Abstract
For the ∼1% of the human genome in the ENCODE regions, only about half of the transcriptionally active regions (TARs) identified with tiling microarrays correspond to annotated exons. Here we categorize this large amount of “unannotated transcription.” We use a number of disparate features to classify the 6988 novel TARs—array expression profiles across cell lines and conditions, sequence composition, phylogenetic profiles (presence/absence of syntenic conservation across 17 species), and locations relative to genes. In the classification, we first filter out TARs with unusual sequence composition and those likely resulting from cross-hybridization. We then associate some of those remaining with proximal exons having correlated expression profiles. Finally, we cluster unclassified TARs into putative novel loci, based on similar expression and phylogenetic profiles. To encapsulate our classification, we construct a Database of Active Regions and Tools (DART.gersteinlab.org). DART has special facilities for rapidly handling and comparing many sets of TARs and their heterogeneous features, synchronizing across builds, and interfacing with other resources. Overall, we find that ∼14% of the novel TARs can be associated with known genes, while ∼21% can be clustered into ∼200 novel loci. We observe that TARs associated with genes are enriched in the potential to form structural RNAs and many novel TAR clusters are associated with nearby promoters. To benchmark our classification, we design a set of experiments for testing the connectivity of novel TARs. Overall, we find that 18 of the 46 connections tested validate by RT-PCR and four of five sequenced PCR products confirm connectivity unambiguously.
In recent years there have been a number of experiments using genomic tiling microarrays that have found significantly more transcribed DNA sequences in the human genome than had been previously annotated as genes (see Kapranov et al. 2002, Rinn et al. 2003, Bertone et al. 2004; Cheng et al. 2005). The biological functions of this vast quantity of additional transcribed RNA are not yet fully understood. There have been independent experiments using complementary sequencing technologies that have also detected large amounts of previously unidentified transcription (Carninci et al. 2005). Genome tiling arrays have also been used for transcript mapping in a variety of different organisms besides human: Arabidopsis thaliana (Yamada et al. 2003), Drosophila melanogaster (Stolc et al. 2004, Manak et al. 2006), Saccharomyces cerevisiae (David et al. 2006), and Oryza sativa (Li et al. 2006).
One of the goals of the ENCODE (ENCyclopedia of DNA Elements) project (The ENCODE Project Consortium 2004) is to map out and determine the function of these unannotated transcripts for the 1% of the human genome selected for the pilot phase of the project. For the selected ENCODE regions, RNA transcript maps were constructed for a variety of cell lines and biological conditions (The ENCODE Project Consortium 2007).
Consistent with earlier studies, a large fraction of the sequences identified as transcribed are not in annotated genomic regions. An important result obtained from these experiments was the discovery of tissue-specific alternative transcription start sites (TSSs), found by conducting 5′ RACE extensions from exons of known transcripts. Many of the TSSs were found to be >100 kb upstream of the annotated start site. Although these alternate long transcripts account for some of the novel transcribed regions detected, the majority remain unexplained. These long transcripts demonstrate that gene loci are quite complex and that there is probably a multiplicity of alternative isoforms that are transcribed from most complex loci. Even in the set of well-curated genes for the ENCODE regions (the GENCODE/HAVANA annotation) (Harrow et al. 2006), we see on average 5.4 alternative isoforms per locus. This number is most likely a significant underestimate of the number of distinct transcripts arising from an average locus in all cell lines, especially when all cellular conditions are considered.
Transcribed regions detected by genomic tiling arrays are known as TARs (transcriptionally active regions [ARs]) (see Rinn et al. 2003) or alternatively as transfrags (transcribed fragments) (see Kapranov et al. 2002). Although novel transcribed regions have been observed and analyzed in previous works, in this article we present an overall characterization and systematic classification of novel TARs. Some of this classification is briefly discussed by The ENCODE Project Consortium (2007) where novel TARs are categorized on the basis of their vicinity to known genes. We extend this analysis by grouping the novel TARs into a number of distinct possible categories: (1A) novel TARs with peculiar sequence composition, (1B) novel TARs that are probably caused by cross-hybridization on the microarray, (2) novel TARs that are associated with known gene loci, and (3) novel TARs not associated with known genes but that can be grouped into clusters which may be novel transcribed loci.
Data sets for novel TARs and their associated information should not be thought of as regular gene annotation, since unlike genes, properties such as the connectivity between novel TARs, which potentially form spliced transcripts, are not very well defined. Moreover, TARs have additional information such as the fluorescent array signal that is not usually associated with gene annotation. Thus, existing databases such as the UCSC Genome Browser (Kent et al. 2002), Gene Expression Omnibus (Edgar et al. 2002), or ArrayExpress (Brazma et al. 2003) do not have the flexibility to store sets of TARs with all the associated information and make them accessible in an efficient manner. For this reason we have constructed a database (DART: Database of Active Regions and Tools) for encapsulating our classification. The database is optimized for browsing sets of TARs discovered in tiling microarray experiments. In addition, the database allows the storage of sites of transcription factor binding and modifications called BARs (binding active regions), which are important to associate with TARs. We have also constructed a set of tools (Active Region Comparer [ARC]) that can be used for the comparison of multiple sets of ARs with each other and with annotations from Ensembl (Birney et al. 2006). Both the database and tools are connected with the UCSC Genome Browser for automated visualization of custom tracks.
The DART methodology developed in this article is a first-pass analysis of the novel TAR data sets and transcript maps that are available today as part of the pilot phase of the ENCODE project. An optimal approach to understanding the biological role of the multitude of novel TARs is to couple array experiments with medium-scale follow-up experiments. As an initial iteration of this process, we used the results of our classification to design some small-scale experiments that investigated the connectivity of novel TARs to exons of known genes and the connectivity between novel TARs clustered into novel transcribed loci. This validation experiment demonstrates that ∼40% of both the novel TARs tested for association with an exon of a known gene and the pairs of novel TARs tested for inclusion in a novel transcribed loci can be confirmed to be connected in a transcribed RT-PCR product. When the next phase of the ENCODE project scales to the whole genome, the resulting experimental data can be used to optimize the classification procedure in future iterations
Results
Novel transcribed regions
Transcript maps were constructed across the 44 ENCODE regions using genomic tiling microarrays for 11 different cell lines and conditions (The ENCODE Project Consortium 2007). The 44 ENCODE regions span 30 Mb of genomic sequence, half of which comes from manually selected gene loci (e.g., HOX cluster and CFTR locus) and half comes from 500-kb regions chosen to stratify differing levels of both gene density and nonexonic conservation with mouse. The 11 different cell lines and conditions were a combination of both poly(A)+ and total RNA samples. Transcript maps were constructed by hybridizing reverse transcribed, double-stranded cDNA to a high-density oligonucleotide tiling array that covered one strand of the ENCODE regions.
TARs were determined by locating stretches of oligonucleotide probes with high hybridization signals compared to background. The signal thresholds used to identify these transcribed genomic regions were determined using bacterial control sequences included on the Affymetrix tiling microarrays (Kampa et al. 2004). We note that the amount of transcription detected and the fraction that is in annotated regions are dependent on the signal threshold used (see Royce et al. 2005; Emanuelsson et al. 2007). Using a more stringent threshold, we detect fewer overall TARs. However, the percentage that corresponds to annotated exons increases because novel TARs tend to be transcribed at lower levels than exonic TARs. A threshold was determined such that the false-positive rate from bacterial negative controls was only 5% for each of the cell lines and conditions mapped. There has been an ongoing debate in the genomics community as to the fraction of the human genome that is transcribed. In The ENCODE Project Consortium (2007), it has been determined that ∼90% of the human genome is transcribed as primary transcripts. However, the use of a stringent threshold for tiling microarray signal selects for genomic regions that are transcribed as part of processed (spliced) RNAs. Thus, the large number of novel TARs detected as part of the ENCODE project’s pilot phase are more likely due to components of processed transcripts rather than due to the basal level of transcribed genomic DNA. Our DART classification procedure attempts to categorize these novel transcribed regions as part of known genes and into potential novel transcribed loci. Although many of the TARs that were detected correspond to exons of known genes, this study focuses on the novel TARs that do not match exonic sequences. These novel, unannotated TARs lie either within the introns of known genes or within the intergenic regions between known genes. Here we will use the set of GENCODE/HAVANA annotation (Harrow et al. 2006), which is a comprehensive set of all the well-curated transcripts contained within the ENCODE regions.
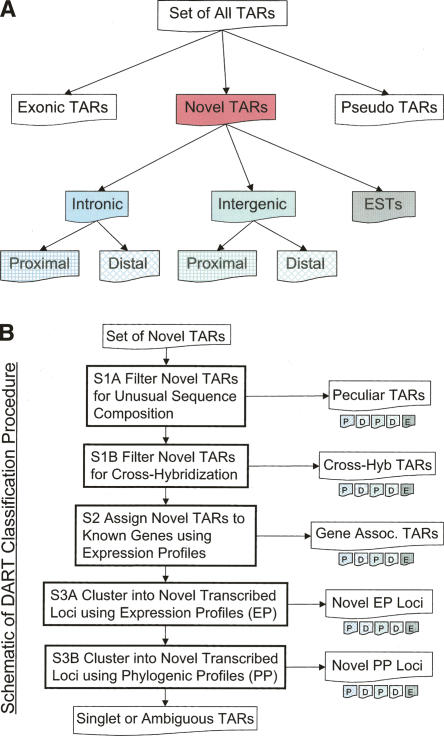
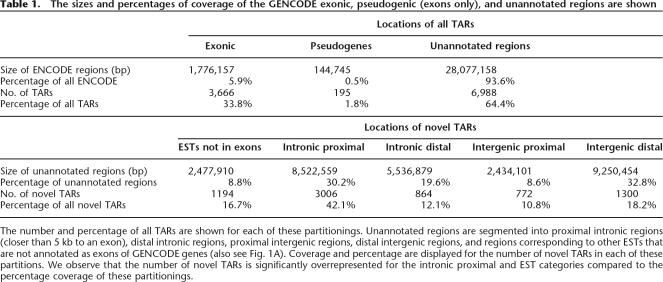
The initial set of all TARs generated can be classified into three basic categories: TARs corresponding to known or putative GENCODE genes, TARs overlapping annotated pseudogenes, and novel TARs in unannotated regions. TARs overlapping pseudogenes are ambiguous given the homology of the pseudogene sequence to its parent gene, both of which are potentially transcribed. Other more detailed studies of pseudogene transcription have determined that a small but significant fraction are transcribed and can be distinguished from parental gene transcription (Zheng et al. 2005). However, in order to avoid these ambiguities, for the purposes of this analysis the sets of TARs are filtered for those that intersect low complexity repeats or any annotated pseudogene in the ENCODE regions. Novel TARs are then classified into one of the following categories: (1) intronic TARs, (2) intergenic TARs, and (3) TARs that match other ESTs that were not part of the GENCODE annotation (typically unspliced ESTs that do not contain a polyadenylation signal). The intergenic and intronic TAR sets are further subdivided into those that are proximal subsets that are within 5 kb of GENCODE exons and distal subsets that are further than 5 kb (for a diagram of this classification, see Fig. 1A). The distribution of TAR locations can be seen in Table 1, where we observe that nearly half of the novel TARs are in intronic regions proximal to exons of known genes. The table also includes the 195 TARs that intersect pseudogenes prior to their removal. In Figure 1B we develop a strategy for a more detailed classification of sets of novel TARs, which will be described in more detail in the steps below. Each of these classified sets can also be individually partitioned as per Figure 1A, on the basis of proximity to annotation.
Figure 1.
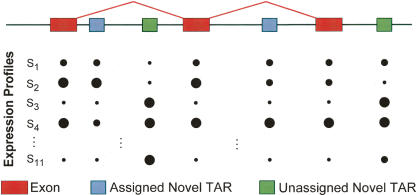
(A) Schema for the partitioning of TARs on the basis of location relative to GENCODE genes and pseudogenes (also see Table 1). Proximal regions are located within 5 kb of the nearest GENCODE exon. (B) Outline of the DART classification procedure of novel TARs. Novel TARs are first filtered on the basis of sequence composition (step 1), and then a fraction of the remaining novel TARs are associated with known genes (step 2). A portion of the remaining novel TARs are clustered in novel transcribed loci on the basis of expression profiles (EPs) and phylogenetic profiles (PPs) (step 3). See Table 2 for the numbers of novel TARs classified by each of these steps. The singlet and ambiguous TARs are what remains at the end of the classification procedure.
Table 1.
The sizes and percentages of coverage of the GENCODE exonic, pseudogenic (exons only), and unannotated regions are shown
The number and percentage of all TARs are shown for each of these partitionings. Unannotated regions are segmented into proximal intronic regions (closer than 5 kb to an exon), distal intronic regions, proximal intergenic regions, distal intergenic regions, and regions corresponding to other ESTs that are not annotated as exons of GENCODE genes (also see Fig. 1A). Coverage and percentage are displayed for the number of novel TARs in each of these partitions. We observe that the number of novel TARs is significantly overrepresented for the intronic proximal and EST categories compared to the percentage coverage of these partitionings.
Each novel TAR has a number of distinct features: expression profile across the biological samples mapped, genomic location relative to known GENCODE annotation, sequence composition, sequence conservation, and phylogenetic profile of conservation (Fig. 2). In the following analysis, we shall make use of some of these features when grouping the sets of novel TARs. As mentioned earlier, novel TARs will be grouped into the following distinct categories (1) potentially artifactual TARs that are caused by peculiar sequence composition or cross-hybridization, (2) novel TARs that can be associated with known gene loci, and (3) novel TARs that can be clustered into groups forming potential novel transcribed loci. For the remaining unclassified novel TARs with above average array signal, additional clustering is performed on the basis of vicinity and phylogenetic similarity. See Figure 1B for a schematic of the stepwise classification procedure. Many of the DART classification steps use the expression profiles of individual novel TARs across the eleven different cell lines and conditions. For each novel TAR, we also construct a phylogenetic profile across the species sequenced by the ENCODE Consortium (The ENCODE Project Consortium 2007). These profiles identify which of these species contain the novel TAR in a syntenic region. The classification uses other information as well, such as the sequence composition of novel TARs and their location relative to known genes. We also study the protein coding potential for novel TARs by searching for homologous protein sequences, and we investigate the likelihood of the various categories of TARs to form structured RNAs (using RNAz) (Washietl et al. 2005). All of these features, as well as the classification sets, are stored in the DART database.
Figure 2.
Summary of the features that are associated with each novel TAR and that are utilized by the classification procedure.
Step 1A: Filtering TARs for peculiar sequence composition
Genomic tiling microarrays interrogate genomic sequences by the use of short oligonucleotide probes that tile the region of interest. There are two main effects that can cause regions to erroneously appear as transcribed. The first effect results from the basic mechanism by which array hybridization works, which is the binding of a sample’s cDNA to its matching reverse complement DNA oligonucleotide probe. The amount of cDNA that hybridizes to a particular spot on the microarray, and the corresponding fluorescent signal measured, are subject to the binding affinity between the cDNA and probe, which is in turn dependent on the sequence composition of the oligonucleotide. Thus probes with higher G/C content tend to bind more tightly and show greater fluorescent signal (SantaLucia 1998). In addition, short sequence motifs that bind with higher affinity would cause many probes to show abnormally high signal in genomic regions not transcribed. Probe sequence effects are dramatically reduced by the use of sliding window scoring, which averages the signal from multiple oligonucleotide probes in a short genomic span (see Cawley et al. 2004; Kampa et al. 2004; Royce et al. 2005). However, biases due to oligonucleotide probe sequence effects are still evident when one compares the G/C content of sequences detected to be transcribed against all annotated sequences.
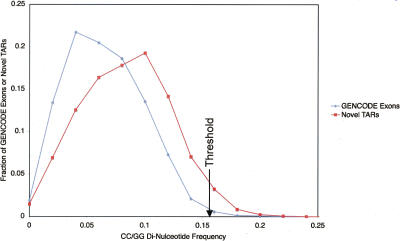
In The ENCODE Project Consortium (2007), the di-nucleotide frequency was compared between novel TARs, exonic TARs, all exons, and randomly selected sequences. This analysis showed that the di-nucleotide frequency of novel TARs was more similar to that for exons than random sequences. However, for the CC/GG and AA/TT di-nucleotides (both the forward and reverse complement di-nucleotides are combined since TARs are not stranded), the average frequency was significantly different from the frequency for annotated exons. Figure 3 illustrates this difference where the distribution of CC/GG frequencies for novel TARs is skewed to higher frequencies than that for GENCODE exons. Thus CC/GGs occur more often in novel TARs than in known exons, while the AA/TT frequency for novel TARs is lower than for exons (see Supplemental Fig. 1). In order to be cautious, we removed novel TARs whose CC/GG frequency was above the top 1% of CC/GG frequencies for GENCODE exons as well as those whose AA/TT frequency was below the bottom 1% for exons. There are 380 novel TARs whose CC/GG frequency is >0.156 (indicated by the black arrow on Fig. 3), as well as 175 novel TARs whose AA/TT frequency is <0.004. Thus 503 novel TARs were excluded from the 6988 total novel TARs, leaving 6485 novel TARs that we shall consider.
Figure 3.
Plot of the distribution of GENCODE exons (blue line) and novel TARs (red line) against CC/GG di-nucleotide frequency. The distribution of novel TARs is skewed to high CC/GG di-nucleotide frequencies. A black arrow indicates the di-nucleotide frequency (0.155) above which only ∼1% of the GENCODE exons are found. This threshold is used to filter novel TARs with peculiar sequence composition (CC/GG di-nucleotide frequency higher than 0.155).
Step 1B: Filtering TARs for cross-hybridization
The second main microarray artifact, which can lead to false-positive detection of transcribed regions, is cross-hybridization. Cross-hybridization happens when oligonucleotide probes on the array hybridize to cDNA from transcripts that have partial sequence complementarity to the probe but the transcripts originate from somewhere else in the genome. One standard approach is to take the sequences of novel transcribed regions and BLAST (Altschul et al. 1990) them against the current build of the genome to identify sites of potential cross-hybridization. However, the limitation of this approach is that once one has located a potential site of cross-hybridization, which could be annotated as either part of a known transcript or an additional putative novel TAR, the true source of transcription remains ambiguous (one or both sites could be transcribed). The approach that we propose would resolve this ambiguity.
Using the method by which novel TARs will be determined to be associated with known gene loci by use of coexpression of novel TARs with exons of known GENCODE genes, we propose the following procedure: We first identify the most likely source for cross-hybridization by using BLAST (we call the matching region a blastTAR). Only TARs that have a significant match are considered (at a BLAST e-value of <10−5 or a bit score of 54.0, which corresponds to ∼40–50 nucleotides with >90% sequence identity). The expression profile of the original novel TAR is then compared against exons of genes in the local genomic vicinity of the blastTAR. If the novel TAR is coexpressed with the blastTAR’s surrounding exons, then the most likely explanation is that the blastTAR is the primary source of transcription and the original novel TAR was detected because of cross-hybridization. Determining the true source of transcription from two genomic locations with a high degree of sequence similarity is thus made possible by using the expression profiles of the novel TARs compared with exons nearby the potential cross-hybridization site.
Of the 6485 filtered novel TARs from step 1A, 658 have matches with an e-value of 10−5 or better. Since the ENCODE regions only cover ∼1% of the genome, a naïve expectation is that only ∼1% of these matches would be located within the ENCODE regions (we can only implement this procedure for blastTARs that are located within ENCODE since we need to compare them with the expression profiles of nearby exons). blastTARs that are located in the same ENCODE region as the original TAR need to be treated separately (this is discussed in further detail later on). However, there are no novel TARs for which a blastTAR is located in a different ENCODE region. Even though we are unable to utilize this approach for the novel TARs in ENCODE, it will be applicable when tiling array studies that cover the entire genome become more abundant.
Step 2: Association of novel TARs with known gene loci
We want to address the question of how many of the novel TARs can be confidently assigned to known gene loci. By this we mean that the novel TARs are transcribed as parts of longer transcripts, which are as yet unannotated isoforms of transcripts from a specific gene locus or of distinct RNAs that are coregulated with the gene of interest. In order to make these assignments, we identify novel TARs that are coexpressed with exons of genes in the vicinity of the novel TARs. We do this by computing the Pearson correlation coefficient between the expression profiles of novel TARs and the expression profiles of nearby exons (Fig. 4). This method is similar to how different genes are determined to be coexpressed. Here, however, we are comparing the expression profiles of individual novel TARs and exons, not those of entire transcripts. For a gene that only encodes a single transcript (i.e., has no alternative isoforms), the expression profile of the gene should be the same as that for each of its constituent exons. However, for a locus that is transcribed as multiple different isoforms, the expression profiles of the different exons may be different. Thus, a novel TAR which is coexpressed with an exon of a known gene can be assigned with some confidence to that locus as part of an alternative isoform or as part of a distinct coregulated RNA.
Figure 4.
Illustration showing how novel TARs can be associated with known genes by identifying novel TARs that are coexpressed with exons of known genes. Coexpression is determined by computing the Pearson correlation of expression profiles of array signals between novel TARs and nearby (closer than 20 kb) exons. The sizes of the circles correspond to the fluorescent signal intensity measured on the tiling arrays for each of the 11 different cell lines indicated by S1 through S11.
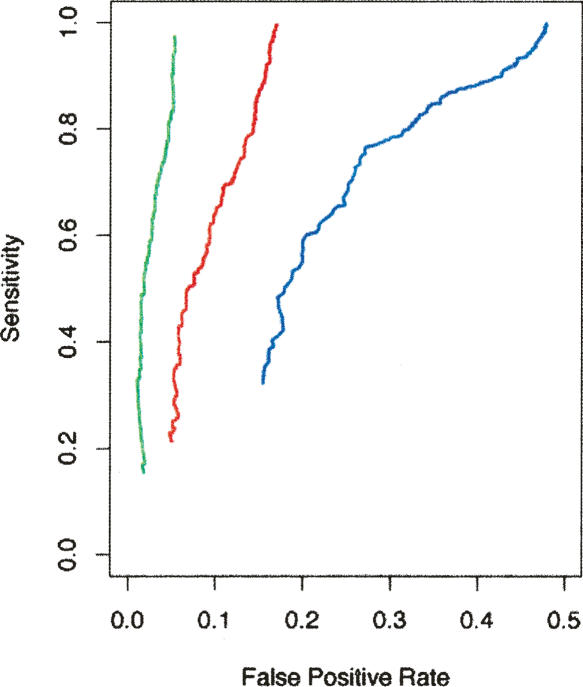
In order to demonstrate that this method works, we first took the set of all known GENCODE genes in the ENCODE regions and computed the expression profiles for all component exons. For each exon, we can test whether we can assign it to the correct gene by comparing its expression profile with the expression profiles of nearby exons. The assignment is made to the target gene which has an exon with the highest correlation. In Figure 5 we plot sensitivity against the false-positive rate using this assignment procedure. The Pearson correlation threshold for making an assignment is what parameterizes each curve. The blue curve represents the assignment to exons for genes anywhere in the ENCODE regions; the red and green curves are for assignment to exons of genes that are within 100 kb and 20 kb of the exon that is being tested. As expected, we see that the accuracy of the assignment is improved by restricting attention to nearby exons. See Supplemental material for a more detailed description of this simulation.
Figure 5.
Plot of sensitivity against false-positive rate for the assignment of exons of known genes to the correct gene on the basis of the exon being coexpressed with other exons. The blue curve is calculated where an exon is allowed to be assigned to any gene in the genome, while the red and green curves are where the assignment is limited to genes which have exons within 100 kb and 20 kb of the target exon, respectively. Restricting assignment to exons of nearby genes reduces the false-positive rate of the assignment. The Pearson correlation of the best possible assignment for each exon is the threshold which parameterizes each curve.
For each novel TAR (for more details, see Methods), we use the above method to find the known exon within a 20-kb window on either side of the TAR and from either strand that has the highest Pearson correlation between its expression profile and that of the novel TAR. We choose to use a Pearson correlation of 0.9 as a threshold, as that corresponds to a P-value of <0.05 (given that the correlation coefficient is computed by comparing expression profiles which have 11 dimensions and on average each novel TAR, has ∼19 known exons within 20 kb).9 Thus, we can associate 955 of the 6485 filtered novel TARs with a known GENCODE exon. From this analysis, we can assign >13% of the original set of 6988 novel TARs as part of new alternative isoforms of known transcripts.
Step 3: Clustering novel TARs into novel transcribed loci
Step 3A: Clusters based on expression profiles
After assigning 955 of the novel TARs to known gene loci, we have 5530 remaining. We cluster coexpressed novel TARs into groups, which we call novel transcribed loci. However, in the assignment of novel TARs to known genes, we only assigned those that were highly correlated with exons of known genes. There are likely many more novel TARs in the remaining group of 5530 that should be assigned to known gene loci but were not because their correlations were below the chosen threshold. In order to focus attention on novel TARs that have a low likelihood of being associated with known gene loci, we first select a subset of the 5530 novel TARs that have at most a Pearson correlation of 0.1 with any GENCODE exons within 20 kb of the novel TAR. Using this criterion, we select a subset of 1846 novel TARs, which we group into novel TAR clusters as described below.
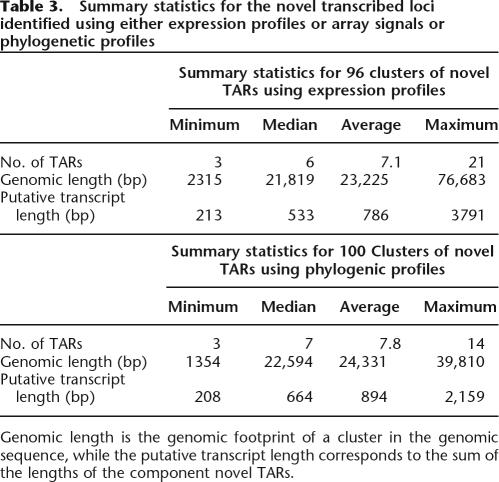
We construct a matrix of correlation coefficients between novel TARs in this set (correlations between novel TARs further than 20 kb apart are set to zero). We use k-means clustering (Hartigan and Wong 1979) with a k of 102, which meets the criterion set by Hartigan (1975) (see Methods for more details). With this value of k, we obtain 96 clusters that have three or more elements and are localized to one ENCODE region. The six remaining clusters, which are not considered, correspond to small groups of only two elements and one large group of novel TARs from multiple chromosomes, which is the set of remaining unclustered TARs. A summary of statistics for these novel TAR clusters is in Table 3.
Table 3.
Summary statistics for the novel transcribed loci identified using either expression profiles or array signals or phylogenetic profiles
Genomic length is the genomic footprint of a cluster in the genomic sequence, while the putative transcript length corresponds to the sum of the lengths of the component novel TARs.
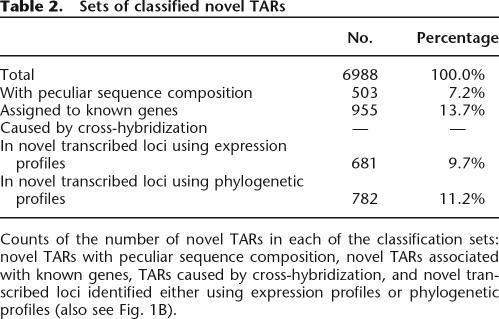
Table 2.
Sets of classified novel TARs
Counts of the number of novel TARs in each of the classification sets: novel TARs with peculiar sequence composition, novel TARs associated with known genes, TARs caused by cross-hybridization, and novel transcribed loci identified either using expression profiles or phylogenetic profiles (also see Fig. 1B).
Step 3B: Clusters based on phylogenetic profiles
Following the preceding steps of the DART classification procedure (steps 1A, 1B, 2, and 3A), we have 4748 novel TARs unassigned. We first partition this group into those with below average array signal, comprising 3122 novel TARs. For the remaining 1626 novel TARs with above average signal, we cluster them in a similar manner to the previous step using the phylogenetic profiles for 17 different species sequenced in the ENCODE regions instead of expression profiles (for more details, see Methods). A correlation matrix is computed between phylogenetic profiles of novel TARs that are within 20 kb of each other. We then use k-means clustering on this matrix and find optimal clustering for a k of 111. This clustering yields 100 clusters of three or more groups of TARs containing a total of 782 novel TARs, with a median cluster size of seven. Summary details of these clusters are also in Table 3. As with the k-means clustering using expression profiles, the majority of the novel TARs are together in one unclustered group.
DART (Database for Active Regions with Tools)
DART (DART.gersteinlab.org) has been developed to facilitate the flexible storage, visualization, and analysis of the growing number of experimentally defined sets of regions detected using genomic tiling microarrays. These are either sets of TARs or sites of transcription factor binding called BARs or more generally ARs. DART has been designed to address a number of challenging issues that arise when attempting to store and analyze this type of data. These challenges will clearly grow in the future, as the ENCODE project expands from the analysis of 1% of the genome to the entire genome, and as more increasingly diverse sets of ARs are experimentally determined. The key aspects of DART include the following:
Dealing with heterogeneous data sets: DART needs to be able to incorporate a rapidly growing number of sets of ARs that have been derived from a wide variety of experimental conditions. The current DART design is a first step toward allowing for multiple sets of ARs to be analyzed in a flexible fashion, including analyzing the unions and intersections of multiple sets and viewing the overlap among ARs in different AR sets.
Flexibility for storing different AR attributes: DART allows the flexible storage of different types of attributes associated with ARs, such as sequence information and array fluorescent signal intensities, as well as the adjustable groupings of ARs into subsets or clusters (potentially forming novel transcribed loci for the case of TARs). To accommodate this diversity, we use the Entity-Attribute-Value (EAV) data storage technique (Nadkarni et al. 1998) to define the attributes of either individual ARs or sets of ARs without modifying the database structure or program. These attributes can be used to search for desired AR sets.
Accommodating new genome builds: DART is designed to handle problems that occur as new builds (versions) of the human genome are defined and as the annotation associated with each AR set is updated to accommodate each new genome build. DART can store multiple values for AR genome locations corresponding to different genome builds. These coordinates are updated using the UCSC liftOver tool, which maps between genome builds (Kent et al. 2002).
Integrated linking to other Web resources for broader visualization and analysis: DART contains a number of capabilities designed to facilitate the integrated visualization and analysis of the data. These include both the ability to pass selected AR sets to the ARC for comparative analysis and annotation and the ability to display overlap among the ARs of different sets. Also, as described above, DART is integrated at several levels with the UCSC Genome Browser (Kent et al. 2002).
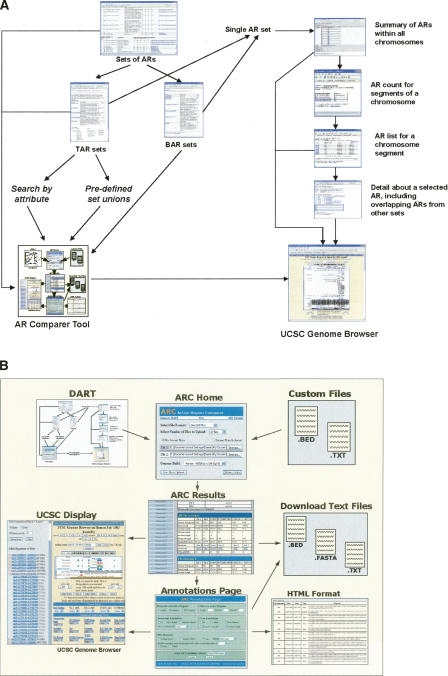
See Figure 6A for an overview of the current implementation of DART’s functionality, more details of which are provided in the Methods section.
Figure 6.
(A) The current functionality of DART is displayed. At the top level, one can access all the sets of ARs (either TARs or BARs) that are in the database. Upon selecting a collection of these sets, one can either transfer sets to the ARC tool or inspect each set individually. At the individual set level, ARs can be viewed either at a complete set level, chromosomal level, or a more local level. Individual ARs can be viewed with all their associated attributes. For an individual AR, DART also displays how it overlaps all other ARs in the database. Additionally, at multiple levels these sets can be visualized via the UCSC Genome Browser. (B) ARC Home accepts data sets from DART and from uploaded text files. Submission of the ARC Home form leads to the ARC Results page, which displays summary statistics for uploaded and newly generated data sets. From the ARC Results page, data sets may be downloaded, annotated in the ARC Annotations page, or visualized in the UCSC Display page. The Annotations page formats its processed data sets for presentation in HTML tables, for download as text files, and for export to the ARC Display page. Data sets sent to the UCSC Display page are loaded in the UCSC Genome Browser as custom tracks. From the UCSC Display page, one can return to the ARC Results page to repeat these analyses for other data sets.
Active Region Comparer tool
The ARC provides a Web-based interface for comparing, filtering, and annotating multiple sets of genomic regions, such as sets of TARs. The tool facilitates the analysis of ARs by determining how the regions in each set overlap those in other sets and by generating summary statistics to describe these relationships.
Thus ARC allows the user to find regions that are common to multiple sets as well as regions that are specific to one set and not another. Additionally, by interfacing with a local Ensembl database (Birney et al. 2006), we can obtain a region’s genomic annotation, which includes the sequence of the region, overlapping or nearby annotated transcripts, and other details such as the lengths and coordinates of overlapping and nearby exons. ARC also has an interface for exporting and visualizing multiple data sets via the UCSC Genome Browser, which displays sets of ARs alongside sets of genomic annotation to provide a graphical overview of the selected region. A diagram of how ARC works and its connectivity with the main DART database is presented in Figure 6B. ARC also has the functionality to view individual ARs together with surrounding TSSs, CpG islands, known transcription factor binding sites, and a local G/C content map using TAR-Vis (Supplemental Fig. 2). See Methods for further details about the inner workings of the ARC tool as well as the TAR-Vis visualizer connected to it.
Observations concerning novel TARs
Tandem duplicated TARs
While attempting to remove novel TARs that were likely caused by cross-hybridization, we found that none of the 658 novel TARs that had a BLAST e-value of 10−5 or better had a corresponding blastTAR located in a different ENCODE region. A naïve expectation would be that given that the ENCODE regions account for 1% of the human genome, ∼1% of the BLAST matches would be within the ENCODE regions. However, we find that there are 396 blastTARs located in the same ENCODE regions as their corresponding TARs. Of these TARs, 64 are located within 1 kb of the original TAR and 144 are located within 20 kb. Of the 396 blastTARs, 249 of them are actually different novel TARs (this makes sense, for if they have similar sequences they would typically also be detected as transcribed by the tiling arrays). These tandem sets of matching TARs come from many of the ENCODE regions, with the following three regions being most overrepresented: ENm006 (chromosome X from 152,635,144 to 153,973,591 with respect to human genome build NCBI Build 35), ENm007 (chromosome 19 from 59,023,584 to 60,024,460) and ENr233 (chromosome 15 from 41,520,088 to 42,020,088). These three ENCODE regions have tandem arrays of paralogs likely arising from segmental duplications (e.g., the ENm007 has a family of immunoglobulin-like receptors).
These tandem sets of novel TARs might be caused by cross-hybridization. However, since they are located in regions arising from local segmental duplication, it is not clear that cross-hybridization is the cause. For this reason, we chose not to remove them from the set of novel TARs under investigation.
Comparison of sets of novel TARs with RACE products
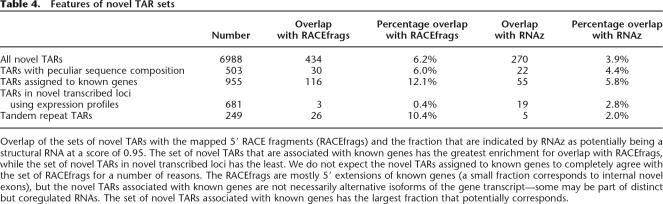
We first compared the different sets of novel TARs against the so-called RACEfrags or RACE (Rapid Amplification of cDNA Ends) fragments generated by hybridization of cloned 5′ RACE products off exons of known genes in the ENCODE regions (for more details, see (The ENCODE Project Consortium 2007). The RACEfrags such as transfrags or TARs are identified as transcribed regions; however, they also indicate the connectivity of the extended 5′ RACE products to the indexed exon from which the primer was selected. Thus, all 5′ RACEfrags upstream of an annotated TSS correspond to a novel 5′ end. The RACE reactions were done using RNAs from 12 tissues, different from the 11 cell lines and conditions that were used in mapping the TARs. We find that the set of all novel TARs has a 6% overlap with the RACEfrags, while the set of novel TARs assigned to gene loci has a 12% overlap (a twofold enrichment). In comparison, the set of novel TARs grouped into novel TAR clusters only has a 0.4% overlap with the RACEfrags, as expected (Table 4). In comparison, a randomly generated set of unannotated regions only has a 1.9% overlap with the RACEfrags (for more details, see Methods).
Table 4.
Features of novel TAR sets
Overlap of the sets of novel TARs with the mapped 5′ RACE fragments (RACEfrags) and the fraction that are indicated by RNAz as potentially being a structural RNA at a score of 0.95. The set of novel TARs that are associated with known genes has the greatest enrichment for overlap with RACEfrags, while the set of novel TARs in novel transcribed loci has the least. We do not expect the novel TARs assigned to known genes to completely agree with the set of RACEfrags for a number of reasons. The RACEfrags are mostly 5′ extensions of known genes (a small fraction corresponds to internal novel exons), but the novel TARs associated with known genes are not necessarily alternative isoforms of the gene transcript—some may be part of distinct but coregulated RNAs. The set of novel TARs associated with known genes has the largest fraction that potentially corresponds.
Structural RNAs
We also investigated the differing potential for the various sets of TARs to form structural RNAs using RNAz (see Methods) (Washietl et al. 2005). The ENCODE companion article in this issue (Washietl et al. 2007) deals with a comprehensive analysis of structural RNAs in the ENCODE regions and discusses approaches for detection of structural RNAs using computational approaches and transcriptional evidence. Here we take a somewhat different focus, investigating what fraction of the classified sets of novel TARs have the potential to form structural RNAs. Using a relatively stringent threshold score from RNAz of 0.95, which corresponds to structural RNA of high confidence, we find that the set of novel TARs that can be associated with known gene loci has the largest fraction with significant scores. We also note that the set of novel TARs with unusual sequence composition has above average enrichment for structural RNAs. This finding most likely reflects the fact that this set of novel TARs tends to have higher G/C content, which can affect the prediction made by RNAz (see Table 4).
Protein homology
By design, the translated sequences of the initial set of 6988 novel TARs do not have strong similarity to known protein sequences, since we filtered out those that have BLASTx matches to annotated genes in the genome (i.e., pseudogenes). However, there may be some novel TARs that have distant homology with gene relics. Using the profile hidden Markov model software HMMER (Eddy 1998), we find that only six of the translated novel TAR sequences have significant matches, all of which are located in intronic regions.
Comparison of novel TAR clusters with TSSs and transcription factor binding sites
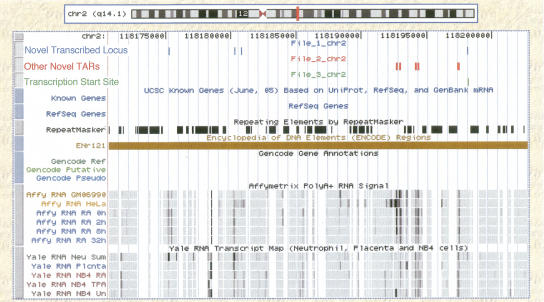
To test the validity of the 96 novel transcribed loci generated using expression profiles, we compare these clusters of novel TARs with two other data sets that were generated in The ENCODE Project Consortium (2007), the set of CAGE tags and paired-end-tags (ditags). These data sets been combined to form a set of 1144 known and putative TSSs. We find that six of the 96 novel TAR clusters have a TSS within 1 kb of either end (since the strandedness of a novel TAR cluster is undetermined). An example of one of these is shown in Figure 7, where we see a novel TAR cluster comprising four novel TARs with the rightmost TAR overlapping a putative TSS. This example is in a region of chromosome 2 (from 118175232–118198192, build NCBI Build 35) where there are no other annotated transcripts. Comparing the set of novel TAR clusters to the composite list of promoters identified in The ENCODE Project Consortium (2007), we find that 23 of the 96 novel TAR clusters have an end that is within 1 kb of a composite promoter.10 When we compare the 100 novel TAR clusters grouped on the basis of similar phylogenetic profiles, we find that 34 have an end within 1 kb of a TSS while 32 have an end within 1 kb of a composite promoter. We performed a simulation for random clusters of similar genomic extent to our novel TAR clusters and found that only 9.2 out of 100 would have an end within 1 kb of a TSS, while 17.5 out of 100 would have an end within 1 kb of a composite promoter (for details of the simulation, see Methods).
Figure 7.
Plot of a novel transcribed locus identified using the expression profile (the clustered TARs are shown in blue). Other novel TARs that are not part of this cluster are shown in red. In green we see the overlap of a putative transcription start site with the likely 5′ end of this cluster. There are no annotated transcripts in the region displayed (chr 2 from 118,175,232 to 118,198,192, NCBI Build 35). We also observe transcript maps for the 11 different cell lines and conditions (not all novel TARs are shown in this region).
Testing connectivity of transcripts using RT-PCR and sequencing
As a small-scale follow-up experiment, we selected 23 novel TARs that were assigned to known gene loci. These were selected such that both the novel TAR and its associated exon are both expressed in placental poly(A)+ RNA. By use of primer pairs generated from the novel TARs and their associated known exons, 23 RT-PCR reactions were performed.
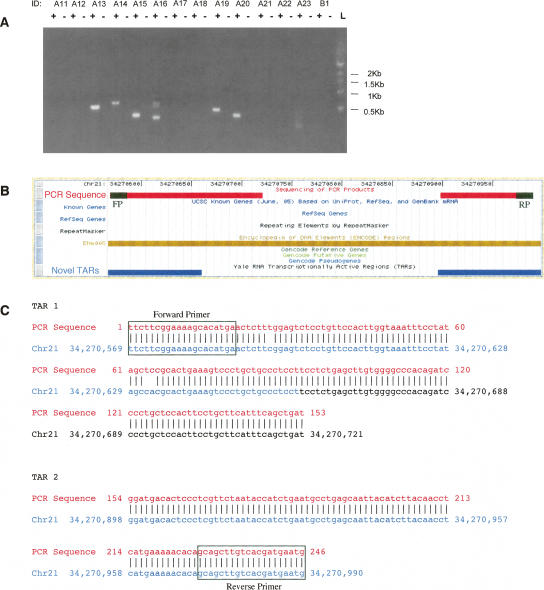
We found that nine out of the 23 primer pairs (39%) yielded a PCR product on the gel (with no band in the absence of RT), which is evidence for a transcribed sequence spanning both the TAR and the known exon. In addition, another 23 pairs of novel TARs that were grouped as being part of a novel TAR cluster were tested for connectivity by selecting a primer from each novel TAR sequence. Of these, again nine out of the 23 (39%) yielded a PCR product that provides experimental support for the connectivity of these novel TARs in a spliced RNA transcript. An additional two pairs of primers were selected as negative controls, neither of which showed any PCR product. The gel for some of these PCR products is presented in Figure 8A. Supplemental Table 1 lists all of the pairs of regions tested for connectivity as well as the presence or absence of a RT-PCR product. When we see a PCR product generated from primers for a pair of novel TARs or for a novel TAR and an exon, it implies that both of the sequences are transcribed and that the product is likely a portion of a spliced transcript that utilizes and connects between both of the sequences. In order to verify the PCR reactions, five PCR products were then directly sequenced using their respective forward and reverse primers. The five PCR products yielded sequences that align to the transcribed sequences tested for connectivity, only one of which could not be counted as confirmed. This PCR product was probably caused by cross-hybridization due to the sequence mapping better to another genomic location. Of the four PCR products that were confirmed by sequencing, one of them yielded a spliced sequence (see Fig. 8B,C) and three produced a sequence that was not spliced and included the intervening sequence between the two regions tested. These results together with the sequences obtained are shown in Supplemental Table 2. Even though not all the sequenced products were spliced, the results do confirm the RT-PCR products. Thus four of the five PCR products that were sequenced unambiguously confirm the connectivity of the associated pairs of novel TARs or novel TARs with known exons.
Figure 8.
(A) Image of an agarose gel of RT-PCR results from testing the connectivity of novel TARs with exons of known genes as well as between pairs of novel TARs clustered as a novel transcribed locus. This was performed using placental poly(A)+ RNA, where + indicates the presence of reverse-transcriptase; –, its absence; and L, the molecular weight ladder. A table of regions tested and their corresponding IDs and primer sequences is located in Supplemental Table 1. (B) Example of a pair of novel TARs (id B15) predicted to be associated with each other, potentially as part of a single transcript. This was confirmed by RT-PCR using placental RNA and was also successfully sequenced. The region displayed is on chromosome 21 from 34,270,568 to 34,270,998 (NCBI build 35). The sequence obtained from the PCR product is shown in red, the two connected novel TARs are in blue, and the forward and reverse primers are in black. (C) Alignment of the sequenced PCR product against the genomic sequence shows the transcript that connects the two novel TARs is spliced.
Discussion
We have developed the DART system for the classification and categorization of the large quantities of novel transcribed regions that have been identified in the human genome. We can group novel TARs with reasonable confidence into one of the following sets: novel TARs that are likely caused by unusual sequence composition or cross-hybridization, novel TARs that can be assigned to known genes, and novel TARs that can be clustered into novel transcribed loci. This last category of novel TARs possibly corresponds to entirely new transcripts.
To encapsulate our classification, we have constructed DART, a database and tool set designed for the storage and visualization of large quantities of TAR sets and all of their additional features. DART is also designed to have a flexible framework that can incorporate any information associated with sets of TARs. DART and its companion tool ARC facilitate the comparison and display of multiple sets of TARs (or a set of ARs such as transcription factor-binding sites) either though its own custom interface or via the UCSC Genome Browser.
We find that the set of novel TARs identified by the ENCODE Consortium has a number of interesting characteristics. There is enrichment in the potential for novel TARs to form structural RNAs compared with random sequences. This trend is especially prominent for the novel TARs that are associated with known gene loci. Some of these might correspond to structural RNAs that are coregulated with genes. We also find a significant overlap between the ends of clusters of novel TARs (novel transcribed loci) derived from either expression or phylogenetic profiles with both TSSs and promoters. There is also a significant enrichment among the novel TARs assigned to known gene loci for overlap with the 5′ RACE extensions of known genes identified in The ENCODE Project Consortium (2007).
We followed up our classification procedure by experimentally testing the connectivity of novel TARs that were assigned to known genes. Using RT-PCR, we found that 39% of the 23 novel TARs tested could be identified as part of a transcript that utilized the sequence of the novel TAR and at least one exon of the known gene. In principle, not all novel TARs that are assigned to known genes must be part of alternative isoforms of known transcripts. Some might correspond to other RNAs that are coregulated with transcripts from the locus. In addition, we tested the connectivity of identified clusters of novel TARs using RT-PCR. Again, we found that 39% of the 23 pairs of novel TARs yielded a PCR product, which is evidence of both the transcription and connectivity of the novel TARs within a single transcript. When a RT-PCR product is obtained from pairs of primers sourced from separated genomic regions (either two novel TARs or a novel TAR and an exon), this confirms that both regions are transcribed as well as that a single spliced transcript exists (of which the PCR product is a small piece) that utilizes the sequence of both regions tested. Of the five PCR products sequenced, four of the sequences match uniquely to the correct genomic location and further verify the results obtain by RT-PCR.
The data sets that were employed in the analysis presented in this article were from the transcript maps derived from 11 different cell lines and conditions for the ∼1% of the human genome included in the ENCODE regions. The statistical power of this procedure will increase nonlinearly as the number and size of the data sets increases: As the number of data sets increases, so will the accuracy with which novel TARs can be associated with known genes. In addition, when transcript maps cover the entire genome, we will be able to more confidently remove novel TARs that are caused by cross-hybridization. In the next phase of the ENCODE project, there will be many more data sets generated that will span the entire genome. The methods developed here can be employed to initially classify the large amount of novel transcription that will be identified. This classification followed by medium-scale experiments will lead to a better understanding of the function of the multitude of RNAs that are transcribed in human cells. This iterative approach, consisting of analysis followed by more detailed experiments that feed back to improve the analytical methods, will lead to a more complete understanding of the diversity of transcripts of the human genome.
Methods
Experimental testing of connectivity of genomic regions by RT-PCT and sequencing
Primer pairs were selected for 23 novel TARs that are expressed in placental RNA and are assigned to known gene loci. The primer sequences were selected from each novel TAR as well as from the exon of the gene with which the novel TAR had the strongest correlation. An additional 23 primer pairs were selected from pairs of different novel TARs that are present in placental RNA and could be clustered together using their expression profiles. An additional two pairs of primers were selected as negative controls from novel TARs that are located on different chromosomes. The regions selected and the corresponding primer sequences are available in Supplemental Table 1. One microgram of human placenta poly(A)+ RNA was used in a final volume of 20 μL RT reaction (50 ng/μL). RT reactions were primed by Oligo dT using Superscript II reverse transcriptase 200U in 20 μL reactions (Invitrogen). In parallel, reactions without reverse transcriptase (RTase minus) were also performed as the negative controls for genomic contamination. RT was followed by PCR amplification using the Advantage 2 PCR Enzyme System (Clontech). The 2 μL RT reaction and the 2 μL RTase minus negative control from the above were used side by side in 50 μL PCR reactions. The PCR program was started at 95°C for 30 sec, followed by 35 cycles of 95°C for 15 sec and 68°C for 1 min, and concluded by an extension cycle of 72°C for 3 min. The PCR products were visualized on a 1% agarose gel. Five of the PCR products were then sequenced using both the forward and reverse primers.
Expression profiles for sets of novel TARs and known exons
For each of the 11 different cell lines and conditions, a transcript map corresponds to fluorescent intensities for 755,457 25mer oligonucleotide probes tiling the nonrepetitive sequence of one strand of the ENCODE regions. The array hybridizations in The ENCODE Project Consortium (2007) were done using double stranded cDNA, thus the signal maps correspond to the signals from both strands. The 11 cells lines and conditions are GM06990 poly(A)+ RNA, HeLa poly(A)+ RNA, HL60 poly(A)+ RNA (0 h after treatment with retinoic acid, 2 h after treatment with RA, 8 h after treatment with RA, 32 h after treatment with RA), placental poly(A)+ RNA, neutrophil total RNA, and NB4 total RNA (untreated, treated with RA, treated with TPA). The transcript maps are first scaled to each other using quantile normalization (Bolstad et al. 2003). An expression profile is then calculated for each novel TAR as well as for each known GENCODE exon by computing the median fluorescent signal from all the oligonucleotide probes contained within the boundaries of the TAR or exon. Exons that are not in the tile path of the Affymetrix ENCODE array are excluded.
Phylogenetic profiles for sets of novel TARs
Phylogenetic profiles were generated using data derived from multi-species sequence alignment constructed by the ENCODE-MSA group (The ENCODE Project Consortium 2007). In this analysis, we surveyed the presence/absence of novel TARs in the orthologous genomic regions of other species. Sixteen mammals (chimp, baboon, macaque, marmoset, galago, rat, mouse, rabbit, cow, dog, rfbat, shrew, armadillo, elephant, tenrec, monodelphis) were selected for this study, since they had received better sequence coverage than the other species used by the MSA group. A TAR was considered as “present” (given a value of one and otherwise zero) in a species if more than one-third of its content was detected in the MSA alignment from that species. We used the alignments constructed by the program TBA (Threaded Blockset Aligner) (Blanchette et al. 2004).
K-means clustering of novel TARs
We use k-means clustering to form groups of nearby novel TARs. The k-means clustering is done using the R statistical package with the default Hartigan and Wong (1979) algorithm. We choose an appropriate value of k for optimal clustering using the rule of thumb of Hartigan (1975), where we find a k such that the weighted ratio of the sum of squares is significantly >10 for (k − 1) compared with k.
where SS is the sum of squares and n is the number of novel TARs being clustered. We find the ratio is 143.4 for k = 102 when clustering with expression profiles and the ratio is 78.3 for k = 111 when clustering with phylogenetic profiles.
Implementation of DART
DART includes a relational database implemented in MySQL on a Linux server. There are tables for recording basic AR information such as chromosome, location, strand, sequence, and genome build number. Other tables and relations define higher-level objects such as sets of ARs, classes of sets, and attributes describing sets.
In Figure 6A we provide an overview of the DART’s current functionality.
At the most general level, the user is presented with a listing of sets of ARs. These AR sets may be searched and selected in various ways and then passed to the ARC tool for further analysis. Alternatively, data about a single AR set may be viewed at successive levels of detail, e.g., (1) a summary of the AR locations by chromosome, (2) a summary of the AR locations by chromosomal segment, (3) a list of ARs found within a selected chromosomal segment, and finally (4) detailed information about a single AR, including a graphical indication of its overlap with ARs in other AR sets. From various DART screens, data can be passed to custom tracks in the UCSC Genome Browser (Kent et al. 2002) so that the DART data can be viewed in a broader context.
Software and Web pages access the DART database through library routines written in Perl. These library routines have a convenient object oriented structure. They support functions such as defining a genome build number, reannotating ARs for a new genome build, inserting ARs, defining sets and their attributes, and defining classes of sets. As objects are entered into DART, the library routines assign a unique accession number to each object created or inserted. Public domain Perl libraries are used to construct and display graphs on certain DART Web pages. URLs are constructed to allow DART data to be sent to public browsers such as the UCSC Genome Browser (Kent et al. 2002).
The current implementation of DART represents a first step in confronting the challenges involved in manipulating and displaying heterogeneous AR data sets. As the amount of data, as well as the heterogeneity of that data, grows rapidly in the future, we will clearly need to extend and augment DART’s capabilities to keep pace with the new challenges that arise. The code base for DART is downloadable from the DART Web site. All the TAR data sets from The ENCODE Project Consortium (2007), as well as the results of this article are available from DART (http://DART.gersteinlab.org).
ARC tool
The ARC site features four pages (see Fig. 6B), the first of which is the ARC Home page. ARC Home accepts formatted files11 and DART data sets for upload, and it offers options for regulating the AR analysis. These options include filtering ARs on length, adding flanking sequences to each AR, grouping ARs by strand identifier, and mapping data sets from one build to another using a local copy of the UCSC liftOver tool.
ARC initiates AR analysis by flattening each file’s genomic intervals onto a single coordinate axis such that any overlapping regions are combined to form a single region. ARC then performs combinatorial operations on these data sets using an algorithm that achieves high efficiency through a hierarchical series of unions and pairwise intersections. These operations may be used to perform one of two types of analysis. The first procedure determines which nucleotides are common to at least k out of n files, where k is a number between 1 and n, while the second procedure determines which nucleotides are common to exactly k out of n files. For each permutation, ARC generates a new data set containing the corresponding genomic intervals. ARC also performs standard subtraction operations on two files, for which it generates new data sets as well.
The combinatorial algorithm described above minimizes run time by reducing the number of intersection operations that ARC must perform. It first takes the union of the genomic intervals in all n data sets to create a file that contains each region present in at least one of the original data sets (i.e., all regions). It then calculates all unique pairwise intersections among the original n data sets to create n choose two new data sets. The union of these data sets yields a file that contains each region present in at least two of the n original data sets. The next iteration of the procedure produces n choose three new data sets whose union produces a file containing regions in at least three of the n original data sets. When carried to completion, the algorithm creates n files (one for each iteration of n choose k). To ensure good performance time, every new group of data sets is clustered as shown in Supplemental Figure 3 so that the fewest possible intersection operations are performed. In the case where a user wishes to see one specific permutation only, instead of all n, ARC uses the algorithm described at http://msdn.microsoft.com/library/default.asp?url=/library/en-us/dv_vstechart/html/mth_lexicograp.asp. This method requires fewer intersection operations when applied to a single permutation.
To present the results of the above computations, ARC displays summary statistics for each data set in the ARC Results page (see Fig. 6B). ARC also creates new sets for the ARs in each chromosome of each full data set, and it provides summary statistics for these subgroups. All sets may be downloaded directly, or they may be further analyzed by the ARC Annotations page and the UCSC Display page.
The ARC Annotations page (see Fig. 6B) annotates and filters AR data sets using a local Ensembl database (Birney et al. 2006). Its options include grabbing features of the interval itself (sequence, G/C content, etc.), identifying overlapping transcripts and exons, and finding neighboring transcripts. The page also filters on AR length, G/C content, and classification (exon, intron, or intergenic). Processed data sets can be downloaded or exported to the UCSC Display page (see Fig. 6B).
The UCSC Display page facilitates the visualization of data sets by exporting them to the UCSC Genome Browser. Each data set received by the Display page is loaded as a custom UCSC track in an in-frame version of the Genome Browser. These tracks can be viewed in the Genome Browser using either UCSC navigation tools or ARC hyperlinks. The tracks can also be analyzed with the UCSC tools. The UCSC Display page retains a history of exported data sets, and selecting multiple data sets from the history loads each one as a custom track in the UCSC browser, allowing for their direct comparison. These features provide a graphical interface for an otherwise abstract set of data points.
TAR-Vis
TAR-Vis is a collection of Perl scripts and modules that uses the open-source Bioperl modules (Stajich et al. 2002) and Ensembl’s Perl API to automatically retrieve, analyze, and display sequences of genomic DNA containing a specific TAR or set of TARs. Given a chromosomal region and a genome build, TAR-Vis fetches the sequence region (including at least 1000 bp upstream and downstream in order to avoid boundary conditions on the subsequent calculations) from Ensembl’s main databases and copies it to the local machine. From there, various calculations are run on the selected region, including Eponine TSS detection (Down and Hubbard 2002), Cluster-Buster (Frith et al. 2003) transcription factor binding site detection (using the JASPAR TFBS database), CpG island detection, and G/C content graphing. Finally, all surrounding gene annotations are collected from Ensembl’s annotation server. The resulting calculations and gene annotations are stored in a GFF3 file and visually presented using the Bio∷Graphics module of Bioperl.
Generation of randomized sets of novel TARs and novel TAR clusters
In order to assess the significance of the overlap of the different sets of novel TARs with the set of RACEfrags, a set of 7000 random TARs was generated (comparable in size to the set of all novel TARs). This set of random TARs was selected so as to avoid intersecting any annotated GENCODE exon, to include only nonrepetitive DNA sequence and to have the same length distribution as the set of all novel TARs detected on the ENCODE tiling arrays.
In order to compare the overlap of the ends of the novel TAR clusters within 1 kb of putative TSSs or composite promoters from The ENCODE Project Consortium (2007), we created a random set of 1000 novel TAR clusters whose length distribution was the same as that for the novel TAR clusters generated using either expression or phylogenetic profiles.
Accessing structural RNA potential of novel TARs using RNAz
We used the following approach to predict structural noncoding RNAs (ncRNAs) with conserved and thus potentially functional secondary structures using the RNAz tool (Washietl et al. 2005): TARs were first collected and extended by 50 nucleotides on either side (this ensures detection of tightly structured ncRNAs, which may hybridize more poorly to microarrays than unstructured RNAs). All sequences were mapped to their corresponding TBA multiple sequence alignment blocks (23-way) constructed for the ENCODE regions. In each case, the human sequence together with the five most distant sequences, each sharing an overall sequence identity of at least 70% with the human sequence, were kept and analyzed using RNAz. Alignment blocks of 120 bp were subjected to analysis by RNAz, using an offset of 40 and considering both DNA strands independently (smaller alignment blocks of a minimum size of 50 bp were analyzed without offset). When comparing different TAR sets, maximum RNAz scores were calculated for each TAR (the RNAz score, from zero to one, denotes the probability for a DNA sequence to encode a structural RNA, calculated based on support vector machine classification, Washietl et al. 2005).
Acknowledgments
We thank The ENCODE Project Consortium for making their data publicly available, specifically to the Genes and Transcripts, Transcription Regulation and Multiple Sequence Alignment groups for providing data. J.R. acknowledges Thomas Royce and Olof Emanuelsson for valuable discussions. This work was supported by grants from the National Institute of Health (NIH).
Footnotes
[Supplemental material is available online at www.genome.org.]
Article is online at http://www.genome.org/cgi/doi/10.1101/gr.5696007
This estimation of a P-value of <0.05 takes into account the multiple testing of the expression profile of a novel TAR with on average 19 known exons within 20 kb. The P-value for obtaining a Pearson correlation of 0.9 for two 11-dimensional vectors is <10−3.
There are 828 putative composite promoters on the list from The ENCODE Project Consortium (2007), which is a set of both known and predicted promoters. Promoters were predicted using multiple ChIP-chip data sets for promoter specific transcription factors and modifications. This set of promoters is available at DART.gersteinlab.org.
ARC accepts files in Browser Extensible Data (BED) format and files containing inclusive intervals. The BED format uses a zero-based, half-open coordinate system. It was developed for the UCSC Genome Browser and is described fully at http://genome.ucsc.edu/FAQ/FAQformat#format1. The inclusive intervals option accepts one-based, closed coordinates as used by Ensembl.
References
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J., Gish W., Miller W., Myers E.W., Lipman D.J., Miller W., Myers E.W., Lipman D.J., Myers E.W., Lipman D.J., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bertone P., Stolc V., Royce T.E., Rozowsky J.S., Urban A.E., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Stolc V., Royce T.E., Rozowsky J.S., Urban A.E., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Royce T.E., Rozowsky J.S., Urban A.E., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Rozowsky J.S., Urban A.E., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Urban A.E., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Tongprasit W., Samanta M., Weissman S., Samanta M., Weissman S., Weissman S., et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–2246. doi: 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- Birney E., Andrews D., Caccamo M., Chen Y., Clarke L., Coates G., Cox T., Cunningham F., Curwen V., Cutts T., Andrews D., Caccamo M., Chen Y., Clarke L., Coates G., Cox T., Cunningham F., Curwen V., Cutts T., Caccamo M., Chen Y., Clarke L., Coates G., Cox T., Cunningham F., Curwen V., Cutts T., Chen Y., Clarke L., Coates G., Cox T., Cunningham F., Curwen V., Cutts T., Clarke L., Coates G., Cox T., Cunningham F., Curwen V., Cutts T., Coates G., Cox T., Cunningham F., Curwen V., Cutts T., Cox T., Cunningham F., Curwen V., Cutts T., Cunningham F., Curwen V., Cutts T., Curwen V., Cutts T., Cutts T., et al. Ensembl 2006. Nucleic Acids Res. 2006;34:D556–D561. doi: 10.1093/nar/gkj133. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette M., Kent W.J., Riemer C., Elnitski L., Smit A.F., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Kent W.J., Riemer C., Elnitski L., Smit A.F., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Riemer C., Elnitski L., Smit A.F., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Elnitski L., Smit A.F., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Smit A.F., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Rosenbloom K., Clawson H., Green E.D., Clawson H., Green E.D., Green E.D., et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad B.M., Irizarry R.A., Astrand M., Speed T.P., Irizarry R.A., Astrand M., Speed T.P., Astrand M., Speed T.P., Speed T.P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Brazma A., Parkinson H., Sarkans U., Shojatalab M., Vilo J., Abeygunawardena N., Holloway E., Kapushesky M., Kemmeren P., Lara G.G., Parkinson H., Sarkans U., Shojatalab M., Vilo J., Abeygunawardena N., Holloway E., Kapushesky M., Kemmeren P., Lara G.G., Sarkans U., Shojatalab M., Vilo J., Abeygunawardena N., Holloway E., Kapushesky M., Kemmeren P., Lara G.G., Shojatalab M., Vilo J., Abeygunawardena N., Holloway E., Kapushesky M., Kemmeren P., Lara G.G., Vilo J., Abeygunawardena N., Holloway E., Kapushesky M., Kemmeren P., Lara G.G., Abeygunawardena N., Holloway E., Kapushesky M., Kemmeren P., Lara G.G., Holloway E., Kapushesky M., Kemmeren P., Lara G.G., Kapushesky M., Kemmeren P., Lara G.G., Kemmeren P., Lara G.G., Lara G.G., et al. ArrayExpress—a public repository for microarray gene expression data at the EBI. Nucleic Acids Res. 2003;31:68–71. doi: 10.1093/nar/gkg091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P., Kasukawa T., Katayama S., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., Kasukawa T., Katayama S., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., Katayama S., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., Oyama R., Ravasi T., Lenhard B., Wells C., Ravasi T., Lenhard B., Wells C., Lenhard B., Wells C., Wells C., et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Cawley S., Bekiranov S., Ng H.H., Kapranov P., Sekinger E.A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Bekiranov S., Ng H.H., Kapranov P., Sekinger E.A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Ng H.H., Kapranov P., Sekinger E.A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Kapranov P., Sekinger E.A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Sekinger E.A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Sementchenko V., Cheng J., Williams A.J., Cheng J., Williams A.J., Williams A.J., et al. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004;116:499–509. doi: 10.1016/s0092-8674(04)00127-8. [DOI] [PubMed] [Google Scholar]
- Cheng J., Kapranov P., Drenkow J., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Kapranov P., Drenkow J., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Drenkow J., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Patel S., Long J., Stern D., Tammana H., Helt G., Long J., Stern D., Tammana H., Helt G., Stern D., Tammana H., Helt G., Tammana H., Helt G., Helt G., et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- David L., Huber W., Granovskaia M., Toedling J., Palm C.J., Bofkin L., Jones T., Davis R.W., Steinmetz L.M., Huber W., Granovskaia M., Toedling J., Palm C.J., Bofkin L., Jones T., Davis R.W., Steinmetz L.M., Granovskaia M., Toedling J., Palm C.J., Bofkin L., Jones T., Davis R.W., Steinmetz L.M., Toedling J., Palm C.J., Bofkin L., Jones T., Davis R.W., Steinmetz L.M., Palm C.J., Bofkin L., Jones T., Davis R.W., Steinmetz L.M., Bofkin L., Jones T., Davis R.W., Steinmetz L.M., Jones T., Davis R.W., Steinmetz L.M., Davis R.W., Steinmetz L.M., Steinmetz L.M. A high-resolution map of transcription in the yeast genome. Proc. Natl. Acad. Sci. 2006;103:5320–5325. doi: 10.1073/pnas.0601091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Down T.A., Hubbard T.J., Hubbard T.J. Computational detection and location of transcription start sites in mammalian genomic DNA. Genome Res. 2002;12:458–461. doi: 10.1101/gr.216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy S.R. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- Edgar R., Domrachev M., Lash A.E., Domrachev M., Lash A.E., Lash A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O., Nagalakshmi U., Zheng D., Rozowsky J.S., Du J., Lian Z., Urban A.E., Stolc V., Weissman S., Snyder M., Nagalakshmi U., Zheng D., Rozowsky J.S., Du J., Lian Z., Urban A.E., Stolc V., Weissman S., Snyder M., Zheng D., Rozowsky J.S., Du J., Lian Z., Urban A.E., Stolc V., Weissman S., Snyder M., Rozowsky J.S., Du J., Lian Z., Urban A.E., Stolc V., Weissman S., Snyder M., Du J., Lian Z., Urban A.E., Stolc V., Weissman S., Snyder M., Lian Z., Urban A.E., Stolc V., Weissman S., Snyder M., Urban A.E., Stolc V., Weissman S., Snyder M., Stolc V., Weissman S., Snyder M., Weissman S., Snyder M., Snyder M., et al. Assessing the performance of different high-density tiling microarray strategies for mapping transcribed regions of the human genome. Genome Res. 2007 doi: 10.1101/gr.5014606. (this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium, The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium, Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007 doi: 10.1038/nature05874. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frith M.C., Li M.C., Weng Z., Li M.C., Weng Z., Weng Z. Cluster-Buster: Finding dense clusters of motifs in DNA sequences. Nucleic Acids Res. 2003;31:3666–3668. doi: 10.1093/nar/gkg540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J., Denoeud F., Frankish A., Reymond A., Chen C.K., Chrast J., Lagarde J., Gilbert J.G., Storey R., Swarbreck D., Denoeud F., Frankish A., Reymond A., Chen C.K., Chrast J., Lagarde J., Gilbert J.G., Storey R., Swarbreck D., Frankish A., Reymond A., Chen C.K., Chrast J., Lagarde J., Gilbert J.G., Storey R., Swarbreck D., Reymond A., Chen C.K., Chrast J., Lagarde J., Gilbert J.G., Storey R., Swarbreck D., Chen C.K., Chrast J., Lagarde J., Gilbert J.G., Storey R., Swarbreck D., Chrast J., Lagarde J., Gilbert J.G., Storey R., Swarbreck D., Lagarde J., Gilbert J.G., Storey R., Swarbreck D., Gilbert J.G., Storey R., Swarbreck D., Storey R., Swarbreck D., Swarbreck D., et al. GENCODE: Producing a reference annotation for ENCODE. Genome Biol. 2006;7(Suppl 1):S4.1–S4.9. doi: 10.1186/gb-2006-7-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartigan J.A. Clustering algorithms. Wiley; New York: 1975. [Google Scholar]
- Hartigan J.A., Wong M.A., Wong M.A. A K-means clustering algorithm. Appl. Stat. 1979;28:100–108. [Google Scholar]
- Kampa D., Cheng J., Kapranov P., Yamanaka M., Brubaker S., Cawley S., Drenkow J., Piccolboni A., Bekiranov S., Helt G., Cheng J., Kapranov P., Yamanaka M., Brubaker S., Cawley S., Drenkow J., Piccolboni A., Bekiranov S., Helt G., Kapranov P., Yamanaka M., Brubaker S., Cawley S., Drenkow J., Piccolboni A., Bekiranov S., Helt G., Yamanaka M., Brubaker S., Cawley S., Drenkow J., Piccolboni A., Bekiranov S., Helt G., Brubaker S., Cawley S., Drenkow J., Piccolboni A., Bekiranov S., Helt G., Cawley S., Drenkow J., Piccolboni A., Bekiranov S., Helt G., Drenkow J., Piccolboni A., Bekiranov S., Helt G., Piccolboni A., Bekiranov S., Helt G., Bekiranov S., Helt G., Helt G., et al. Novel RNAs identified from an in-depth analysis of the transcriptome of human chromosomes 21 and 22. Genome Res. 2004;14:331–342. doi: 10.1101/gr.2094104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P., Cawley S.E., Drenkow J., Bekiranov S., Strausberg R.L., Fodor S.P., Gingeras T.R., Cawley S.E., Drenkow J., Bekiranov S., Strausberg R.L., Fodor S.P., Gingeras T.R., Drenkow J., Bekiranov S., Strausberg R.L., Fodor S.P., Gingeras T.R., Bekiranov S., Strausberg R.L., Fodor S.P., Gingeras T.R., Strausberg R.L., Fodor S.P., Gingeras T.R., Fodor S.P., Gingeras T.R., Gingeras T.R. Large-scale transcriptional activity in chromosomes 21 and 22. Science. 2002;296:916–919. doi: 10.1126/science.1068597. [DOI] [PubMed] [Google Scholar]
- Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Pringle T.H., Zahler A.M., Haussler D., Zahler A.M., Haussler D., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Wang X., Stolc V., Li X., Zhang D., Su N., Tongprasit W., Li S., Cheng Z., Wang J., Wang X., Stolc V., Li X., Zhang D., Su N., Tongprasit W., Li S., Cheng Z., Wang J., Stolc V., Li X., Zhang D., Su N., Tongprasit W., Li S., Cheng Z., Wang J., Li X., Zhang D., Su N., Tongprasit W., Li S., Cheng Z., Wang J., Zhang D., Su N., Tongprasit W., Li S., Cheng Z., Wang J., Su N., Tongprasit W., Li S., Cheng Z., Wang J., Tongprasit W., Li S., Cheng Z., Wang J., Li S., Cheng Z., Wang J., Cheng Z., Wang J., Wang J., et al. Genome-wide transcription analyses in rice using tiling microarrays. Nat Genet. 2006;38:124–129. doi: 10.1038/ng1704. [DOI] [PubMed] [Google Scholar]
- Manak J.R., Dike S., Sementchenko V., Kapranov P., Biemar F., Long J., Cheng J., Bell I., Ghosh S., Piccolboni A., Dike S., Sementchenko V., Kapranov P., Biemar F., Long J., Cheng J., Bell I., Ghosh S., Piccolboni A., Sementchenko V., Kapranov P., Biemar F., Long J., Cheng J., Bell I., Ghosh S., Piccolboni A., Kapranov P., Biemar F., Long J., Cheng J., Bell I., Ghosh S., Piccolboni A., Biemar F., Long J., Cheng J., Bell I., Ghosh S., Piccolboni A., Long J., Cheng J., Bell I., Ghosh S., Piccolboni A., Cheng J., Bell I., Ghosh S., Piccolboni A., Bell I., Ghosh S., Piccolboni A., Ghosh S., Piccolboni A., Piccolboni A., et al. Biological function of unannotated transcription during the early development of Drosophila melanogaster. Nat. Genet. 2006;38:1151–1158. doi: 10.1038/ng1875. [DOI] [PubMed] [Google Scholar]
- Nadkarni P.M., Brandt C., Frawley S., Sayward F., Einbinder R., Zelterman D., Schacter L., Miller P.L., Brandt C., Frawley S., Sayward F., Einbinder R., Zelterman D., Schacter L., Miller P.L., Frawley S., Sayward F., Einbinder R., Zelterman D., Schacter L., Miller P.L., Sayward F., Einbinder R., Zelterman D., Schacter L., Miller P.L., Einbinder R., Zelterman D., Schacter L., Miller P.L., Zelterman D., Schacter L., Miller P.L., Schacter L., Miller P.L., Miller P.L. Managing attribute-value clinical trials data using the ACT/DB client-server database system. J. Am. Med. Inform. Assoc. 1998;5:139–151. doi: 10.1136/jamia.1998.0050139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn J., Euskirchen G., Bertone P., Martone R., Luscombe N.M., Hartman S., Harrison P.M., Nelson F.K., Miller P., Gerstein M., Euskirchen G., Bertone P., Martone R., Luscombe N.M., Hartman S., Harrison P.M., Nelson F.K., Miller P., Gerstein M., Bertone P., Martone R., Luscombe N.M., Hartman S., Harrison P.M., Nelson F.K., Miller P., Gerstein M., Martone R., Luscombe N.M., Hartman S., Harrison P.M., Nelson F.K., Miller P., Gerstein M., Luscombe N.M., Hartman S., Harrison P.M., Nelson F.K., Miller P., Gerstein M., Hartman S., Harrison P.M., Nelson F.K., Miller P., Gerstein M., Harrison P.M., Nelson F.K., Miller P., Gerstein M., Nelson F.K., Miller P., Gerstein M., Miller P., Gerstein M., Gerstein M., et al. The transcriptional activity of human chromosome 22. Genes & Dev. 2003;17:529–540. doi: 10.1101/gad.1055203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royce T.E., Rozowsky J.S., Bertone P., Samanta M., Stolc V., Weissman S., Snyder M., Gerstein M., Rozowsky J.S., Bertone P., Samanta M., Stolc V., Weissman S., Snyder M., Gerstein M., Bertone P., Samanta M., Stolc V., Weissman S., Snyder M., Gerstein M., Samanta M., Stolc V., Weissman S., Snyder M., Gerstein M., Stolc V., Weissman S., Snyder M., Gerstein M., Weissman S., Snyder M., Gerstein M., Snyder M., Gerstein M., Gerstein M. Issues in the analysis of oligonucleotide tiling microarrays for transcript mapping. Trends Genet. 2005;21:466–475. doi: 10.1016/j.tig.2005.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SantaLucia J. A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl. Acad. Sci. 1998;95:1460–1465. doi: 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stajich J.E., Block D., Boulez K., Brenner S.E., Chervitz S.A., Dagdigian C., Fuellen G., Gilbert J.G., Korf I., Lapp H., Block D., Boulez K., Brenner S.E., Chervitz S.A., Dagdigian C., Fuellen G., Gilbert J.G., Korf I., Lapp H., Boulez K., Brenner S.E., Chervitz S.A., Dagdigian C., Fuellen G., Gilbert J.G., Korf I., Lapp H., Brenner S.E., Chervitz S.A., Dagdigian C., Fuellen G., Gilbert J.G., Korf I., Lapp H., Chervitz S.A., Dagdigian C., Fuellen G., Gilbert J.G., Korf I., Lapp H., Dagdigian C., Fuellen G., Gilbert J.G., Korf I., Lapp H., Fuellen G., Gilbert J.G., Korf I., Lapp H., Gilbert J.G., Korf I., Lapp H., Korf I., Lapp H., Lapp H., et al. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 2002;12:1611–1618. doi: 10.1101/gr.361602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolc V., Gauhar Z., Mason C., Halasz G., van Batenburg M.F., Rifkin S.A., Hua S., Herreman T., Tongprasit W., Barbano P.E., Gauhar Z., Mason C., Halasz G., van Batenburg M.F., Rifkin S.A., Hua S., Herreman T., Tongprasit W., Barbano P.E., Mason C., Halasz G., van Batenburg M.F., Rifkin S.A., Hua S., Herreman T., Tongprasit W., Barbano P.E., Halasz G., van Batenburg M.F., Rifkin S.A., Hua S., Herreman T., Tongprasit W., Barbano P.E., van Batenburg M.F., Rifkin S.A., Hua S., Herreman T., Tongprasit W., Barbano P.E., Rifkin S.A., Hua S., Herreman T., Tongprasit W., Barbano P.E., Hua S., Herreman T., Tongprasit W., Barbano P.E., Herreman T., Tongprasit W., Barbano P.E., Tongprasit W., Barbano P.E., Barbano P.E., et al. A gene expression map for the euchromatic genome of Drosophila melanogaster. Science. 2004;306:655–660. doi: 10.1126/science.1101312. [DOI] [PubMed] [Google Scholar]
- Washietl S., Hofacker I.L., Stadler P.F., Hofacker I.L., Stadler P.F., Stadler P.F. Fast and reliable prediction of noncoding RNAs. Proc. Natl. Acad. Sci. 2005;102:2454–2459. doi: 10.1073/pnas.0409169102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washietl S., Pedersen J.S., Korbel J.O., Stocsits C., Gruber A.R., Hackermüller J., Hertel J., Lindemeyer M., Reiche K., Tanzer A., Pedersen J.S., Korbel J.O., Stocsits C., Gruber A.R., Hackermüller J., Hertel J., Lindemeyer M., Reiche K., Tanzer A., Korbel J.O., Stocsits C., Gruber A.R., Hackermüller J., Hertel J., Lindemeyer M., Reiche K., Tanzer A., Stocsits C., Gruber A.R., Hackermüller J., Hertel J., Lindemeyer M., Reiche K., Tanzer A., Gruber A.R., Hackermüller J., Hertel J., Lindemeyer M., Reiche K., Tanzer A., Hackermüller J., Hertel J., Lindemeyer M., Reiche K., Tanzer A., Hertel J., Lindemeyer M., Reiche K., Tanzer A., Lindemeyer M., Reiche K., Tanzer A., Reiche K., Tanzer A., Tanzer A., et al. Structured RNAs in the ENCODE selected regions of the human genome. Genome Res. 2007 doi: 10.1101/gr.5650707. (this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K., Lim J., Dale J.M., Chen H., Shinn P., Palm C.J., Southwick A.M., Wu H.C., Kim C., Nguyen M., Lim J., Dale J.M., Chen H., Shinn P., Palm C.J., Southwick A.M., Wu H.C., Kim C., Nguyen M., Dale J.M., Chen H., Shinn P., Palm C.J., Southwick A.M., Wu H.C., Kim C., Nguyen M., Chen H., Shinn P., Palm C.J., Southwick A.M., Wu H.C., Kim C., Nguyen M., Shinn P., Palm C.J., Southwick A.M., Wu H.C., Kim C., Nguyen M., Palm C.J., Southwick A.M., Wu H.C., Kim C., Nguyen M., Southwick A.M., Wu H.C., Kim C., Nguyen M., Wu H.C., Kim C., Nguyen M., Kim C., Nguyen M., Nguyen M., et al. Empirical analysis of transcriptional activity in the Arabidopsis genome. Science. 2003;302:842–846. doi: 10.1126/science.1088305. [DOI] [PubMed] [Google Scholar]
- Zheng D., Zhang Z., Harrison P.M., Karro J., Carriero N., Gerstein M., Zhang Z., Harrison P.M., Karro J., Carriero N., Gerstein M., Harrison P.M., Karro J., Carriero N., Gerstein M., Karro J., Carriero N., Gerstein M., Carriero N., Gerstein M., Gerstein M. Integrated pseudogene annotation for human chromosome 22: Evidence for transcription. J. Mol. Biol. 2005;349:27–45. doi: 10.1016/j.jmb.2005.02.072. [DOI] [PubMed] [Google Scholar]