Abstract
The regulation of transcriptional initiation in the human genome is a critical component of global gene regulation, but a complete catalog of human promoters currently does not exist. In order to identify regulatory regions, we developed four computational methods to integrate 129 sets of ENCODE-wide chromatin immunoprecipitation data. They collectively predicted 1393 regions. Roughly 47% of the regions were unique to one method, as each method makes different assumptions about the data. Overall, predicted regions tend to localize to highly conserved, DNase I hypersensitive, and actively transcribed regions in the genome. Interestingly, a significant portion of the regions overlaps with annotated 3′-UTRs, suggesting that some of them might regulate anti-sense transcription. The majority of the predicted regions are >2 kb away from the 5′-ends of previously annotated human cDNAs and hence are novel. These novel regions may regulate unannotated transcripts or may represent new alternative transcription start sites of known genes. We tested 163 such regions for promoter activity in four cell lines using transient transfection assays, and 25% of them showed transcriptional activity above background in at least one cell line. We also performed 5′-RACE experiments on 62 novel regions, and 76% of the regions were associated with the 5′-ends of at least two RACE products. Our results suggest that there are at least 35% more functional promoters in the human genome than currently annotated.
The pilot phase of The ENCODE Project Consortium has generated a large volume and variety of functional genomics data (The ENCODE Project Consortium 2004, 2007). Over 150 independent experiments were conducted to characterize transcriptional regulatory elements in human cell lines. The majority of these data sets measure transcription-factor binding and histone modifications using the technique of chromatin immunoprecipitation combined with genomic microarrays (ChIP-chip) or tag sequencing. Other data sets include high-throughput promoter reporter assays. Many of these experiments were conducted on factors known by previous studies to mark sites of transcription initiation, such as TAF1, methylation of lysine 4 on histone H3, and RNA polymerase II. This compendium of data thus provides an unprecedented collection of experimental observations characterizing transcription start sites (TSSs) and their associated promoters in 1% of the human genome.
With this set of transcriptional regulatory element data, we aimed to map transcriptional promoters and regulatory regions throughout the ENCODE-defined regions independent of mRNA to genomic DNA sequence alignments. We used an integrated approach that evaluated the data as a whole in a quantitative manner rather than studying each data set individually. One of the most significant analytical challenges with microarray-based functional genomics is the continuous nature of the data. Specifically in the case of ChIP-chip, a discreet biochemical event (e.g., histone modification) is usually not reflected as a binary experimental output. Therefore, invoking a threshold for calling a site bound or unbound by a transcription factor in an individual data set is often arbitrary, and individual data points near the threshold can be easily misclassified depending on whether the emphasis is placed on specificity or sensitivity. These shortcomings can be overcome when a number of experiments are analyzed together, as a modest signal that is reproduced across a number of experiments can become much more significant than it would be in a single experiment.
To this end, we have implemented four complementary methods to integrate the compendium of ENCODE transcriptional regulatory element data. First, a “naïve Bayes” method computes a score that combines the ChIP signals in different experiments, which are thresholded and weighted according to how well they perform on a set of known promoters. Second, we developed a “tree-weighting” (TW) method that computes a weighted sum of counts for a given region, where the weights account for both the TSS enrichments of individual experiments and the correlation between experiments. Third, a “majority-voting” method determines the level of experimental support for each genomic position, defined by the number of cross-laboratory, cross-platform, or cross-factor experiments that designate that position above some statistical threshold. Last, we developed a “Z-score method” that generates a cumulative score by summing over the Z-scores of a genomic interval across multiple experiments.
These methods predict regions of 0.6- to 1.5-kb sizes, dictated by the resolution of the underlying ChIP data sets. The regions do not indicate the direction of transcription or connectivity of exons in the vicinity, because the methods do not use sequence as input. Our main goal is to identify novel sites of transcription initiation from evidence other than existing cDNA sequences. We, therefore, take a promoter-centric approach in designing validation experiments.
To evaluate the effectiveness of these different methods, we compared their predictions with TSSs identified by other independent experiments and genome annotations, many of which have been produced by the ENCODE project. We also conducted extensive experimental validation of novel regions that were not part of existing promoter annotation. We experimentally validated 85 novel promoters with transient transfection assays and rapid amplification of cDNA ends (5′-RACE) experiments, and demonstrated the power of an analytical approach that integrates the data from many genome-scale experiments. Extrapolating from these results, we estimate that there are at least 35% more novel promoters than currently annotated.
Results
Promoter regions predicted by the four methods
The four complementary approaches make different assumptions and therefore have unique advantages and disadvantages. For example, the Z-score assumes that each experiment has the same predictive power for promoters, but it makes no assumption on how a promoter should look. In contrast, naïve Bayes uses a training set of known promoters to determine which experiments have the highest predictive power and weighs the experiments accordingly. Voting explicitly takes into account the finding that experiments performed by the same laboratory or on the same microarray platform tend to identify similar genomic regions as significant. TW determines this laboratory or platform bias automatically via correlating the data sets.
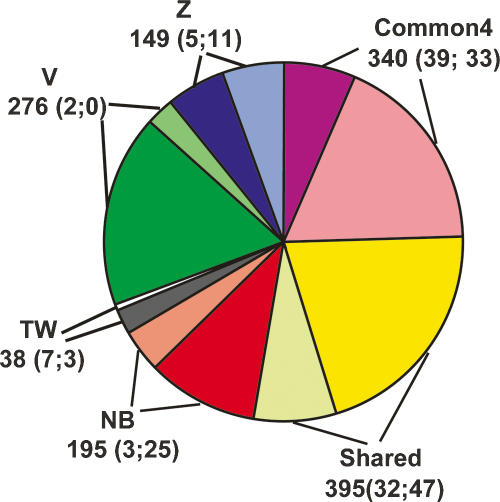
The number of regions predicted by each method and the agreement between them are shown in Figure 1 (for a full listing, see also Supplemental Table 1). Z-score identified the smallest number of regions (580), followed by naïve Bayes (689), TW (714), and voting (985). There are 340 regions that are predicted by all four methods, and these are likely the highest confidence promoter regions. Interestingly, Z-score, naïve Bayes, and voting had a similar percentage of unique regions (26%, 28%, and 28%, respectively); however, TW had only 5% unique regions, with 92% of its regions included in the voting list. These comparisons indicate that all four methods are identifying a significant number of the same regions but also many regions unique to that particular method, and that TW and voting perform more similarly to each other than the others. In addition, the near twofold variation in the absolute number of regions identified by the four different methods (from 580 to 985) suggests that some of the approaches may be more specific than others.
Figure 1.
Summary of predicted regions and experimental validation by transient transfection assays and 5′-RACE experiments. Numbers in parentheses indicate the number of validated and tested but unvalidated regions in each category. A region is considered tested if it was tested by either transient transfection assays or 5′-RACE experiments; the validated status is similarly defined. Common4 are regions common to all four methods. Shared are regions predicted by two or three methods. NB, Z, V, and TW indicate regions uniquely predicted by naïve Bayes, Z-score, voting, and tree-weighting methods, respectively. Each class is represented by two pieces of the pie, with the darker colored one indicating novel regions and the lighter colored one indicating known regions. Note that due to the substantially different validation rates of the two experimental approaches and the uneven selection of method-unique regions, there are not sufficient data to directly compare the performances of the methods.
The different methods also tend to predict regions of varying length (Supplemental Fig. 1). Z-score and TW predict regions that are on average 1.5 ± 0.8 kb long, while naïve Bayes and voting predict regions roughly half the size (0.8 ± 0.3 kb and 0.6 ± 0.3 kb, respectively). The resolution of our predictions is limited by the underlying data sets—the genomic DNA produced in the fragmentation process of ChIP is ∼0.05–1 kb long. Regions that are predicted by all methods are longest (3.8 ± 2 kb; called Common4) as we merge the overlapping predictions by the four methods together. Shared regions (predicted by two or three methods) are affected by merging in the same way (1.6 ± 0.9 kb). The difference in length distribution impacts the region-based accounting of validation rate described below, as longer regions have a higher chance of being validated.
Comparison of predicted promoter regions with other data sets and annotations
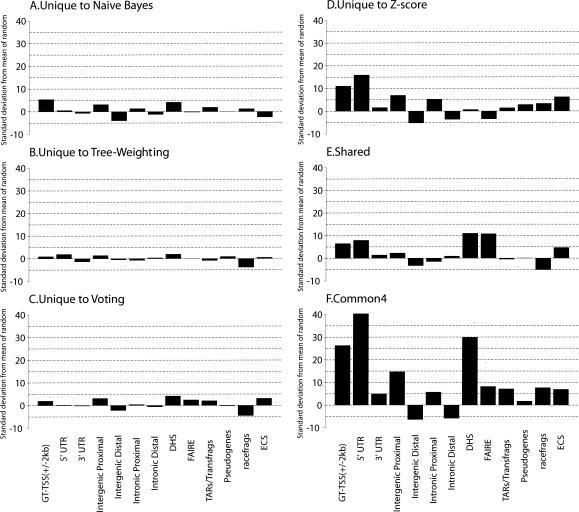
As one way to assess the accuracy of the promoter predictions by each approach, we compared the 340 regions common to the four lists along with the regions unique to each list with other experimental data sets and genomic annotations that independently mark sites of transcription initiation. In order to assess the significance of these overlaps, we randomly placed the same number of size-matched regions 100 times in ENCODE regions for each comparison to determine the mean amount of overlap by chance, and the actual observed overlap is expressed as the number of standard deviations away from the mean. The other data sets and genomic annotations we compared against included a high-confidence set of TSSs defined by the Genes and Transcripts Analysis Group of the ENCODE consortium (GT-TSS), which is an integration of GENCODE (Harrow et al. 2006), annotated TSSs and 5′-cap analysis of gene expression (CAGE) and gene identification signature paired-end ditag (GIS-PET) defined 5′-ends (Shiraki et al. 2003; Ng et al. 2005), regions of nucleosome displacement assayed by FAIRE (Lee et al. 2004; Giresi et al. 2007), regions of DNase I hypersensitivity (Sabo et al. 2006), 5′-UTRs, 3′-UTRs, and coding sequences of known genes (Fig. 2).
Figure 2.
The significance of the overlap of predicted regions in different categories with various genomic features. See Methods for their definitions and origins, as well as details on randomization. The significance is given in terms of the number of standard deviations away from the mean number of overlaps between a set of predicted regions and a set of randomly placed, size-matched regions corresponding to the genomic features. (A) Regions unique to the naïve Bayes method. (B) Regions unique to the tree-weighting method. (C) Regions unique to the voting method. (D) Regions unique to the Z-score method. (E) Regions shared by two or three methods (Shared). (F) Regions supported by all the methods (Common4).
As shown in Figure 2F, the intersection of all four methods shows the highest degree of overlap with all markers, supporting the hypothesis that these regions are more likely to be promoters than those identified by any of the individual methods alone. Not surprisingly, GT-TSSs and 5′-UTRs were two of the top three categories that showed the highest degree of overlap with the intersection of the four lists. Interestingly, regions of DNase I hypersensitivity have the second highest degree of overlap, perhaps because the ChIP-chip and the DNase I hypersensitivity experiments both identify the most active promoters in the cell lines tested. Further support for the regulatory potential of the predicted regions comes from the significant enrichment with data sets of active transcription (TARs/transfrags and RACEfrags) (The ENCODE Project Consortium 2007) and with those of nonexonic regions that are proximal to known genes (intergenic proximal and intronic proximal), as well as the significant depletion of nonexonic regions that are distal to genes (intergenic distal and intronic distal). In addition, there is a significant enrichment of evolutionarily constrained sequences (Karolchik et al. 2003), indicating that on average the predicted regions are under selective pressure. There is also a slight enrichment of pseudogenes, which could be accounted for by the actual transcriptional activities of some pseudogenes (Balakirev and Ayala 2003; Zheng et al. 2005) or could be due to the cross-hybridization of microarray probes targeting pseudogenes with genomic regions from the parental genes.
Panels A–D of Figure 2 show the degree of overlap of the same categories with the regions unique to each of the four methods. The regions unique to Z-score (Fig. 2D) and unique to naïve Bayes (Fig. 2A) show the highest degree overlap with GT-TSSs, suggesting that these two approaches are more specific than TW and voting. TW shows the least significant overlap with the other categories but also has the smallest number (38) of unique regions. Naïve Bayes and voting show the most overlap with categories that potentially indicate novel regulatory regions (DNase I hypersensitivity and FAIRE). Figure 2E shows the results for regions predicted by two or three methods, with significant overlaps with GT-TSS, 5′-UTR, DNase I hypersensitivity, and FAIRE.
The significant overlaps with independent data sets are highly encouraging and indicate that we are indeed identifying promoters with an integrated analysis of ENCODE ChIP-chip data. Interestingly, some of the regions that we identified do not overlap with known promoters and are thus putative novel promoters. When we began this project, the GENCODE annotation was not fully developed, and we defined a novel promoter as one that was >2 kb away from the TSS of a GenBank cDNA. All of the promoters that we chose for experimental validation were novel based on that definition. Upon completion of the GENCODE annotations, we revised our definition of novel promoters to those that were ±2 kb surrounding GENCODE-annotated TSSs. Consequently, some of the regions we previously designated “novel promoters” are now part of the GENCODE annotation and are thus categorized as “known” below.
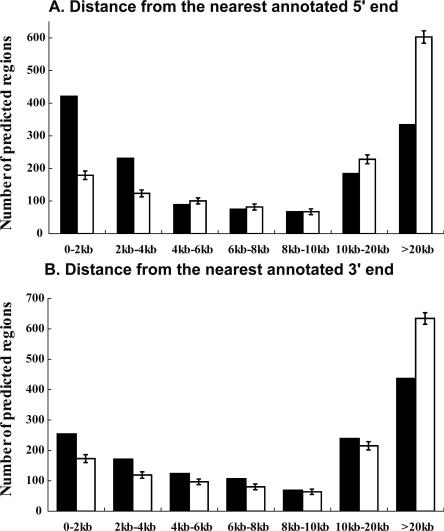
Ninety of the 340 regions (26%) predicted by all four methods and 861 (62%) of the 1393 regions predicted by at least one method were thus deemed novel based upon the GENCODE criteria. Of the predicted regions, a significant proportion is localized to the boundaries of GENCODE-annotated transcripts (Fig. 3 shows the distance distribution in comparison to randomly placed regions of equal sizes). Yet 319 regions are >20 kb away from the 5′-end of an annotated transcript.
Figure 3.
Distance of predicted regions from annotated transcripts. Black bars indicate the number of regions at various distances from 5′-ends (A) and 3′-ends (B) for all predicted regions. White bars indicate the number of regions expected by a randomization process as used in Figure 2.
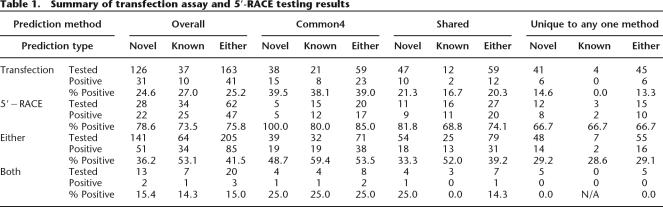
In order to assess whether some of the predicted regions >2 kb away from the 5′-end of a cDNA were indeed active promoters, we tested 163 regions (126 novel regions based on the GENCODE definition) by transient transfection reporter assays and 62 regions (28 remain novel) by 5′-RACE experiments.
Transient transfection assays validated 41 of 163 predicted regions
We cloned 250 genomic fragments that are within 1 kb of 163 predicted promoter regions and tested them for promoter activity in four human cell lines (HT1080, HeLa, HCT116, and CRL1690) using high-throughput transient transfection reporter assays (Table 1). Nearby CAGE and GIS-PET (Shiraki et al. 2003; Ng et al. 2005) were used to determine the direction of the fragment when available, otherwise the region was cloned in both directions. An independent set of 24 randomly chosen genomic regions was previously cloned to establish the background of luminescent signal (Cooper et al. 2006), and a tested fragment is deemed active if its signal is three or more standard deviations away from the mean of these negatives. Thus ∼0.1% of randomly chosen genomic regions are positive by chance. We call a promoter validated by this method if any one of its cloned fragments is positive in at least one cell line.
Table 1.
Summary of transfection assay and 5′-RACE testing results
Overall, 41 tested putative promoters were functional out of the 163 tested, corresponding to a validation rate of 25%. Encouragingly, the validation rates for the novel promoters were only lower by 2% than that of the known promoters, suggesting that a similar validation rate would be observed for the remaining novel predictions if they were also tested. Regions predicted by multiple methods clearly had the highest validation rate. Specifically, predictions common to all four methods had a validation rate of 39%, followed by predictions made by two or three methods (20%), and only 13% of regions unique to one method were validated.
We compared sequence features of the predicted regions that were validated and the ones that were not. The former had a higher tendency of overlapping with a CpG island (36% versus 9%) or containing a TATA-box (12% versus 9%). This is in agreement with our previous study, which showed that promoter fragments active in transient transfection assays tended to be GC rich (Cooper et al. 2006). When we grouped the predicted regions in that study by the number of cell lines in which they were active, we observed a strong linear correlation between this number and the percentage of the regions in the group that overlap CpG islands (R2 = 0.84) (Supplemental Fig. 3). The regions tested in this study followed the same trend (squares in Supplemental Fig. 3). The correlation also explains the apparently lower validation rate in our current study (25% active in at least one of four cell lines) compared with that in our previous study (40% and 60% active in at least one of the same four cell lines, or one of 16 cell lines, respectively). Only 36% of the predicted regions in this study overlap with CpG islands, compared with the much higher 66% in the previous study. Accordingly, 40% of validated regions in this study are active in only one of four cell lines, compared with the much lower 18.7% in the previous study. Regions that overlap CpG islands have similar validation rates in the two studies (80% in this study versus 83% in the study by Cooper et al. 2006). We argue that the present set of predicted regions contains a higher proportion of noncanonical promoters (i.e., promoters that do not overlap CpG or TATA), which are weaker and active in fewer cell lines and thus more difficult to detect with transfection assays. It is likely that many of the unvalidated predictions in this study are actually functional if more cell lines are used or if a less stringent threshold is used for calling a fragment active.
5′-RACE validated 47 of 62 predicted regions
We performed 5′-RACE in one cell line (NB4) to test 62 predicted regions. 5′-RACE experiments provide sequence-based evidence of the 5′ end of endogenous transcripts in living cells and thus complement transient transfection reporter assays that measure promoter activity in the context of plasmid construct. In total, we designed 149 pairs of nested primers targeting a ±1-kb window around the predicted regions. Multiple designs were made for some regions, depending upon neighboring TAR evidence (Bertone et al. 2004). If at least two sequenced clones map to within 1 kb of a predicted region (regardless of the strand), it is deemed as a validated promoter. The results are summarized in Table 1. Of the 62 regions we set out to test, 47 (76%) were thus validated. Interestingly, the validation rate is even slightly higher for the 28 novel regions (79%) compared with the 34 regions that got annotated as a GENCODE TSS (74%). Clearly the GENCODE annotation provides additional evidence to validate the positive RACE results and indicates the robustness of our predictions.
Transposable elements have been suggested to play a role in the evolution of regulatory regions by dispersing novel promoters throughout the genome (Jordan et al. 2003). To determine whether repeat-containing promoters were more likely to be 5′-RACE validated, we compared the overlap with SINE and LINE repeats of the promoters that were 5′-RACE validated with those that were not, and found them to be indistinguishable (Fisher’s exact test P-value = 0.12) (Supplemental Table 4). Moreover, for repeat-containing promoters, the distributions of the distances between the mapped RACE regions and the nearest repeats were not different between validated and unvalidated promoters (Wilcoxon ranked sum test P-value = 0.74). A similar analysis for low-complexity regions did not detect any bias for RACE validation toward these regions (Fisher’s exact test P-value = 0.11) (Supplemental Table 4).
We also analyzed whether segmental duplications affected the validation by 5′-RACE. Out of 47 promoters that were validated by 5′-RACE, five of them (three novel) overlapped segmental duplications. For all these promoters, we examined BLAT alignments of the RACE fragments to the vicinity of the tested promoters and to the duplicated regions. In every case, there were at least two RACE products with better alignments to the tested region than to the duplication (Supplemental Table 5).
The number of promoters validated by 5′-RACE generally correlated with the number of methods used to predict the promoter. Regions predicted by all four methods had a validation rate of 85%, while the ones predicted by only one method had a validation rate of 67%, and the ones predicted by two or three methods had an intermediate rate of 74%. Among the 15 tested predictions made by only one method, 10 were by the TW method and seven were validated by the RACE experiment. Unfortunately there are not enough RACE data on regions unique to other methods. The validation rate was not correlated with whether or not a CAGE/GIS-PET was present near the predicted promoter (77% for tag absent and 72% for tag present; the overall rate was 75%).
We manually inspected the promoters validated by 5′-RACE with respect to GENCODE-annotated transcripts. Most of them are associated with existing genes. Only two did not overlap known transcripts; nevertheless, they seemed to interact with yet unannotated transcripts, as they fell within the boundaries of novel transcripts defined by a GIS-PET cluster. Some of them initiate transcription of products that are embedded in an intron (as sense or anti-sense), others provide an alternative TSS (and hence a new variant), and the remaining are anti-sense to an exon (typically the 5′-UTR or 3′-UTR and less frequently an internal exon) of the associated gene. Figure 4 and Supplemental Figure 4 show three examples of anti-sense transcripts represented by our RACE products. Interestingly, in many of the intron embedded and alternative TSS cases, a SINE or LINE (indicated by RepeatMasker; http://ftp.genome.washington.edu/RM/RepeatMasker.html) was found at or near the promoter region. Additionally, in two of the 3′-UTR anti-sense cases, the transcripts appeared to be spliced.
Figure 4.
Anti-sense example of RACE products. UCSD genome browser graph for prediction 1.4, a region on Chr.11 identified by all four methods. The “Predicted Promoters” track show the regions predicted by any of the four methods. The “Designed Primers” track shows the nested primer pairs used to perform the 5′-RACE experiments. For 5′-RACE, the transcripts are oriented opposite to the primers and end at the nested primer. The “Mapped RACE products” track shows the validated results of sequencing the RACE products. Only the properly oriented RACE products are considered fully valid and the TSSs should be at their 5′-ends (for more details, see Methods). Other standard tracks from the May 2004 (hg17) assembly are shown to give the context of the promoter. Note the empty pseudogene tracks indicating that the identified transcripts are unlikely to be pseudogenes, and the histone modification tracks that constitute a large fraction of the experiments used to make the predictions. Two clusters were found. The cluster on the right appears to be a 3′-UTR anti-sense transcript to GENCODE putative AC051649.7. The cluster on the left appears to be a 5′-UTR anti-sense transcript to a novel gene currently only identified by a GIS-PET.
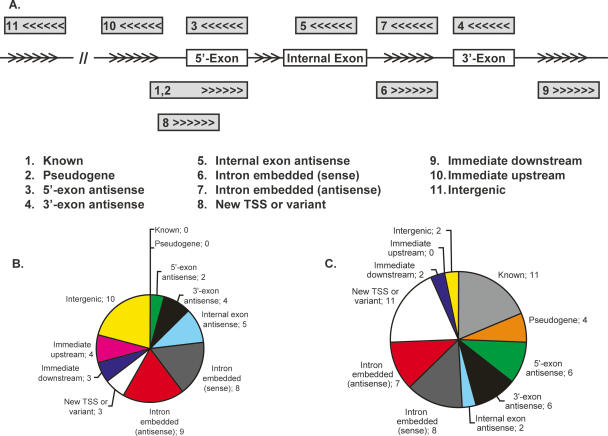
We systematically classified the transcripts associated with the 41 promoters validated by transient transfection assays and the 47 promoters validated by 5′-RACE experiments (inferred for the former and the RACE products for the latter) into 11 categories, depending upon the relative positions of the transcripts with respect to the nearest GENCODE-annotated gene (Fig. 5). The total number of cases is summed to 48 for transfection and 59 for RACE, as some classes (notably intron embedded) can be interpreted as other classes (e.g., new TSS or anti-sense). The two sets both have large representations of 5′-exon anti-sense, 3′-exon anti-sense, and intron embedded; however, the transfection set has 10 intergenic regions, while the RACE set has 11 known promoters and four pseudogenes. The discrepancy could be due to different criteria for region selection. Such classification should be helpful for inferring the biological functions of newly validated promoters.
Figure 5.
Classification of validated promoters with respect to the nearest GENCODE-annotated gene. Exons are indicated by boxes and arrows indicate the 5′ to 3′ direction. (A) (1) Known: the promoter appears within 1 kb upstream of the first exon of any variant of the gene and the transcribed strand is the same as the gene. In case of RACE, the sequence, if spliced, should match the gene splice sites. (2) Pseudogene: like known, but the gene is a pseudogene. (3) 5′-Exon anti-sense: The promoter is within 1 kb of the first exon of some variant of the gene and the transcription is anti-sense to the gene. The transcripts if not present or if short, should at least have a reasonable potential to overlap the exon. (4) 3′-Exon anti-sense: like 5′-exon anti-sense but for the last exon of some variant. (5) Internal exon anti-sense: like 5′-exon anti-sense but for an internal exon. (6) Intron embedded (sense): The promoter overlaps the gene span and transcription is on the same strand as the gene, but the transcripts do not appear to interact with any exons from any variant. (7) Intron embedded (anti-sense): like (5) but for anti-sense direction. (8) New TSS or variant: Transcription is on the same strand as the gene, and the transcribed product overlaps one or more exons of some variant but does not share the same splice sites. (9) Immediate downstream: The promoter is within 2 kb downstream of the last exon and transcription is on the same strand, but the transcripts do not overlap any exons of any variant of the gene. (10) Immediate upstream: The promoter is within 2 kb upstream of the first exon and transcription is on the opposite strand and the transcripts cannot overlap with the first exon of any variant. (11) Intergenic: >2 kb away from any annotated transcript. (B) The 41 regions validated by transient transfection assays. The total number of cases is 48, as some classes (notably intron embedded) can be interpreted as other classes (e.g., new TSS or anti-sense). (C) The 49 regions validated by 5′-RACE. The total number of cases is 59.
Discussion
In this study we have identified 1393 putative promoter regions in 1% of the human genome (44 ENCODE regions totaling 30 Mb) by integrating the results of many transcription-factor binding and histone modification ChIP-chip data sets. The results of this analysis provide an alternative way to map TSSs and promoters independent of aligning cDNA sequences to the genome. Approximately 52% of the promoters annotated by GENCODE in ENCODE regions were identified by our approach. Because the ChIP experiments were carried out in a limited number of cell lines under only a few conditions, we did not expect all GENCODE promoters to be identified. The observed overlap was highly significant and gave us confidence that we were able to identify many of the previously known promoters.
Of the regions we identified without cDNA support, we experimentally validated 85 novel promoters from a total of 205 tested (41.5%), with 41 of 163 validated by transient transfection reporter assays and 47 of 62 by 5′-RACE experiments. Twenty regions were tested by both methods, and 18 (90%) were validated by one or both of the methods (13 were validated by 5′-RACE uniquely, two by transfection uniquely, and three by both methods). If we extrapolate the validation rate of 41.5% (85 of 205) to 861 novel regions, we estimate that there are 357 functional novel promoters in the ENCODE regions. By extrapolation, we conclude that there are at least 35% more functional promoters than those currently annotated in the human genome. Because a limited number of cell lines were used for the experimental validation and because of other inherent limitations of these experiments, this is likely an underestimate.
By examining these validated promoters individually, we observed that 13% of the novel promoters are alternative promoters that start downstream of the most 5′ TSS of previously characterized genes, or extend the 5′-end of previously known genes. Approximately 11% of the novel promoters are in intergenic regions and may represent the TSSs of new genes. A reason that the intergenic class may be underrepresented in the RACE-validated set is likely due to the requirement of an index exon for RACE experiments. It would be difficult to design index primers to an exon of a new gene associated with a novel promoter. Meanwhile, a surprisingly high proportion (23%) of the novel promoters are on the anti-sense strand of previously identified transcripts (mostly terminal exons), potentially driving transcription of an anti-sense transcript (Fig. 5).
It will require additional experimental work to determine the structure of the transcripts originating at these functional promoters and, consequently, whether these are alternative promoters of existing genes or promoters of new genes yet to be identified. Deep sequencing efforts (Carninci et al. 2005) are invaluable in providing such information. Thus, of our predictions a large portion is awaiting the confirmation of high-throughput transcriptome projects. Some predictions, however, in particular the ones that function in specific cell types under specific conditions, will require targeted experiments that link the new 5′-ends to existing or novel gene transcripts.
While we are confident stating that the validated novel promoters are bound by proteins frequently associated with active transcription and are able to drive transcription in transient transfection assays or produce a transcript detectable by 5′-RACE, the biological relevance of these sequences remains to be determined. In vivo experiments such as targeted knockout of these sequences or in vivo reporter assays need to be performed to further characterize the roles of these sequences in living organisms. While these sequences may indeed promote transcription, the possibility exists that this may represent inconsequential transcriptional activity that has neither a positive nor a detrimental effect on the organism. In this capacity, these sequences may serve as reservoirs of regulatory potential that may be utilized in the course of evolution to positively select new genes or regulate existing genes in different ways. Thus, some or all of the novel regulatory sequences we have identified in this project may represent a snapshot of the equilibrium that has been reached between the creation and erosion of regulatory sequences in the evolving human genome.
Four integrative methods were applied in this study to identify promoters because promoter-related factors were the focus of the available experimental data sets. There is no reason, however, why these approaches could not be applied to other sets of functional data to identify other types of functional genomic elements. Specifically, identifying long-range transcriptional regulatory elements such as enhancers and insulators has proven to be very difficult. With appropriate types of experimental data, a similar analysis as was conducted here could be applied to identify certain classes of long-range elements. In fact, some of the data sets we used were not restricted to promoters, e.g., mono-methylation of the lysine 4 residue on histone H3 and the binding of sequence specific factors such as TP53 and STAT1. Thus some of our predicted regions may be functional long-range elements.
The major strength of our approach is that sensitivity can be improved by integration without sacrificing specificity, as integrating weak scores in multiple data sets can lead to a reliable prediction by our approach. It was clear that regions predicted by multiple methods had a higher validation rate than regions predicted by a single method, and this was seen for both experimental validation approaches. This highlights the value of using multiple methods. It would also be important to compare the performances of the different methods. The experimental results for regions predicted by only one method (Supplemental Table 1) do not support a statistically robust comparison in this work. This particular aspect of our study is an important future direction. Certainly, these analyses will become more powerful as more genome-wide functional data become available. Another potential future direction of this work would be to combine the unique advantages that the different methods afford to create a hybrid method that eliminates the shortcomings of the individual methods. For example, the experimental weightings derived by the Bayesian approach could be used to weight the contribution of the different experiments in the Z-score approach. Then, the regions identified by the Z-score approach could be added to the Bayesian training set to refine the weights of the individual experiments, and an iterative process could be invoked by this cycle.
Methods
ChIP-chip data sets
Among the data generated by the ENCODE consortium, the genomic regions targeted by 18 sequence-specific transcription factors, six histone modifications, POLR2A, TAF1, and GTF2B (formerly TFIIB) were determined by ChIP using antibodies to these components and either genomic tiling array (high-density oligonucleotide or PCR products) or sequencing-based analyses (ChIP-PET and STAGE). In total there are 129 data sets on 11 different cell lines. Some of these experiments were performed at four time points after retinoic acid stimulation, and some were performed before and 30 min after interferon γ treatment. The raw data of these experiments were obtained from the UCSC genome browser (the ENCODE consortium; http://genome.ucsc.edu/ENCODE/).
In addition, thresholded target lists (or hits) reported for each data set at several false discovery rate (FDR) cutoffs (1%, 5%, and 10% FDR) were obtained from the Transcriptional Regulation Analysis Group (The ENCODE Project Consortium 2007). These hits were used by both TW and voting methods as described below.
The naïve Bayes method
Training set
CAGE and GIS-PET clusters (Shiraki et al. 2003; Ng et al. 2005) were used to identify positive examples of TSSs. Clusters of CAGE tags with less than four tags were removed to get 797 examples. Among these, 223 that overlapped with the 5′-ends of GIS-PETs in either HCT116 or MCF7 cell lines were used as the positive training set. Additionally, 225 regions spanning ∼450 kb based on deep introns (third or deeper) and the CDS parts of deep exons were used to build the negative training set. The introns that were overlapping with exons from other transcripts, TARs, or transfrags were filtered out. A set of 1365 negative examples was collected by extracting all possible uniformly distributed and nonoverlapping windows of 300 base pairs (bp).
Training of the Bayesian model
Each TSS training example was associated with a ChIP-chip enrichment score profile from different ChIP-chip experiments. The average enrichment score within a 1-kb window around the TSS was used. The average scores were binarized at a cutoff that maximized the correlation between the training set and the binarized ChIP-chip data set. After this binarization, the training set consisted of positive and negative examples of a TSS, and each TSS had a binary profile of various ChIP-chip data sets. Using this training set, we can write the log-odds of a TSS given the data as
Assuming that the data sets are conditionally independent of each other (hence the name naïve Bayes), the log-odds of a TSS consist of two terms:
The first term, which is data independent, is the prior expectation of a TSS. The second term, which is based on the data, gives the log-odds of a TSS given the data. Di is a binary variable associated with the ith data set. If Di equals 1, we denote the contribution to the second term positive log-likelihood (PLL) ratio, and if it equals 0, we denote it the negative log-likelihood (NLL) ratio. For each ChIP-chip data set, PLL and NLL were estimated empirically using the binarized profiles for the training set.
Scanning of ENCODE regions with the Bayesian model
The naïve Bayes model consists of PLL and NLL scores for each data set, which give the Bayesian contribution of each data set to the prediction of TSS. New regulatory regions were predicted by scanning ENCODE regions using the naïve Bayes model. For each base pair in the ENCODE regions, the Bayesian contributions (PLL if the score is over the binarization cutoff from training, NLL otherwise) from each data set are summed. A chosen cutoff was defined to binarize the final scores. All the contiguous base pairs that scored 1 at the end of the binarization were clustered. These regions were further filtered by pruning all the regions <300 bp and by joining regions separated by <200 bp. The score cutoff was calculated based on the expected prevalence of TSSs in the entire ENCODE region but was later made more stringent to obtain a higher confidence set of predictions. As expected, the 223 positive examples were predicted by the algorithm. These represent easy cases and were also predicted by at least one other method. Thus we chose to keep these predictions in the downstream analysis.
The TW method
For each ChIP-chip experiment (i), we first computed the fold-enrichment (Fi) of its hits (determined at 1% FDR) near a TSS, defined as the number of observed hits near TSS (−2 kb–200 bp) divided by the expected number derived from simulation in which size-matched DNA fragments were randomly distributed back into individual ENCODE regions (excluding repeats). Subsequently, a tree was constructed to cluster all ChIP experiments, according to their correlation coefficients with respect to the genomic distribution of hit and nonhit regions. Using a branch-length division method (Gerstein et al. 1994), we then assigned a weight (Wi) to each experiment in order to minimize the bias introduced by the same factors being tested in several conditions and multiple platforms. Within this scheme, the overall weight for a factor would be shared by individual experiments with ratios between 1/n and 1 (n is the number of experiments that tested this factor). The hits from all experiments were then merged to generate a list of nonoverlapping regions, with hits that overlapped by ≥50 bp joined. This resulted in 3227 regions with an average length of 1.1 kb. A score (Sj) was subsequently assigned to each of these regions defined as ∑ (Ni × Fi × Wi), where Ni was the number of hits within this region j from experiment i, Fi and Wi were the fold-enrichment and weight computed for experiment i, respectively. We thresholded Sj at 0.05 to generate a final integrative list of 828 regions, which had a mean length of 1.7 kb. This cutoff approximately corresponded to two ChIP hits per region.
The Z-score method
All of the ChIP-chip data sets have a resolution much lower than a single base. In addition, different methods have different resolutions and also probe somewhat different subregions of the ENCODE regions. We thus needed to match corresponding data points between data sets, so we divided the ENCODE region into ∼24,000 reference intervals that largely corresponded to the probes from the two types of PCR tiling arrays. We then fit the normalized ChIP intensity data from each experiment to these reference intervals by taking the average value over the interval.
With all the data sets aligned to one reference interval set, we did a Z-score transformation (number of standard deviations away from the mean) of each individual data set to normalize for variation between data sets. This is appropriate because each experimental data set is dominated by negative results; therefore, the distribution of each data set is approximately normal. The normalized scores allow comparing the same genomic interval between data sets in a consistent framework.
For each interval, the score assigned is simply the sum of all the normalized scores of the different data sets at that interval. To determine the significance of the score, we produced a background distribution of score sums by shuffling the values of each individual data set over the ∼24,000 intervals and summed the scores at each interval. By repeating the process 10 times, we obtained a background set of ∼240,000 scores against which the real score sums can be assigned a P-value. We define an interval as part of a putative promoter if it had a positive score and a P-value <0.001. Putative promoter intervals within 100 bp were merged together.
The voting method
The voting method is based on weights that take into account the number of different laboratories that performed the experiments on a particular factor or histone modification and the number of different experimental platforms used in these studies. Supplemental Table 2 shows the weights used for each experiment. For each experiment, all the base pairs within a hit list were assigned the same weight. Thus every position in ENCODE regions was assigned a score: zero if the position was not part of any hit, and otherwise the sum of the weights of all experiments that included that position in their hits. The weights were selected so that the score was above 1 if the base was supported by at least two experiments performed on the same platform by different laboratories or on different platforms by the same laboratory. A continuous stretch of positions with scores above 1 was clustered together to define a genomic region whose score was the mean score of all the positions contained within it.
Merging of the predicted regions by the four methods
The four sets of predicted regions from the four methods were pooled and two regions merged if they overlap by one or more base pairs. This resulted in 1393 regions with length distribution shown in Supplemental Figure 1. These regions were then intersected with the original four sets of regions to determine which methods predicted each region. Each region was consequently assigned to one of the six categories: “Common4” consisted of regions supported by all four methods, “Shared” consisted of regions supported by two or three methods, and four categories each consisted of regions “unique to” an individual method.
Overlap of the predicted regions with genomic annotations (Fig. 2)
Each category of the predicted regions defined in the previous paragraph was intersected with the following 13 genomic annotation data sets: (1) GT-TSS (±2kb), a high-confidence set of TSSs that has evidence for one or more complete transcripts from GENCODE (Harrow et al. 2006) and/or five or more tags from CAGE or GIS-PET; (2) 5′-UTR, which is a 5′-untranslated region defined by GENCODE transcripts; (3) 3′-UTR, which is a 3′-untranslated region defined by GENCODE transcripts; (4) intergenic distal, which is the intergenic region >5 kb away from a GENCODE transcript; (5) intergenic proximal, which is the intergenic region within 5 kb of a GENCODE transcript; (6) intronic distal, which is the intronic region >5 kb away from a GENCODE exon; (7) intronic proximal, which is the intronic region within 5 kb of a GENCODE exon; (8) DHS (DNase I hypersensitive sites) determined by the Chromatin and Replication Analysis Group of the ENCODE Consortium (Sabo et al. 2006); (9) FAIRE (Lee et al. 2004; Giresi et al. 2007); (10) TARs and transfrags, which are transcribed regions determined by hybridizing mRNA to genomic tiling oligonucleotide arrays; (11) pseudogenes; (12) RACEfrags (downloaded from http://encode.g2.bx.psu.edu/) (Giardine et al. 2005), which are transcribed regions generated by hybridizing RACE products to genomic tiling arrays; and (13) evolutionarily constrained sequences (ECS) based on the most conserved track at the UCSC Genome Browser (Karolchik et al. 2003). GT-TSS, TAR, 5′-UTR, 3′-UTR, intergenic distal, intergenic proximal, intronic distal, intronic proximal, transfrags, and RACEfrags were produced by the Genes and Transcripts Analysis Group of the ENCODE Consortium (The ENCODE Project Consortium 2007). We randomly placed each of the 13 genomic data sets in ENCODE regions (excluding RepeatMask-ed regions for TARs/transfrags, RACEfrags, and FAIRE as the tiling arrays did not tile over repeats). The number of the predicted regions that overlapped a genomic annotation was calculated for each randomization trial, and 100 trials were performed. The significance of the overlap is reported as the number of standard deviations away from the mean number of overlapping regions in the random trials.
Distance distributions of predicted regions with respect to transcript boundaries (Fig. 3)
For each predicted region, the distance from its start or end to the nearest GENCODE-annotated transcript on either strand was calculated. There were two ENCODE regions that did not contain any annotated transcripts. Twelve predicted regions fell within these ENCODE regions and were excluded from the analysis. There were 3794 GENCODE-annotated first exons and 2608 last exons. Overlapping first exons were merged into 1372 representative first exons, and overlapping last exons merged into 1254 representative last exons. All the regions that contained exons from different transcripts were removed. Based on this processing, 1339 5′-ends and 1227 3′-ends were defined, upon which the distance calculations were based.
Sequence analysis of the validated and unvalidated regions
The fraction of regions that overlap with CpG islands (UCSC Genome Browser’s CpG islands track) was calculated. For motif search, an in-house motif scanning algorithm called Possum was used (http://zlab.bu.edu/~mfrith/possum/) using TRANSFAC matrices (Wingender et al. 2000) for TATA-box (M00216, M00252, M00471). The fraction of the promoters with at least one Possum hit (score ≥8) was reported. As expected, CpG-island-enriched and TATA-containing validated promoters represent two different groups with insignificant overlap (Supplemental Table 3).
Fragment cloning for testing promoter activity using transfection assays
From the full set of predicted promoters, we randomly selected a mixture of promoters representing cases that were identified by one method or multiple methods, were high scoring, were low scoring (near threshold), fell in gene-rich regions, and fell in gene-poor regions. For each of the putative promoters to be tested, we determined the presence of at least one CAGE or GIS-PET supporting a TSS in that region. If a region had CAGE or GIS-PET support, we used the 5′-end of the CAGE or GIS-PET sequence as the predicted TSS and used Primer3 software to design primers by inputting 600 bp of upstream sequence and 100 bp downstream of the predicted TSS (Trinklein et al. 2003). Each primer pair was required to flank the TSS. For the promoters that lacked nearby transcripts, we designed primers to amplify a 1000-bp fragment so that we could clone it in both directions. A putative promoter was thus possibly tested by more than one fragment. We added 16-bp tails to the 5′-end of each primer to facilitate cloning by the Infusion Cloning System (BD Biosciences, Clontech catalog no. 639605; left primer tail: 5′-CCGA GCTCTTACGCGT-3′, right primer tail: 5′-CTTAGATCGCAGA TCT-3′). We amplified the fragments using the touchdown PCR protocol previously described (Trinklein et al. 2003) and Titanium Taq Enzyme (BD Biosciences, Clontech catalog no. 639210). To clone our PCR amplified fragments using the Infusion Cloning System, we combined 2 μL of purified PCR product and 100 ng of linearized pGL3-Basic vector (Promega). We added this mixture to the Infusion reagent and incubated for 30 min at 42°C. After incubation, the mixture was diluted and transformed into competent cells (Clontech catalog no. 636758). We screened clones for insert by PCR, and positive clones were prepared as previously described. We quantified DNA with a 96-well spectrophotometer (Molecular Devices, Spectramax 190) and standardized concentrations to 50 ng/μL for transfections.
Cell Culture, transient transfection, and reporter gene activity assays
Transfection was performed in four cultured human cell lines (HeLa, HCT116, HT1080, and CRL1690) as previously described (Trinklein et al. 2003). The four cell lines were chosen for the promoter reporter assays because they transfect reliably and represent a large fraction of the cell-type specific activity that we demonstrated in a previous study. We seeded 5000–10,000 cells per well in 96-well plates. Twenty-four hours after seeding, we cotransfected 50 ng of each experimental luciferase plasmid with 10 ng of Renilla luciferase control plasmid (pRL-TK, Promega catalog no. E2241) in duplicate using 0.3 μL of FuGene (Roche) transfection reagent per well. We also transfected 24 random genomic fragments as negative controls for each cell line separately. Cells were lysed 24–48 h post-transfection, depending on cell type. We measured luciferase and Renilla luciferase activity using the PE Wallac Luminometer and the Dual Luciferase Kit (Promega catalog no. E1960). We followed the protocol suggested by the manufacturer with the exceptions of injecting 60 μL each of the luciferase and Renilla luciferase substrate reagents and reading for 5 sec.
Identification of active promoters
All activity data were reported as a transformed ratio of luciferase to Renilla luciferase. We determined the mean ratio and standard deviation of the 24 negative controls in the four cell lines independently. Fragment activity was then expressed as the number of standard deviations from the mean for each fragment in each cell line. We called a fragment significantly positive if it was three standard deviations above the mean ratio of the negatives. We called a putative promoter active if any of its tested fragments were significantly positive in at least one cell line.
Selection of putative promoters for RACE validation
We tested 62 predicted promoters for activity in one cell line NB4 with RACE experiments. We chose this cell line because of available transcript data for it that aided our design of the RACE primers. The selection of the test regions was mainly designed around the TW method and we selected a roughly equal number of regions from each of the following groups: unique to TW, shared between TW and only one method, shared with two methods, and shared with three methods. The promoter regions were extended to be the union of the regions identified by individual methods, as described above.
In all cases, only promoters with some evidence of transcriptional activity nearby (such as a TAR, a CAGE tag, or a GIS-PET) were selected, and one active region was used as the index for the 5′-RACE design. In cases where the transcriptional activity was based only on TARs, two indices were selected: one upstream and one downstream of the promoter. To determine the design basis, we constructed a matrix for describing all the putative promoter regions. It summarized the relationship between each promoter and various transcription data. A promoter was considered to be putatively novel if it was not near (from −2kb to 200 bp) the 5′-end of a gene in the known genes track on the UCSC genome browser. We also computationally assessed each promoter’s functional potential based on its distance to nearby transcriptional activity as detected by transfrags/TARs, CAGE tags, and GIS-PET. A promoter was considered to be functional if a transfrag, a CAGE tag, or the 5′ tag of a GIS-PET was detected within this promoter region or in its close proximity (±1.5 kb). This comparison clearly separated our predicted promoters into lists with or without transcriptional support.
Some of the putative promoters were then chosen for experimental validation based on the above matrix describing an individual promoter’s relationship with transcriptional data (including known TSS) and the number of methods predicting it. Whenever possible, the candidates from each group were selected randomly with one half predicted to be highly novel (i.e., not near GENCODE TSS).
5′-RACE experiments
We selected primers in two TARs within 3 kb of the distance to the putative novel promoter sites predicted via the above method. We designed four primers for each TAR—two gene specific primers (GSP1, GSP2) and two nested gene specific primers (NGSP1, NGSP2) on both plus and minus strands. When there was CAGE or GIS-PET information, the strand information of the gene expression was known, and in these cases, only two primers were picked for each TAR. The primers were mapped against the genome to make sure they mapped to only one location (with identity <80% to other locations). The primers are 23–28 nucleotides (nt) long, with GC content of 50%–70% and with Tm >70°C, optimally 73°C–74°C.
Total RNA from human NB4 cell line was used in cDNA amplification by SMART RACE kit (Clontech). First-strand cDNA was synthesized using PowerScript Reverse Transcriptase. A total of 1 μg RNA was used in a final volume of 10 μL of reverse transcription (RT) reaction (100 ng/μL). RACE was followed by PCR amplification using Advantage 2 PCR Enzyme System (Clontech); 0.5 μL RT reaction from the above was used in 50 μL of PCR reaction. Nested PCRs were performed using 1 μL of RACE PCR product in 50 μL reaction. The PCR program was 30 sec at 94°C and 3 min at 72°C for five cycles; then 30 sec at 94°C, 30 sec at 70°C, and 3 min at 72°C for five cycles; followed by 25 cycles of 30 sec at 94°C and 30 sec at 68°C; concluded by an extension cycle of 3 min at 72°C. PCR products were gel-purified with QIAquick 96-well PCR purification kit (Qiagen) and subsequently treated with Taq polymerase to add “A” overhang. These PCR products were then cloned into TOPO XL PCR cloning vectors (Invitrogen). Transformation was performed with One Shot Top10 ultracompetent cells (Invitrogen) in 96-well format. Five to six subclones were produced for each specific RACE PCR product. The DNA of each subclone was prepared and digested with EcoRI. The digestions were analyzed by agarose gel electrophoresis in order to determine the approximate size of the insert. All subclones were end-sequenced using M13 forward and reverse primers. Supplemental Figure 2 shows examples of RACE PCR products. All the sequenced RACE PCR products are available as Supplemental Materials.
Assignment of RACE products to putative promoters
For each tested promoter, multiple primer sets were often used; each primer set typically produced three to four bands on the gel, and each band produced five to six clones and then sequenced in both directions. Each promoter therefore contributed between 30 and 100 individual RACE sequences. Due to the large number of clones obtained and the multiplicity of products obtained from each experiment, the manually kept record of direct connection between a RACE-cDNA sequence and the promoter it was testing was prone to potential annotation errors. Thus we decided to determine the relationship directly from the sequence data. The sequence data themselves come as read pairs (forward and reverse) from each clone and are the raw sequencing product (containing the parts of the sequencing vector, various primers, and the actual insert).
To evaluate the activity success rate of the predicted promoters, we first constructed a genomic promoter-vicinity library by extracting the genomic DNA sequence from 5 kb upstream to 5 kb downstream around each of the 62 promoters, from the hg17 release of the human genome (NCBI build 35). All further mapping used BLAT (Kent 2002) against this library. We attempted to align the sequence of the RACE product in the local region surrounding the target promoter since the goal of the experiment was to validate a transcript that is produced from the promoter region. In addition, none of the primers designed were from a repeat region, and no repeats overlapped the mapped inserts that were used in prediction validation. The default settings of BLAT have been tuned for high specificity and speed, considering its primary application on mapping a query sequence against large vertebrate genomes (Kent 2002). Applying default setting failed to map many of the raw sequences from 5′-RACE, due to a combination of their short length, sequencing error, and the inclusion of nonhuman sequences from the cloning vector and RACE primers. Thus, we used nondefault settings of BLAT, aiming at maximizing sensitivity and sequencing error tolerance. The decreased specificity is compensated by applying a filtering algorithm (below).
We then mapped all the RACE-cDNA sequences against the library and also confirmed the position and orientation of the primers by mapping them to the library. In addition, we mapped three essential features of the RACE product onto the cDNA sequence itself: The linker/adaptor and the two regions of the TOPO XL cloning vector immediately upstream and downstream of the insert.
Finally, we applied a filtering algorithm to validate the association between a RACE-cDNA sequence and a promoter by requiring that the mapped part of the sequence start at the primer site and extend toward the promoter. The algorithm also ensures that the mapped part of the sequence was the full length of the insert by requiring that the TOPO XL sequences be immediately adjacent to the portion of the sequence that BLAT could map to the genomic region and in the correct orientation relative to each other and to the primer site. This end is taken as the TSS of the transcript. The filtering algorithm utilizes the presence of the forward and reverse reads and combines them to reconstruct the RACE insert. This is important since the insert can be long and the two complementary reads might not overlap but only cover the two ends of the insert, leaving the actual length of the insert unknown without using additional cues. A clone is considered positive evidence for promoter activity if the TSS falls within the region of the predicted promoter plus 1 kb on either end. All the processed sequences were deposited in GenBank under accession nos. EL582345–EL585325.
Acknowledgments
We thank The ENCODE Project Consortium for making their data publicly available, the Genes and Transcripts Analysis Group for providing transcription data sets, the Transcriptional Regulatory Analysis Group for providing ChIP-chip and ChIP-sequencing data sets, and the ENCODE Chromatin/Replication Analysis Group for providing the DHS and FAIRE data set. This work was funded by ENCODE grant R01HG03110 from NHGRI, NIH to Z.W.; ENCODE grant U01HG003162 from NHGRI, NIH to R.M.M.; and ENCODE grant U01HG003156 from NHGRI, NIH to M.S. P.J.C. was supported by Stanford Genome Training Program grant T32HG00044 from NHGRI.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article is online at http://www.genome.org/cgi/doi/10.1101/NA
References
- Balakirev E.S., Ayala F.J., Ayala F.J. Pseudogenes: Are they “junk” or functional DNA? Annu. Rev. Genet. 2003;37:123–151. doi: 10.1146/annurev.genet.37.040103.103949. [DOI] [PubMed] [Google Scholar]
- Bertone P., Stolc V., Royce T.E., Rozowsky J.S., Urban A.E., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Stolc V., Royce T.E., Rozowsky J.S., Urban A.E., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Royce T.E., Rozowsky J.S., Urban A.E., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Rozowsky J.S., Urban A.E., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Urban A.E., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Tongprasit W., Samanta M., Weissman S., Samanta M., Weissman S., Weissman S., et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–2246. doi: 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- Carninci P., Kasukawa T., Katayama S., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., Kasukawa T., Katayama S., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., Katayama S., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., Oyama R., Ravasi T., Lenhard B., Wells C., Ravasi T., Lenhard B., Wells C., Lenhard B., Wells C., Wells C., et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Cooper S.J., Trinklein N.D., Anton E.D., Nguyen L., Myers R.M., Trinklein N.D., Anton E.D., Nguyen L., Myers R.M., Anton E.D., Nguyen L., Myers R.M., Nguyen L., Myers R.M., Myers R.M. Comprehensive analysis of transcriptional promoter structure and function in 1% of the human genome. Genome Res. 2006;16:1–10. doi: 10.1101/gr.4222606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium, The ENCODE (ENCyclopedia of DNA Elements) Project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium, Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007 doi: 10.1038/nature05874. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein M., Sonnhammer E.L., Chothia C., Sonnhammer E.L., Chothia C., Chothia C. Volume changes in protein evolution. J. Mol. Biol. 1994;236:1067–1078. doi: 10.1016/0022-2836(94)90012-4. [DOI] [PubMed] [Google Scholar]
- Giardine B., Riemer C., Hardison R.C., Burhans R., Elnitski L., Shah P., Zhang Y., Blankenberg D., Albert I., Taylor J., Riemer C., Hardison R.C., Burhans R., Elnitski L., Shah P., Zhang Y., Blankenberg D., Albert I., Taylor J., Hardison R.C., Burhans R., Elnitski L., Shah P., Zhang Y., Blankenberg D., Albert I., Taylor J., Burhans R., Elnitski L., Shah P., Zhang Y., Blankenberg D., Albert I., Taylor J., Elnitski L., Shah P., Zhang Y., Blankenberg D., Albert I., Taylor J., Shah P., Zhang Y., Blankenberg D., Albert I., Taylor J., Zhang Y., Blankenberg D., Albert I., Taylor J., Blankenberg D., Albert I., Taylor J., Albert I., Taylor J., Taylor J., et al. Galaxy: A platform for interactive large-scale genome analysis. Genome Res. 2005;15:1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giresi P.G., Kim J., McDaniell R.M., Iyer V.R., Lieb J.D., Kim J., McDaniell R.M., Iyer V.R., Lieb J.D., McDaniell R.M., Iyer V.R., Lieb J.D., Iyer V.R., Lieb J.D., Lieb J.D. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res. 2007 doi: 10.1101/gr.5533506. (this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J., Denoeud F., Frankish A., Reymond A., Chen C.K., Chrast J., Lagarde J., Gilbert J.G., Storey R., Swarbreck D., Denoeud F., Frankish A., Reymond A., Chen C.K., Chrast J., Lagarde J., Gilbert J.G., Storey R., Swarbreck D., Frankish A., Reymond A., Chen C.K., Chrast J., Lagarde J., Gilbert J.G., Storey R., Swarbreck D., Reymond A., Chen C.K., Chrast J., Lagarde J., Gilbert J.G., Storey R., Swarbreck D., Chen C.K., Chrast J., Lagarde J., Gilbert J.G., Storey R., Swarbreck D., Chrast J., Lagarde J., Gilbert J.G., Storey R., Swarbreck D., Lagarde J., Gilbert J.G., Storey R., Swarbreck D., Gilbert J.G., Storey R., Swarbreck D., Storey R., Swarbreck D., Swarbreck D., et al. GENCODE: producing a reference annotation for ENCODE. Genome Biol. 2006;(Suppl 1) 7:S4.1–S4.9. doi: 10.1186/gb-2006-7-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan I.K., Rogozin I.B., Glazko G.V., Koonin E.V., Rogozin I.B., Glazko G.V., Koonin E.V., Glazko G.V., Koonin E.V., Koonin E.V. Origin of a substantial fraction of human regulatory sequences from transposable elements. Trends Genet. 2003;19:68–72. doi: 10.1016/s0168-9525(02)00006-9. [DOI] [PubMed] [Google Scholar]
- Karolchik D., Baertsch R., Diekhans M., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Baertsch R., Diekhans M., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Diekhans M., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Schwartz M., Sugnet C.W., Thomas D.J., Sugnet C.W., Thomas D.J., Thomas D.J., et al. The UCSC Genome Browser Database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W.J. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.K., Shibata Y., Rao B., Strahl B.D., Lieb J.D., Shibata Y., Rao B., Strahl B.D., Lieb J.D., Rao B., Strahl B.D., Lieb J.D., Strahl B.D., Lieb J.D., Lieb J.D. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat. Genet. 2004;36:900–905. doi: 10.1038/ng1400. [DOI] [PubMed] [Google Scholar]
- Ng P., Wei C.L., Sung W.K., Chiu K.P., Lipovich L., Ang C.C., Gupta S., Shahab A., Ridwan A., Wong C.H., Wei C.L., Sung W.K., Chiu K.P., Lipovich L., Ang C.C., Gupta S., Shahab A., Ridwan A., Wong C.H., Sung W.K., Chiu K.P., Lipovich L., Ang C.C., Gupta S., Shahab A., Ridwan A., Wong C.H., Chiu K.P., Lipovich L., Ang C.C., Gupta S., Shahab A., Ridwan A., Wong C.H., Lipovich L., Ang C.C., Gupta S., Shahab A., Ridwan A., Wong C.H., Ang C.C., Gupta S., Shahab A., Ridwan A., Wong C.H., Gupta S., Shahab A., Ridwan A., Wong C.H., Shahab A., Ridwan A., Wong C.H., Ridwan A., Wong C.H., Wong C.H., et al. Gene identification signature (GIS) analysis for transcriptome characterization and genome annotation. Nat. Methods. 2005;2:105–111. doi: 10.1038/nmeth733. [DOI] [PubMed] [Google Scholar]
- Sabo P.J., Kuehn M.S., Thurman R., Johnson B.E., Johnson E.M., Cao H., Yu M., Rosenzweig E., Goldy J., Haydock A., Kuehn M.S., Thurman R., Johnson B.E., Johnson E.M., Cao H., Yu M., Rosenzweig E., Goldy J., Haydock A., Thurman R., Johnson B.E., Johnson E.M., Cao H., Yu M., Rosenzweig E., Goldy J., Haydock A., Johnson B.E., Johnson E.M., Cao H., Yu M., Rosenzweig E., Goldy J., Haydock A., Johnson E.M., Cao H., Yu M., Rosenzweig E., Goldy J., Haydock A., Cao H., Yu M., Rosenzweig E., Goldy J., Haydock A., Yu M., Rosenzweig E., Goldy J., Haydock A., Rosenzweig E., Goldy J., Haydock A., Goldy J., Haydock A., Haydock A., et al. Genome-scale mapping of DNase I sensitivity in vivo using tiling DNA microarrays. Nat. Methods. 2006;3:511–518. doi: 10.1038/nmeth890. [DOI] [PubMed] [Google Scholar]
- Shiraki T., Kondo S., Katayama S., Waki K., Kasukawa T., Kawaji H., Kodzius R., Watahiki A., Nakamura M., Arakawa T., Kondo S., Katayama S., Waki K., Kasukawa T., Kawaji H., Kodzius R., Watahiki A., Nakamura M., Arakawa T., Katayama S., Waki K., Kasukawa T., Kawaji H., Kodzius R., Watahiki A., Nakamura M., Arakawa T., Waki K., Kasukawa T., Kawaji H., Kodzius R., Watahiki A., Nakamura M., Arakawa T., Kasukawa T., Kawaji H., Kodzius R., Watahiki A., Nakamura M., Arakawa T., Kawaji H., Kodzius R., Watahiki A., Nakamura M., Arakawa T., Kodzius R., Watahiki A., Nakamura M., Arakawa T., Watahiki A., Nakamura M., Arakawa T., Nakamura M., Arakawa T., Arakawa T., et al. Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. Proc. Natl. Acad. Sci. 2003;100:15776–15781. doi: 10.1073/pnas.2136655100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinklein N.D., Aldred S.J., Saldanha A.J., Myers R.M., Aldred S.J., Saldanha A.J., Myers R.M., Saldanha A.J., Myers R.M., Myers R.M. Identification and functional analysis of human transcriptional promoters. Genome Res. 2003;13:308–312. doi: 10.1101/gr.794803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingender E., Chen X., Hehl R., Karas H., Liebich I., Matys V., Meinhardt T., Pruss M., Reuter I., Schacherer F., Chen X., Hehl R., Karas H., Liebich I., Matys V., Meinhardt T., Pruss M., Reuter I., Schacherer F., Hehl R., Karas H., Liebich I., Matys V., Meinhardt T., Pruss M., Reuter I., Schacherer F., Karas H., Liebich I., Matys V., Meinhardt T., Pruss M., Reuter I., Schacherer F., Liebich I., Matys V., Meinhardt T., Pruss M., Reuter I., Schacherer F., Matys V., Meinhardt T., Pruss M., Reuter I., Schacherer F., Meinhardt T., Pruss M., Reuter I., Schacherer F., Pruss M., Reuter I., Schacherer F., Reuter I., Schacherer F., Schacherer F. TRANSFAC: An integrated system for gene expression regulation. Nucleic Acids Res. 2000;28:316–319. doi: 10.1093/nar/28.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D., Zhang Z., Harrison P.M., Karro J., Carriero N., Gerstein M., Zhang Z., Harrison P.M., Karro J., Carriero N., Gerstein M., Harrison P.M., Karro J., Carriero N., Gerstein M., Karro J., Carriero N., Gerstein M., Carriero N., Gerstein M., Gerstein M. Integrated pseudogene annotation for human chromosome 22: Evidence for transcription. J. Mol. Biol. 2005;349:27–45. doi: 10.1016/j.jmb.2005.02.072. [DOI] [PubMed] [Google Scholar]