therefore possible that the positive slope in Fig. 2 arises from this sampling artifact. We used a resampling procedure to recreate this artifact as a null expectation for the relationship between population niche breadth and diet variation for each of our case studies (dotted lines in Fig. 2; see Materials and Methods). Raw data were not published in the Anolis study (26), so the null model could not be applied. In the remaining four case studies, the null model did indeed predict a positive relationship between diet variation and population niche breadth (Fig. 2). However, a generalized linear model confirmed that the observed trends were significantly steeper than the null model would predict in each case (Table 2), indicating that the increase in diet variation was more than a sampling artifact. In addition, the observed level of diet variation was significantly greater than could be explained by random sampling from a common diet distribution (exceptions were one sample in perch and one sample in the frogs).
Discussion
Our results confirm that more generalized populations also tend to be more ecologically heterogeneous. This trend appears to be quite general, holding across a diverse set of taxa: two teleost fishes, lizards, several species of tropical frogs, and intertidal gastropods. It also holds at multiple scales of comparison. The two fish studies reflect more and less generalized samples from within single populations because of temporal variation in perch (27) and experimentally altered population density in stickleback (28). The Anolis data set, in contrast, reflects geographic populations that are more or less generalized (26). The Nucella and frog data sets include among-species as well as temporal and geographic variation in population niche width (25, 29–31).
Of the five case studies evaluated here, three involved populations that may reasonably be expected to have experienced some degree of ecological release. The three-spine stickleback and perch populations studied both have no major fish competitors. Variation in total niche width (TNW) in these populations was a result of population density differences, with larger population niche breadth when intraspecific competition was strong. Niche width differences among Anolis populations were also a result of competitive release, being more generalized on islands with fewer congeneric competitors (26). The Brazilian frogs, by contrast, inhabit a species-rich community where ecological release and niche variation would be unexpected. Different levels of niche variation in the frogs are due to seasonal variation in prey abundance rather than the number of competitors (31). We hypothesize that competitive release could occur even in this species-rich system if interspecific competition were strong in the dry season, when prey are scarce, but weak in the wet season, when prey may be so abundant that competing species would have little effect. The ecological reasons for niche width differences among Nucella samples is unknown.
Our results thus suggest that the NVH may be more widely applicable than generally appreciated. This conclusion also implies that foraging tradeoffs are widespread. In the absence of tradeoffs, we would expect all individuals in a panmictic population to use essentially the full array of available resources (19). Instead, we observe individual niche widths remaining roughly constant within a given study system, even as the population niche width expands. The implication is that there are constraints on individual niche width, most readily explained by biomechanical, cognitive, or physiological tradeoffs that prevent individuals from efficiently by using a diverse set of resources (23).
The patterns we describe here may have been overlooked previously because past tests of the NVH have focused exclusively on morphological variation (2, 10). However, the association between morphological and functional variation can be quite weak because of complex many-to-one mapping of morphological structures onto biomechanical functions (32) and because of the influence of behavioral variation (24). For example, in three-spine stickleback, morphology has a strong and consistent effect on diet within a given population. Individuals with deeper bodies, larger gape, and fewer/shorter gill rakers consumed more benthic invertebrate prey and fewer zooplankton (refs. 28 and 33; M.S.A., P. R. Guimarães, D.I.B., R. S., A. Pinheiro, E. G. Martins, and S. F. Reis, unpublished results). Despite this correlation between morphology and diet within each group of individuals, groups with greater TNW were not more morphologically variable [regression of morphological variance on TNW: t = −1.28, P = 0.233; morphometric details have been described previously (28)]. How, then, was the increased niche breadth achieved? The answer is that, although morphological variation was equal across the groups of stickleback, the correlation between diet and morphology was stronger in populations with higher TNW (28). Behavior thus moderates the relationship between morphological variation and diet variation. Because morphological variation in stickleback is known to be heritable (34, 35), this amounts to behavioral control of the heritability of resource use. Consequently, selection acting on resource use (36) will be most effective in high-TNW populations, in which resource use is most closely associated with a heritable morphological trait.
Morphology–diet correlations are also well established in the perch system, resulting from experimentally demonstrated tradeoffs in foraging efficiency for different prey (37, 38). The morphological variation underlying these tradeoffs is largely the result of phenotypic plasticity following behavioral shifts in the microhabitat use of individuals (39). However, there is a significant, if small, genetic component to the variation as well. In Anolis, TNW is correlated with a morphological character (head size). Because variance increases with size, it is possible that the niche variation in the lizards is a simple consequence of growth trajectories (18). In contrast, the frog data set exhibited no significant morphological correlates of diet variation (31). Hence, it is not surprising that we found no relationship between morphological variance and TNW (P = 0.117). No morphological measurements were taken in the snail study.
Although we cannot rule out the possibility that one or more unmeasured morphological traits were responsible for diet variation in the frog and snail systems, it is also possible that niche variation was purely behavioral. Such variation could nonetheless be heritable (40, 41), or may arise as a simple consequence of cognitive constraints. For instance, in some species, individuals can retain search images for only a few prey at a time (42, 43). Although individuals specialize on a limited subset of the available prey, the particular prey used by an individual may be determined randomly, as a result of chance encounters early in the development of the consumer. Without more extensive breeding studies, we cannot tell to what extent there is a genetic basis to the behavioral component of variation in the case studies examined here. Nonetheless, our results suggest an often underappreciated role for behavior in driving niche variation (24, 28). We therefore emphasize that natural selection can favor ecological character release via phenotypic or behavioral plasticity, as well as the heritable morphological diversification that has been the primary focus of the NVH in the past.
From an ecological standpoint, it may not matter whether niche variation arises via evolutionary shifts in genetic variance, as opposed to morphological or behavioral plasticity, or simple cognitive constraints. In any of these cases, populations are subdivided into groups of individuals that compete largely among themselves and differ in their interspecific interactions. Theory suggests that such niche variation within populations may lead to increased carrying capacities (44) and possibly alter the dynamics of interactions with other species. For example, niche variation can stabilize predator–prey dynamics (45) when predation risk is highest for individuals using a particular resource or microhabitat. In this case, only the subset of the prey population that uses the risky resource will be subject to predation, creating an ecological refuge for the rest of the prey species and thus weakening the predator–prey interaction. However, no models to date have evaluated whether genetic versus plastic niche variation impart different ecological dynamics.
When considering the evolutionary consequences of increased niche variation, it becomes quite important that the variation be heritable. Because individual specialists tend to compete with only a subset of their population, intraspecific competition may lead to frequency-dependent disruptive selection (36, 44). This selection can only actuate evolutionary changes such as polymorphism (44, 46), sexual dimorphism (47), or speciation (48), if the variation has a heritable basis. Of the five case studies examined here, there is evidence for heritable variation in stickleback and perch. Interestingly, the more ecologically variable samples in these species were not more genetically variable. It therefore remains uncertain whether greater niche variation in generalist populations imparts a consistently greater capacity for future frequency-dependent evolution. However, because increased diet variation in stickleback occurred via a behavioral strengthening of the morphology-diet association (36), it is likely that selection on morphology would be more effective in the more generalized but variable samples. We also emphasize that purely behavioral diet variation may also be evolvable. This is obvious when differences in foraging behavior are genetic (40, 41), but they also can occur via cultural evolution. In sea otters (Enhydra lutris), females teach their offspring to forage, so behavioral variation in prey preferences among parents is transmitted to the following generation (49). If some foraging preferences (or diet breadths) confer higher fitness, the degree of individual specialization could therefore change over generations, despite being nongenetic.
In conclusion, our results indicate that there is a repeated tendency for more generalized populations to exhibit higher niche variation, holding for a diverse set of study organisms. This niche variation may result from behavior, morphology, or an interaction between the two and may be heritable or plastic to varying degrees. Although these different mechanisms have distinct evolutionary causes and consequences, it is not known whether they impart different population or community dynamics. What is clear is that niche variation appears to be a widespread phenomenon (23), and documenting the incidence and degree of this variation is a first step toward understanding its basis and implications.
Materials and Methods
Data.
For four of the case studies, we analyzed raw diet data in the form of counts of the number of each prey type consumed by each individual. Raw data are available on request. Prey were categorized to the lowest feasible taxonomic level (25, 29–31) or into functional categories (27, 28). In the fifth case study (Anolis), we used published values for niche breadth and diet variation (26). The Nucella data set contains longitudinal rather than cross-sectional diet data (29, 30), whereas, in three other systems, the cross-sectional gut content variation has been corroborated by independent lines of evidence. Specifically, in stickleback and perch, diet differences among individuals can be attributed to morphological variation (refs. 28 and 33; M.S.A., P. R. Guimarães, D.I.B., R.S., A. Pinheiro, P. Guimarães, E. G. Martins, and S. F. Reis, unpublished results; D.I.B., unpublished results) and is reflected in significant stable isotope variation among individuals (D.I.B., unpublished results; R.S., unpublished results). Such evidence rules out purely stochastic sampling artifacts, such as the spatial location of individuals immediately before capture for stomach content analyses. In the Brazilian frogs, stable isotope analyses have demonstrated that stomach contents yield roughly accurate quantitative measures of the degree of diet variation within populations (25).
Within each case study, there were multiple groups of individuals from different species (25, 29–31), geographic populations (26, 29, 30), sampling times (25, 27, 29–31), or experimental enclosures (28). Niche breadth and the level of among-individual diet variation were calculated for each group, which provided individual data points for linear regressions within each study system.
Quantifying Population Niche Breadth and Diet Variation.
The TNW of each group was quantified by using the Shannon–Weaver diversity index, following Roughgarden (20). This index will yield a value of 0 when the entire population uses only a single category of prey, increasing with both the number of prey categories and the evenness with which they are used. To quantify diet variation among individuals, we calculated the proportional similarity between the diet proportions of each individual and the averaged population diet distribution. Proportional similarity (51) is calculated as
where pij is the frequency of prey type j in the diet of individual i, and qj is the frequency of the prey type in the overall diet distribution of the population (50). The mean PSi in the population represents the average level of diet overlap between individuals and the population as a whole (IS) (50). To make this measure more intuitive, we used an index V = 1 − IS (where V indicates diet variation), which ranges from 0 when all individuals use the full range of resources used by the population, toward higher decimal values when individuals are more heterogeneous and use smaller subsets of the population diet distribution. Both TNW and V were calculated in IndSpec1.0 (50). We then regressed V on TNW within each case study. We could not calculate V for Anolis lizards, because raw data were not available. Instead, we used published values of the fraction of TNW attributed to between-individual variation, between-individual component/TNW. This measure is highly correlated with V, although less appropriate for the categorical data used to evaluate the other four case studies (50).
Null Model.
For each group of individuals, we first pooled all prey counts and determined the frequency of each prey category in the summed population diet. Each individual, observed to have consumed some number n of prey items, was then randomly reassigned n items via multinomial sampling from the population diet frequencies. The null degree of diet variation (V) was calculated once all individuals were assigned random diets. For each group, we carried out 1,000 such resampling estimates, implemented in IndSpec1.0 (50). We then regressed the mean resampled V against the observed TNW, to evaluate the null hypothesis that limited individual diet data also generate a positive relationship between these measures. To evaluate whether our observed trend can be explained by this null model alone, we used a general linear model to test for a difference between the slopes of the observed and simulated V against TNW. The regression and general linear model analyses were performed in SYSTAT 11 (Systat Software, San Jose, CA). For any given sample of individuals, we also evaluated whether the observed degree of diet variation was greater than expected by chance, by determining the number of Monte Carlo resampled values of V that were larger than we observed empirically.
Acknowledgments
This work was supported by the University of Texas and a National Science Foundation grant (to D.I.B.), the Swedish Research Council (R.S. and L.P.), and the CAPES Foundation (M.S.A).
Abbreviations
- NVH
niche variation hypothesis
- TNW
total niche width.
Footnotes
The authors declare no conflict of interest.
References
- 1.Van Valen L. Am Nat. 1965;99:377–389. [Google Scholar]
- 2.Soule M, Stewart BR. Am Nat. 1970;104:85–97. [Google Scholar]
- 3.Hamilton S, Johnston RF. Auk. 1978;95:313–323. [Google Scholar]
- 4.Werner TK, Sherry TW. Proc Natl Acad Sci USA. 1986;84:5506–5510. doi: 10.1073/pnas.84.15.5506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galeotti P, Rubolini D. Biol J Linn Soc. 2004;82:237–248. [Google Scholar]
- 6.Ebenman B, Nilsson SG. Am Nat. 1982;119:331–343. [Google Scholar]
- 7.Dayan T, Simberloff D. Ecology (Tempe) 1994;75:1063–1073. [Google Scholar]
- 8.Dennison MD, Baker AJ. Evolution (Lawrence, Kans) 1991;45:29–39. doi: 10.1111/j.1558-5646.1991.tb05263.x. [DOI] [PubMed] [Google Scholar]
- 9.Diaz M. Oecologia. 1994;99:1–6. doi: 10.1007/BF00317076. [DOI] [PubMed] [Google Scholar]
- 10.Patterson BD. Evolution (Lawrence, Kans) 1983;37:375–388. doi: 10.1111/j.1558-5646.1983.tb05546.x. [DOI] [PubMed] [Google Scholar]
- 11.Meiri S, Dayan T, Simberloff D. Ecology. 2005;86:1432–1440. doi: 10.1111/j.1461-0248.2005.00825.x. [DOI] [PubMed] [Google Scholar]
- 12.Blondel J, Perret P, Anstett M-C, Thebaud C. J Evol Biol. 2002;15:440–450. [Google Scholar]
- 13.Grant PR. Biol J Linn Soc. 1979;11:103–129. [Google Scholar]
- 14.Dennison MD, Baker AJ. Evolution (Lawrence, Kans) 1991;45:29–39. doi: 10.1111/j.1558-5646.1991.tb05263.x. [DOI] [PubMed] [Google Scholar]
- 15.Feinsinger P, Swarm LA. Ecology. 1982;63:1574–1587. [Google Scholar]
- 16.Simberloff D, Dayan T, Jones C, Ogura G. Ecology. 2000;81:2086–2099. [Google Scholar]
- 17.Roughgarden J. Am Nat. 1974;108:429–441. [Google Scholar]
- 18.Lister BC, McMurtrie RE. Am Nat. 1976;110:311–314. [Google Scholar]
- 19.Taper ML, Case TJ. Ecology. 1985;66:355–371. [Google Scholar]
- 20.Roughgarden J. Am Nat. 1972;106:683–718. [Google Scholar]
- 21.Ackermann M, Doebeli M. Evolution (Lawrence, Kans) 2004;58:2599–2612. doi: 10.1111/j.0014-3820.2004.tb01614.x. [DOI] [PubMed] [Google Scholar]
- 22.Svanbäck R, Bolnick DI. Evol Ecol Res. 2005;7:993–1012. [Google Scholar]
- 23.Bolnick DI, Svanbäck R, Fordyce JA, Yang LH, Davis JM, Hulsey CD, Forrister ML. Am Nat. 2003;161:1–28. doi: 10.1086/343878. [DOI] [PubMed] [Google Scholar]
- 24.Werner TK, Sherry TW. Proc Natl Acad Sci USA. 1987;84:5506–5510. doi: 10.1073/pnas.84.15.5506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Araújo MS, Bolnick DI, Machado G, Giaretta AA, Reis SF. Oecologia. 2007 doi: 10.1007/s00442-007-0687-1. in press. [DOI] [PubMed] [Google Scholar]
- 26.Lister BC. Evolution (Lawrence, Kans) 1976;30:677–692. doi: 10.1111/j.1558-5646.1976.tb00948.x. [DOI] [PubMed] [Google Scholar]
- 27.Svanbäck R, Persson L. J Anim Ecol. 2004;73:973–982. [Google Scholar]
- 28.Svanbäck R, Bolnick DI. Proc R Soc London Ser B. 2007;274:839–844. doi: 10.1098/rspb.2006.0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.West L. Ecology. 1986;67:798–809. [Google Scholar]
- 30.West L. Ecology. 1988;69:1839–1854. [Google Scholar]
- 31.Araújo MS, Reis SF, Giaretta AA, Machado G, Bolnick DI. Copeia. 2007 in press. [Google Scholar]
- 32.Alfaro M, Bolnick DI, Wainwright PC. Evolution (Lawrence, Kans) 2004;58:495–503. [PubMed] [Google Scholar]
- 33.Robinson BW. Behavior. 2000;137:865–888. [Google Scholar]
- 34.Day T, Pritchard J, Schluter D. Evolution (Lawrence, Kans) 1994;48:1723–1734. doi: 10.1111/j.1558-5646.1994.tb02208.x. [DOI] [PubMed] [Google Scholar]
- 35.Hermida M, Fernandez C, Amaro C, San Miguel E. Can J Zool. 2002;80:532–541. [Google Scholar]
- 36.Bolnick DI. Evolution (Lawrence, Kans) 2004;87:608–618. [PubMed] [Google Scholar]
- 37.Svanbäck R, Eklöv P. Oikos. 2003;102:273–284. [Google Scholar]
- 38.Svanbäck R, Eklöv P. Oecologia. 2002;131:61–70. doi: 10.1007/s00442-001-0861-9. [DOI] [PubMed] [Google Scholar]
- 39.Svanbäck R, Eklöv P. Evol Ecol Res. 2006;8:37–49. [Google Scholar]
- 40.Latshaw JS, Smith BH. Behav Ecol Sociobiol. 2005;58:200–207. [Google Scholar]
- 41.Gibbons ME, Ferguson AM, Lee DR. Anim Behav. 2005;69:721–732. [Google Scholar]
- 42.Lewis AC. Science. 1986;232:863–864. doi: 10.1126/science.232.4752.863. [DOI] [PubMed] [Google Scholar]
- 43.Persson L. Oecologia. 1985;67:338–341. doi: 10.1007/BF00384938. [DOI] [PubMed] [Google Scholar]
- 44.Bürger R. J Math Biol. 2005;50:355–396. doi: 10.1007/s00285-004-0294-2. [DOI] [PubMed] [Google Scholar]
- 45.Doebeli M. J Theor Biol. 1997;188:109–120. [Google Scholar]
- 46.Bürger R. Am Nat. 2002;160:661–682. doi: 10.1086/342813. [DOI] [PubMed] [Google Scholar]
- 47.Bolnick DI, Doebeli M. Evolution (Lawrence, Kans) 2003;57:2433–2449. doi: 10.1111/j.0014-3820.2003.tb01489.x. [DOI] [PubMed] [Google Scholar]
- 48.Dieckmann U, Doebeli M. Nature. 1999;400:354–357. doi: 10.1038/22521. [DOI] [PubMed] [Google Scholar]
- 49.Estes JA, Riedman ML, Staedler MM, Tinker MT, Lyon BE. J Anim Ecol. 2003;72:144–155. [Google Scholar]
- 50.Bolnick DI, Yang LH, Fordyce JA, Davis JA, Svanbäck R. Ecology. 2002;83:2936–2941. [Google Scholar]
- 51.Schoener TW. Ecology. 1968;49:704–726. [Google Scholar]