Abstract
This report presents systematic empirical annotation of transcript products from 399 annotated protein-coding loci across the 1% of the human genome targeted by the Encyclopedia of DNA elements (ENCODE) pilot project using a combination of 5′ rapid amplification of cDNA ends (RACE) and high-density resolution tiling arrays. We identified previously unannotated and often tissue- or cell-line-specific transcribed fragments (RACEfrags), both 5′ distal to the annotated 5′ terminus and internal to the annotated gene bounds for the vast majority (81.5%) of the tested genes. Half of the distal RACEfrags span large segments of genomic sequences away from the main portion of the coding transcript and often overlap with the upstream-annotated gene(s). Notably, at least 20% of the resultant novel transcripts have changes in their open reading frames (ORFs), most of them fusing ORFs of adjacent transcripts. A significant fraction of distal RACEfrags show expression levels comparable to those of known exons of the same locus, suggesting that they are not part of very minority splice forms. These results have significant implications concerning (1) our current understanding of the architecture of protein-coding genes; (2) our views on locations of regulatory regions in the genome; and (3) the interpretation of sequence polymorphisms mapping to regions hitherto considered to be “noncoding,” ultimately relating to the identification of disease-related sequence alterations.
Annotation of the current working draft of the euchromatic portion of the human genome revealed that it contains 20,000–25,000 protein-coding genes (Lander et al. 2001; Venter et al. 2001; International Human Genome Sequencing Consortium 2004), a figure not dramatically higher than the estimated number of protein-coding genes in yeast, fly, and worm genomes (Goffeau et al. 1996; C. elegans Sequencing Consortium 1998; Adams et al. 2000). It was hypothesized that functional diversification of this limited number of genes is required in order to create the highly elaborated systems necessary for mammalian life. This diversity might occur via the production of different protein-coding and noncoding transcripts from a single locus through alternative splicing. Though currently estimated to be rare in invertebrates (10%–20% of genes affected; Misra et al. 2002; Reboul et al. 2003), alternative splicing is common in mammalian genomes. Recent manual annotation of 1% of the human genome showed that this phenomenon occurs in up to 86% of multi-exon gene loci and generates >5.4 transcript variants per locus on average (Harrow et al. 2006). In addition, at least half of the mammalian genes are regulated by more than one promoter (Carninci et al. 2006; Kimura et al. 2006).
The National Human Genome Research Institute launched The ENCODE Project (Encylopedia of DNA Elements) to identify all the functional elements in the human genome. During its pilot phase, the project has focused on 44 regions totaling 30 Mb or ∼1% of the human genome sequence (The ENCODE Project Consortium 2004). In this framework we sought to map the transcription start sites (TSS) of transcripts emanating from these regions and to identify novel exons of all the coding genes mapping in the ENCODE regions (Harrow et al. 2006). Strikingly, we observed by using a combination of rapid amplification of 5′ cDNA ends (5′ RACE) and tiling array readouts that more than half of the protein-coding genes mapping in the ENCODE regions utilize a tissue-specific and often unannotated set of exons outside the current boundaries of the annotated genes. In this study we report on the characterization of these previously unannotated exons, the transcripts that contain them, and the implications of such hitherto undetected RNA structures.
Results
Discovery of unannotated distal and proximal exons using RACE and tiling arrays
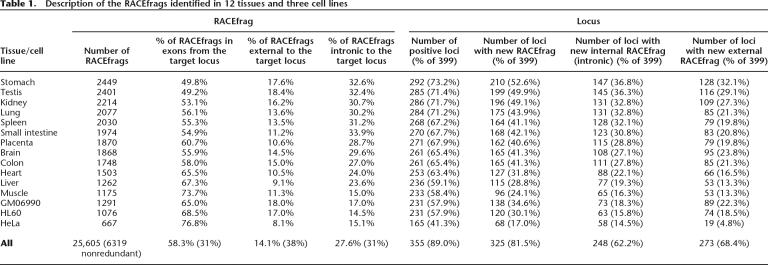
A combination of 5′ RACE with high-density tiling microarrays was used to empirically annotate 5′ transcription start sites (TSS) and internal exons of all 410 annotated protein-coding loci across the 1% of the human genome targeted by the Encyclopedia of DNA elements (ENCODE) pilot project. RACE allows detection of low-copy-number transcripts/isoforms and high-resolution analysis of genes individually, while pooling strategies and array hybridization permit high-resolution characterization of RACE products and high-throughput sample readout. The 5′ RACE reactions were performed with oligonucleotides mapping to a coding exon common to most of the average 5.4 transcripts of a protein-coding gene locus annotated by GENCODE (Harrow et al. 2006) on polyA+ RNA from 12 adult human tissues (brain, heart, kidney, spleen, liver, colon, small intestine, muscle, lung, stomach, testis, placenta) and three cell lines (GM06990, HL60, and HeLaS3). The RACE reactions were pooled to achieve maximal distance between neighboring genes and then hybridized to 20-nucleotide resolution on average tiling arrays covering the nonrepeated regions of the 44 ENCODE regions as described in Kapranov et al. (2005). The detected RACE reactions generated fragments specifically linked to the assayed coding locus (index locus; see Methods) and were named “RACEfrags” following the coining of the term “transfrags” (transcribed fragments), which denotes array-detected regions of transcription (Kampa et al. 2004; Cheng et al. 2005). They are schematically compared with annotated and unannotated transcripts in Figure 1. The mapping position of all identified RACEfrags can be retrieved from the UCSC genome browser (http://genome.ucsc.edu/ENCODE/). A successful amplification (i.e., detection of at least one RACEfrag overlapping annotated exons of target genes) was found for 89% of the interrogated loci (364 positives out of 410 loci) and 89% of the loci completely mapping into ENCODE regions (355 positives of 399 loci). This approach is suitable to identify potential 5′ TSS of genes as revealed by detection of GENCODE-annotated first exons in 92% of the RACE-positive genes (336 out of 364). It should be emphasized that although novel distal 5′ RACEfrags were detected, these RACEfrags may not serve as the ultimate TSS for that gene since the lengths of most ENCODE regions are 500 kb and the positions of some of the interrogated genes are situated proximal to the boundaries of the ENCODE regions (The ENCODE Project Consortium 2004, 2007). The transcriptome of stomach, kidney, testis, and lung showed the highest complexity (highest number of RACEfrags and >70% of tested genes expressed), while muscle and the three cell lines were less complex, in accordance with previous reports (Reymond et al. 2002b; Table 1).
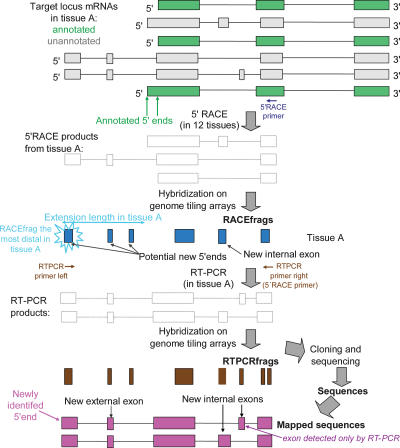
Figure 1.
Schematic comparison of RACEfrags and RT-PCRfrags with annotated and unannotated transcripts. The locus to be interrogated is transcribed in alternatively spliced annotated (green) and unannotated (gray) isoforms. Rapid amplification of 5′ cDNA ends (5′ RACE) with a primer (blue arrow) mapping to a coding exon common to most of the transcripts (the index exon) results in a mix of cDNAs (ghost transcripts), which are hybridized to high-resolution tiling arrays to detect “RACEfrags” (blue boxes). RACEfrags are transcribed fragments specifically linked to the targeted coding locus. The connectivity between a RACEfrag overlapping an unannotated exon and the index exon can be verified by RT-PCR with two specific primers (brown arrows). This reaction produces a combination of overlapping alternatively spliced transcripts (ghost transcripts) that identify “RT-PCRfrags” upon hybridization to the same tiling array (brown boxes). Thus, RT-PCRfrags are transcribed fragments that link two targeted exons. Alternatively, these transcripts can be cloned and sequenced to precisely determine the beginning and the end of the novel exons and the exon composition of the transcripts (purple boxes). Because tiling arrays interrogate only nonrepeated regions and as they have a 20-bp resolution, RACEfrags and RT-PCRfrags do not fully overlap exons.
Table 1.
Description of the RACEfrags identified in 12 tissues and three cell lines
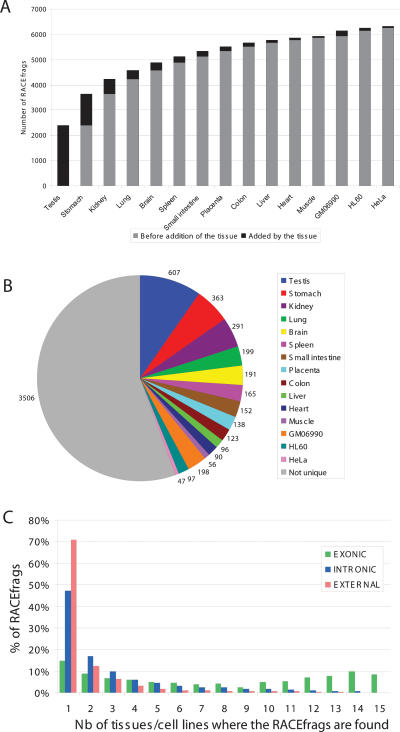
More than 50% of RACEfrags (2324 out of 4573 projected RACEfrag) did not correspond to GENCODE-annotated exons (Harrow et al. 2006) of the interrogated gene (see Methods). About two-thirds of the interrogated loci (68.4%; 273 out of 399) were shown to have unannotated 5′ extensions, while a similar fraction (62.2%; 248 out of 399) of genes have alternative internal RACEfrags. Three hundred twenty-five (81.5%) loci were found to have at least one new exon (Table 1). The number of genes with new intronic exons and new extensions in a specific tissue varied from 14.5% (HeLaS3) to 37% (stomach) and 5% (HeLaS3) to 32% (muscle), respectively (Table 1). The majority (58%: 47% of intronic and 71% of external) of these newly identified RACEfrags are tissue- or cell-specific, with no tissue/cell line providing the vast majority of unique RACEfrags. Testis and HeLaS3 are the largest and smallest source of unique RACEfrags, respectively (Fig. 2; Table 1). The unexpectedly high frequency of novel RACEfrags raised the possibility that technical artifacts had troubled the microarray experiments. However, this possibility seems unlikely because we were able to validate the newly identified exons by RT-PCR amplification followed by hybridization, or by cloning and sequencing (see below). Thus, our results highlight that the transcript complexity of a defined locus of the human genome has not yet been fully surveyed through cDNA sequencing.
Figure 2.
A large proportion of RACEfrags are tissue-specific. (A) Cumulative number of RACEfrags identified in the 12 tissues and three cell lines; (B) numbers of RACEfrags specific to a single tissue; (C) proportion of exonic (green), intronic (blue), and external (orange) RACEfrags identified by one, two, three, or more tissues.
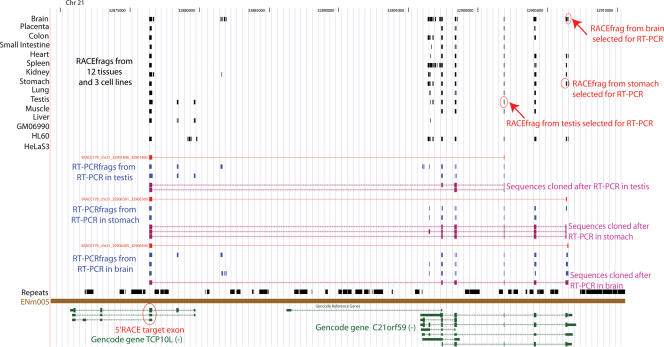
The 5′ distal RACEfrags map on average 186 kb (median 85 kb) upstream of the most 5′ annotated exons. Since there is on average an annotated protein-coding gene every 62 kb in the ENCODE regions (Harrow et al. 2006; The ENCODE Project Consortium 2007), these RACEfrags often map to an upstream locus (an example is shown in Fig. 3), sometimes even creating transcripts with exons mapping to loci separated by multiple coding genes (Kapranov et al. 2005; The ENCODE Project Consortium 2007). In 87% of the loci extended at their 5′ end, at least one of the identified RACEfrags reaches across an upstream-positioned gene locus (238 out of 273; 92%, 195/212 if we remove the target loci that are in gene clusters; see Methods). In more than half of these cases (57%, 136/238 if all; 56%, 110/195 if we remove loci in gene clusters) these RACEfrags correspond to annotated exons of an upstream-positioned gene, thus creating transcripts that possibly encode chimeric versions of already annotated proteins. Such fusions have been recently reported (Carninci et al. 2005; Kapranov et al. 2005; Akiva et al. 2006; Parra et al. 2006), but our results show that the extent of this phenomenon is greater than previously anticipated. We checked whether the genes linked by transcription-induced chimeras were part of the same pathways by comparing the Gene Ontology terms that characterize these loci (Ashburner et al. 2000), but we failed to find any obvious association. More features of the transcription-induced chimeras are described in the Supplemental section.
Figure 3.
Example of a transcription-induced chimera between C21orf59 and TCP10L. The results of a 5′ RACE/tiling array analysis of the HSA21 TCP10L gene are presented. The GENCODE-annotated transcripts of this section of the ENCODE region ENm005 are shown (green, at the bottom). The index exon where the primer used for the 5′ RACE maps is indicated. RACEfrags-positive regions obtained upon hybridization of the tiling array by the RACE reactions performed in 12 human tissues and three cell lines are shown (black boxes, upper part). Red boxes joined by thin red lines depict connectivity between index exons and RACEfrags selected to be independently verified by RT-PCR. The corresponding RACEfrags are highlighted in the upper part of the panel. The hybridization of these RT-PCR reactions to the same tiling arrays allowed us to identify RT-PCRfrags (blue boxes, see text for details). Note that some of the RT-PCRfrags do not intersect RACEfrags, denoting that not all transcripts were detected by the RACE reactions. The cloning and sequencing of the RT-PCR reactions amplimers’ revealed the exon composition and chimeric nature of transcripts containing the targeted RACEfrags (purple transcripts).
Sequence analysis of RACEfrags
In order to further characterize these novel exons, a set of 538 RACEfrags corresponding to 261 extended loci was selected for independent verification of their connectivity with the index-annotated protein-coding gene (see Supplemental Methods for the selection procedure). The hybridization of these reactions to the ENCODE tiling arrays allowed us to identify RT-PCRfrags (Figs. 1, 3). We confirmed connectivity between the RACEfrags and the index exon chosen to design the RACE oligonucleotide in 314 cases (58.4%). No significant differences in the success of the RT-PCR studies were reported between the different laboratories and the different strategies used to prepare the cDNA (see Supplemental Methods).
To further characterize the protein-coding transcripts that possess unannotated proximal and distal exons, we subsequently attempted to clone and sequence 309 and 76 of these RT-PCR products with confirmed or unconfirmed connectivity, respectively, by the array hybridization approach (see above). These 385 RT-PCR reactions correspond to 199 distinct genomic loci in the ENCODE regions and are enriched for RACEfrags that are the most distally observed by RACE/array (244 reactions). Eighty-nine RT-PCR reactions (69 loci) produced at least one cDNA clone with a sequence unambiguously mapping to the target region. None of these sequences belongs to the set of RT-PCR unconfirmed by the array approach, suggesting that this approach is efficient to classify the RT-PCR reactions. Obtaining full-length cDNA clones from the RT-PCR has proved to be challenging, as illustrated by the low success rate in the positive RT-PCR set (89/309 = 28.8%). One hundred thirty-two nonredundant sequences were obtained for the 89 RT-PCR reactions, mapping to 69 distinct loci. It is notable that some RT-PCRfrags do not overlap any RACEfrag, indicating that not all transcripts present in a sample were detected during the RACE reactions (see Fig. 1, which schematically compares RACEfrags and RT-PCRfrags; Fig. 3, which shows an example; and Supplemental Fig. S1 for coverage of the novel exons by the tiling array). They were submitted to GenBank under accessions DQ655905-DQ656069 and EF070113-EF070122 and used to further upgrade the GENCODE annotation (Harrow et al. 2006).
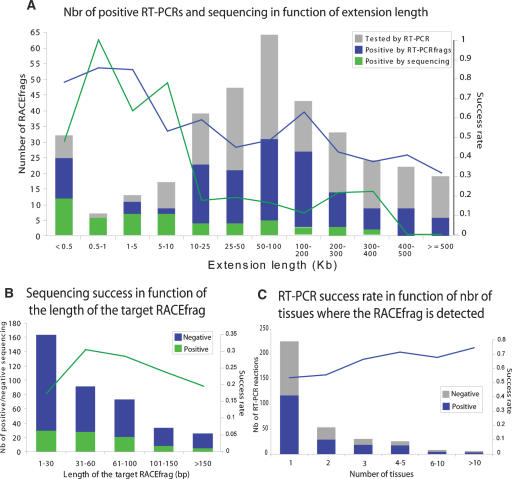
The success rate of cloning and sequencing of the RT-PCR reactions correlates with the number of tissues in which a RACEfrag was identified (Fig. 4C), but does not seem to be significantly affected by the level of expression of a RACEfrag (see below and Supplemental Fig. S2). On the other hand, it diminishes as the distance between the targeted exon and the RACE-identified putative 5′ TSS increases. The success rate among the most distal exons per tissue was 18% (43/244), while that from internal alternative exons was 34% (48/141; Fig. 4A). The increasing lengths of cDNAs to be cloned and the relatively small number of clones sequenced for each of the RACE extension reactions contribute to this relatively low yield of full-length cDNAs. However, we isolated and sequenced several clones that represent transcripts whose RACE-identified putative alternative 5′ TSS sites were in excess of 50,000–100,000 bp from the originally annotated 5′ TSS. We also observe a correlation between the size of the targeted RACEfrag and the success rate of the RT-PCR reactions (Fig. 4B), suggesting that longer RACEfrags, i.e., those covered by a larger number of probes on the tiling array, are more likely to represent bona fide exons. Another alternative explanation for this result is methodological, as we sometimes had to artificially extend RACEfrags on their 3′ end to be able to design the 25mer oligonucleotides with sufficient specificities (see Methods). Hence, we may have sometimes designed oligonucleotides that do not map to exons.
Figure 4.
Characteristics of RACEfrags subjected to RT-PCR, cloning, and sequencing and success rates. Distributions of RACEfrags selected to be independently verified by RT-PCR according to the genomic distance separating them from their index exon (A), their lengths (B), or the number of tissues where they were detected (C). The histograms (Y-axis scale on the left) show the fractions of RACEfrags successfully confirmed only by RT-PCRfrags (blue, see text for details), or by RT-PCRfrags, cloning, and sequencing (green). The curves (Y-axis scale on the right) indicate the success rate by hybridization (blue curve) or by hybridization, cloning, and sequencing (green curve).
The cloned sequences correspond to novel intronic exons (15 loci) and to extensions (54 loci) ranging from <100 bp to >200,000 bp of genomic space. Interestingly, 28 sequences correspond to chimeric transcripts; i.e., they link exons of the index genes with annotated exons of other 5′-positioned same-strand protein-coding genes (13 loci). Sixty-five sequences correspond to new 5′ exons upstream of the current GENCODE annotation (34 loci); 24 are 5′ extensions of the first GENCODE-annotated exon (18 loci), while 15 uncover new intronic unannotated exons (15 loci). Multiple new sequences were obtained for some loci, placing them in more than one of these categories. More than half of the RT-PCR-produced and sequenced unique exons are novel; either they overlap with known exons but have new alternative splice sites (123 exons), or they map entirely in GENCODE introns (17 exons) or intergenic regions (85 exons). The vast majority of them appear to be UTR exons, as 27% (36 out of 132) of the RT-PCR sequences were assigned an already annotated coding sequence (CDS). However, 18% (24 out of 132) of the RT-PCR sequences (mapping to 16 different loci) show a novel CDS (see Supplemental Table S1 for summary and Supplemental Table S2 for detailed description of the sequences). Interestingly, 14 transcripts (six loci) join exons of neighboring genes, creating transcription-induced chimeras (Carninci et al. 2005; Kapranov et al. 2005; Akiva et al. 2006; Parra et al. 2006) while maintaining the open reading frame, thus putatively encoding fusion proteins (an example is presented in Fig. 3). As the GENCODE “gold standards” (Harrow et al. 2006) are very conservative in assigning a CDS, we also used an automatic pipeline to detect potential new CDSs. We predict that 50 additional sequences could correspond to novel CDSs (Supplemental Tables S1, S2). More features of the sequenced RT-PCR fragments, e.g., exon length and GC content, intron length, and strength of donor-acceptor sites, are described in the Supplemental section and can be viewed in Supplemental Figures S3 and S4.
Evolutionary conservation of new sequences
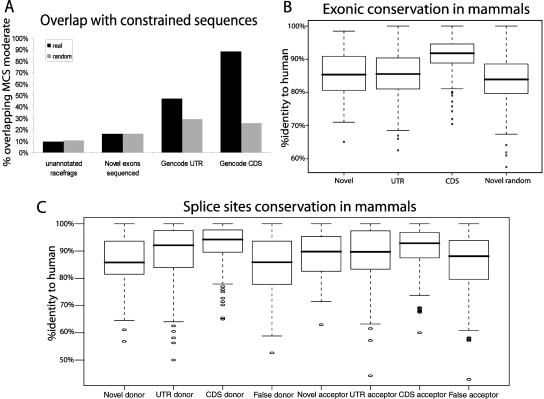
Having demonstrated that these new transcript isoforms are biochemically validated does not necessarily imply that they are biologically functional, as they might result from erroneous transcription, for example. To further assess their role, we examined whether they show evidence of purifying selection. We took advantage of the multi-species alignments and conservation analyses available in ENCODE (The ENCODE Project Consortium 2007; Margulies et al. 2007) to evaluate the conservation of the novel exons. First, we measured the overlap of 86 entirely novel exons (not a single nucleotide belonging to an annotated exon) with the set of Multi-species Conserved Sequences (MCS) identified by several approaches (The ENCODE Project Consortium 2007; Margulies et al. 2007; Fig. 5A). In contrast to the GENCODE CDS or UTR exons, neither the novel sequenced exons nor the unannotated RACEfrags overlap constrained sequences more than expected by chance. Second, we defined conservation through 23 mammalian species as the percent identity to human, ignoring all gap characters using the MAVID alignment tool (Bray and Pachter 2004; Margulies et al. 2007). The conservation of the novel exons is not significantly different from that of the GENCODE UTRs, but is significantly higher than that of mock novel exons, i.e., randomly distributed exons mimicking the novel exons (p = 0.03238), and is significantly lower than that of GENCODE CDS exons (p = 1.003 × 10−10) (Fig. 5B). Thus, novel exons do not overlap MCS more than randomly expected, but they appear to be conserved across the mammalian lineage at a rate similar to what is reported for UTRs, consistent with the 5′ UTR nature of almost all of the sequenced novel exons (see above). Third, we assessed the conservation of the 90 novel acceptor and 48 novel donor splice sites (positions −2 to +6 and −6 to +2, respectively) (Fig. 5C). Novel donor splice sites are significantly less constrained than UTR and CDS donors (p = 0.001 and 2.3 × 10−9, respectively) and not more than randomly picked false donors (random GT). On the other hand, conservation of novel acceptors is not significantly different from either UTR or false acceptors (random AG), but is significantly reduced compared with that of CDS acceptors (p = 0.03). Hence, novel exons are overall relatively poorly conserved, i.e., at a rate similar to that observed for UTR exons. Their splice sites, however, tend not to be as constrained as GENCODE UTR splice sites. Nevertheless, their strength, which is similar to that of GENCODE splice sites, argues for their genuineness (see Supplemental text and Fig. S4).
Figure 5.
Evolutionary conservation of RACEfrags. (A) Overlap of four data sets with constrained sequences. For each dataset, the percentage of projected (black) and random objects (gray; same sizes as real objects but randomly distributed in nonrepeated regions and unannotated for RACEfrags or novel exons) overlapping MCS (Multi-species Conserved Sequences)-constrained sequences by at least one nucleotide are represented on the Y-axis. Please note that GENCODE UTR and GENCODE CDS show an overlap with MCS significantly greater than random sequences. (B) Exonic conservation in mammals. For each dataset, a boxplot depicting the distribution of nucleotide conservation scores is shown. Conservation is computed as the percent identity to the human sequence for the entire length of the feature. The heavy black line marks the median score, the box contains the second and third quartiles, and whiskers mark the fifth and ninety-fifth percentiles. Novel random features are randomly chosen from unannotated nonrepetitive regions that exhibit the same size distribution as novel exons. For CDS features, a random nonredundant subset of GENCODE-annotated known coding exons was used. The CDS exons are significantly more conserved than the other features. Note that the novel sequenced exons and GENCODE UTR exons are significantly more conserved than random sequences (Novel random). (C) Splice sites conservation in mammals. For each data set, donor sequences (−2 to +6 with respect to the 5′ splice junction) and acceptor sequences (−6 to +2 with respect to the 3′ splice junction) were scored for conservation to the human splice site sequence. Boxplots were produced as in B. False splice sites were picked at random from the set of all GT or AG dinucleotides in ENCODE regions that do not overlap GENCODE-annotated exons or repeats. UTR and CDS donors and CDS acceptors are significantly more conserved than false splice sites (random GT or AG). Novel splice sites do not exhibit elevated conservation over background.
Expression levels of RACEfrags
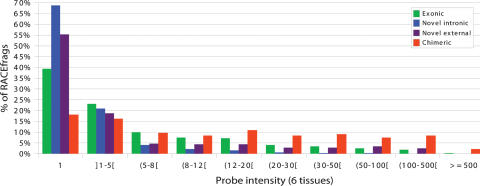
None of the observations reported above allowed us to unequivocally conclude the functionality of the newly identified transcripts, indicating the need for other lines of evidence. One such kind of support might come from the abundance of the transcripts that incorporate the new exons, because it is likely that functional transcripts will be present in at least one copy in multiple cells of a given tissue. To assess how often the exons corresponding to the distal RACEfrags are transcribed compared with exons that form the index protein-coding gene, transcriptome maps were generated from polyA+ RNA from brain, kidney, small intestine, colon, liver, and stomach used for the RACE reactions using the same arrays (see Methods). These maps were used to measure intensity signals of the probes overlapping four different sets of RACEfrags: (1) those mapping to the GENCODE-annotated exons of the RACE-interrogated locus (“exonic”); (2) unannotated RACEfrags mapping into introns of the RACE-interrogated locus (“intronic”); (3) unannotated RACEfrags mapping externally to the RACE-interrogated locus (“external”); (4) annotated RACEfrags mapping externally to the RACE-interrogated locus, i.e., linking the RACE-interrogated locus to a 5′-positioned locus into a transcription-induced chimera (“chimeric”). The results are summarized in Figure 6 and detailed in Supplemental Table S3. In each tissue, considering all loci, “chimeric” RACEfrags appear to be expressed at a higher level than “exonic” RACEfrags, while the latter are more highly expressed than the “intronic” RACEfrags. The fourth category, “external” RACEfrags, shows levels of expression similar to the ones measured for “exonic” RACEfrags; however, a larger fraction of them appear not to be expressed (Fig. 6; Supplemental Table S3).
Figure 6.
Intensity signal registered for RACEfrags. Distribution of exonic (green columns), novel intronic (blue columns), novel external (purple columns), and chimeric (red columns) RACEfrags according to the intensity signals measured on probes overlapping the regions where they map in six tissues. Intensity values are represented on the X-axis. Values of 1 mean no signal (ratio of 1 compared with control), as positive probes have intensity >1. The percentage of RACEfrags in each intensity bin is given on the Y-axis.
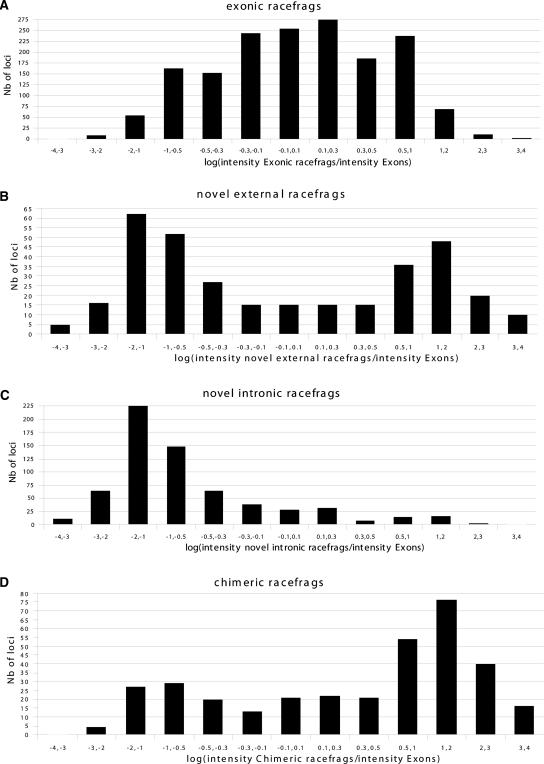
To get an estimate of the abundance of the unannotated RACEfrags relative to the known exons, we compared the expression of the target locus with the expression of the RACEfrags (see Methods) in each tissue. First, to control the validity of this approach, we verified that the exonic RACEfrags have levels of expression close to the ones showed by all the exons from the targeted locus. Convincingly, we found that 65.5% of the ratios of intensities of exonic RACEfrags over all exons are between 0.5- and twofold (Fig. 7A). Of the “external” RACEfrags, 50.3% and 38.2% of showed intensities between 0.1- and onefold and one- and 10-fold, respectively, that of exons from the target locus, respectively (Fig. 7B). In some loci, the expression is lower for the “external” RACEfrags than for the target loci, while in other loci it is higher. These results suggest that a substantial proportion of the distal novel exons identified by the combination of RACEs and arrays are not part of rare splice forms. On the other hand, the novel internal RACEfrags have consistently lower expression levels than the target gene exons and appear to be less frequently incorporated in transcripts (Fig. 7C), but most of the differences in expression levels are usually <10-fold. Similar to “external” RACEfrags, “chimeric” RACEfrags show ratios between 0.1- and 10-fold (84% of the ratios; 55% of the ratio >1) (Fig. 7D). Again, differences from locus to locus are evident, but in the majority of investigated loci the “chimeric” RACEfrags appear to be incorporated in more transcripts than the target locus. A probable explanation for this trend is that the exons corresponding to the chimeric RACEfrags tend to be incorporated into more than one type of transcript, the “chimeric” transcripts, as well as the “classical” transcripts from that locus.
Figure 7.
Expression levels of RACEfrags. Distribution of ratios of intensity signals measured for probes overlapping different subsets of RACEfrags: exonic (A), novel external (B), novel intronic (C), and chimeric (D). The expression levels in the different sets were calculated by averaging the median intensities of positive probes in each RACEfrags/exons among all the exons/RACEfrags in the set. The ratios are calculated as the intensity level obtained in the considered set of RACEfrags divided by the intensity level obtained for exons from the target locus. The bins on the X-axis represent the log of the ratios (logs between −0.3 and 0.3 correspond to ratios between 0.5- and twofold).
Overlapping of RACEfrags with biochemically identified regions
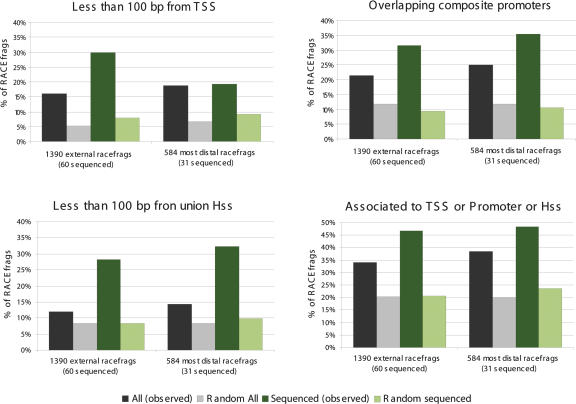
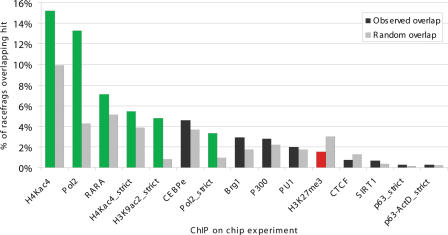
To further support the notion that the 5′ ends of the newly identified transcripts correspond to bona fide TSS, we explored whether they were associated with TSS hallmarks such as chromatin remodeling. We compared the mapping position of the external RACEfrags not overlapping annotated first exons to the mapping positions of the sets of TSS, composite promoters positions, open-chromatin sites, and the union of these, which were established by the ENCODE project (The ENCODE Project Consortium 2007). TSS were alternatively determined by massive sequencing of CAGE (5′-specific cap analysis gene expression) tags and 5′ PETs (paired-end 5′ and 3′ ditags) (Carninci et al. 2005; Ng et al. 2005). Promoter regions were identified by tiling array-coupled chromatin-immunoprecipitation (ChIP-on-chip) with different antibodies recognizing multiple members of the transcription machinery, while opening of the chromatin was assessed by hypersensitivity to DNAseI (The ENCODE Project Consortium 2007). We found that both external and 5′-most distal RACEfrags significantly overlap more with these three different regions than expected by chance (all p < 0.01), providing independent evidence supporting their proximity to transcription initiation (Fig. 8). Interestingly, the overlap is proportionally increased for the subset of 5′-most distal RACEfrags, suggesting that these are more likely to be definitive TSSs. Similarly, we observe that the overlap is proportionally larger for the RACEfrags supported by sequenced RT-PCR products (Fig. 8). We then assessed the overlap between the external RACEfrags and the regions bound by diverse transcription factors or by modified histones (The ENCODE Project Consortium 2007). We limited the analysis to RACEfrag and ChIP-on-chip data obtained for the HL60 cell line and the same tiling array. First, we assessed the overlap between the coordinates of the 161 HL60 external RACEfrags not overlapping annotated first exons and the ChIP-on-chip-identified regions. We found a significant enrichment of external RACEfrags (p < 0.05) in POLR2A-bound regions, further supporting the notion that these distal RACEfrags are close to transcription initiation (not shown). To increase the power of our analysis we evaluated the overlap of the 791 RACEfrags identified in HL60 and non-overlapping annotated first exons. We observe a significant enrichment of RACEfrags in regions bound by POLR2A, Retinoic acid receptor alpha (RARA), tetra-acetylated histone H4, and di-acetylated histone H3 (all p < 0.05) (Fig. 9). Conversely, we found that the K27 tri-methylated histone H3-bound regions were significantly depleted of RACEfrags (Fig. 9). Thus, as expected of the regions representing true sites of transcription and transcription initiation, the RACEfrags appear to be associated with open chromatin regions marked by tetra-acetylated histone H4 and K9, K14 di-acetylated histone H3 (Jenuwein and Allis 2001). Conversely, closing of the chromatin as assessed by binding of K27 tri-methylated histone H3 (Martin and Zhang 2005) results in fewer RACEfrags emanating from these regions.
Figure 8.
Overlap of RACEfrags with 5′ ends related data sets. Proportion of RACEfrags (gray) and sequence-validated RACEfrags (green) in the real (dark color) and random (light color) sets at <100 bp from transcription start sites (TSS; top left), overlapping composite promoters (top right), at <100 bp from DNase I hypersensitive sites (Hss; bottom left), and their union (bottom right). The data are shown for the 1390 external RACEfrags and 584 5′ most distal RACEfrags and their sequenced subsets on the left- and righthand side, respectively.
Figure 9.
Overlap of RACEfrags with protein-binding sites and chromatin modifications. Proportion of RACEfrags in the real (dark gray) and random (light gray) sets overlapping protein-binding or chromatin modification sites. Significant enrichments (green) and reductions (red) (p < 0.05) are highlighted. The data are shown for the 791 RACEfrags, protein-binding, and chromatin modification sites identified in HL60 cells.
Discussion
We have attempted to determine whether the current collection of annotated exons and transcription start sites (TSS) of the genes mapping to the 44 regions selected for the ENCODE pilot phase (The ENCODE Project Consortium 2004) was comprehensive. By specifically interrogating each of the protein-coding genes using a combination of 5′ RACE and tiling arrays, >2300 sites of transcription that do not overlap GENCODE-annotated exons were observed (51% of the sites identified; Harrow et al. 2006; The ENCODE Project Consortium 2007). The majority (>60%) of interrogated loci present potential new exons mapping in their introns, while two-thirds (68%) of the investigated loci show potential new putative TSS upstream of their annotated first exon, often reaching into neighboring genes.
Several lines of evidence suggest that the TSS and novel exons identified in this report correspond to bona fide exons. First, the 5′ distal RACEfrags exhibit a statistically significant trend to map in the vicinity of TSS identified using independent methods such as CAGE tags (5′-specific cap analysis gene expression), 5′ PETs (5′ paired-end ditags) (Carninci et al. 2005; Ng et al. 2005; The ENCODE Project Consortium 2007), promoter mapping, and/or sensitivity to DNase (The ENCODE Project Consortium 2007; Fig. 8). Second, the splice site strength of the novel exons appears as high as that of GENCODE UTRs and CDSs (Supplemental Fig. S4). Third, the transcripts that contain novel exons could be independently isolated. Fourth, these novel exons show some conservation in the mammalian lineage.
Why were these 5′ extensions of known transcripts and novel internal exons not identified before? A possible explanation would be that they are expressed at relatively low levels. While true for some, it appears that a significant fraction of the new exons are expressed at levels comparable to the level measured for GENCODE exons (Fig. 6; Supplemental Table S3). Alternatively, they may have been missed because they are expressed only in a restricted set of conditions. Support for this explanation comes from the fact that these novel sites of transcription tend to be tissue- or cell-line-specific. They might even be restricted to a specific cell type within a given tissue. Consistently, we observe that the three cell lines used in this study appear to possess less complexity than the interrogated tissues (Table 1; Fig. 2). Thirdly, they might have eluded identification because they present characteristics that make it problematic to clone and propagate them in bacteria. It is noteworthy that the cloning effort was challenging for this group of transcripts, indicating that they may have yet unrecognized properties, not necessarily related to their lower GC contents, that render their cloning difficult (Supplemental Fig. S3). The most plausible explanation, which is consistent with recent results (Carninci et al. 2005, 2006; Kimura et al. 2006), might be that until now ESTs and full-length cDNA sequencing efforts never reached the coverage required to truly explore the transcriptome complexity.
A sizable fraction of the novel exons and distal extensions was not identified by direct hybridization of cDNA to tiling arrays, as few transcribed fragments (transfrags), a.k.a transcriptionally active regions (TAR), overlap with them (The ENCODE Project Consortium 2007). On average, only 31% (from 18% in stomach to 52% in colon) of RACEfrags were scored as positive by direct hybridization (≥50% coverage by transfrags identified in the same tissue), emphasizing the expected differences in the sensitivity of the two approaches. Direct hybridization methods detect the entire transcriptional output of the genome, while RACE/array is limited to the transcripts containing the index region. Thus, this difference in target complexity may preclude the two methods from detecting the same transcripts; unnannotated transfrags detect predominantly noncoding RNA, while unannotated RACEfrags identify novel exons of coding transcripts.
It appears that multiple transcriptionally active regions alternatively associate to form a large variety of transcripts at a given locus. Many of these are noncoding alternative transcripts of known protein-coding genes (see Results section; Harrow et al. 2006). Multiple different hypotheses not necessarily mutually exclusive can be considered for the role of these variants and for expressed pseudogenes (Marques et al. 2005; Vinckenbosch et al. 2006; Zheng et al. 2006), antisense transcription (Kampa et al. 2004; Katayama et al. 2005), and structured RNAs (Washietl et al. 2006). They might regulate transcription and/or translation directly, like miRNA precursors, or indirectly by maintaining an open chromatin status, for instance. Additionally, some might have no functional role per se because they represent the outcome of consistent and deterministic transcription of genomic regions. This last class of transcripts would not require being under purifying selection. Consistently, many noncoding RNAs, transfrags, and RACEfrags do not appear to be under strong selective constraints (Fig. 5A; The ENCODE Project Consortium 2007; Margulies et al. 2007). Primary transcripts would emanate from regions of open chromatin and would then be spliced in a manner that depends solely on the presence of sequences that can be recognized as donor and acceptor sites. Consistently, the splice site strength of the novel exons appears as high as that of GENCODE UTRs and CDSs (Supplemental Fig. S3). This strength does not strongly correlate with splice site conservation, neither among novel nor amid GENCODE splice sites. Similarly, premature termination codon-containing splice variants were shown to be expressed at low levels across diverse mammalian cells and tissues, independently of the action of nonsense-mediated mRNA decay (Pan et al. 2006).
Some RACEfrags, however, are part of transcript variants that putatively encode novel polypeptides by modifying the ORF (Supplemental Tables S1, S2). Chimeric transcripts, for example, fuse two different ORFs to potentially generate a new protein (see example in Fig. 3). This recently described phenomenon (Kapranov et al. 2005; Akiva et al. 2006; Parra et al. 2006) appears to be widespread, affecting more than half of the loci investigated (and 25% of the novel extensions for which we obtained sequences; 13 loci out of 52 for which an extension was targeted). It might represent a means to increase protein diversity from a limited number of genes and exons.
Our results also suggest that genes are using the promoter(s) of other neighboring genes in specific cells and developmental stages, as was recently reported for Drosophila (Manak et al. 2006). Consistently, we observe that (1) 6.2% (46/738) of the new 5′ ends identified by RACEfrags are shared by several genes, a proportion very likely to be underestimated because of the stringent filterings of non-pool-specific RACEfrags in the assignment procedure (5.3% [319/5954] of all RACEfrags are shared by several genes); and (2) that several RACEfrags are as conserved as GENCODE UTRs (Fig. 5B) and part of transcript variants that modify only the 5′ UTR, while maintaining the same CDS (Supplemental Tables S1, S2). Furthermore, we find RACEfrags suggesting 5′ extensions that increase gene territories by >300 kb (median 85 kb). Provocatively, one may argue that enhancers that map hundreds of kilobases away from a gene are in fact positioned close to the true, as yet unrecognized TSS. Would this hypothesis be correct, it requires that primary nuclear transcripts traverse long genomic distances. The issues associated with such long-distance transcription events are numerous and may argue for alternative mechanisms to create spliced transcripts that incorporate distal exons, such as trans-splicing (Horiuchi and Aigaki 2006).
The notion that mammalian transcriptomes are made of a swarming mass of different overlapping transcripts sometimes originating from both strands (Kapranov et al. 2002, 2005; Bertone et al. 2004; Kampa et al. 2004; Carninci et al. 2005; Cheng et al. 2005; Katayama et al. 2005; Engstrom et al. 2006; The ENCODE Project Consortium 2007), together with the findings reported here suggesting that we have uncovered only a congruent portion of its complexity, have important implications for medicine and the study of model organisms. First, they increase the size of the genomic regions that might harbor causative polymorphisms and pathogenic mutations predisposing to a complex common phenotype and associated with a Mendelian disease, respectively. Second, they may impair positional cloning strategies pursued to identify genes implicated in these pathologies. Third, they suggest that one should use extra caution when associating a phenotype with a gene knock-out or knock-in, as it appears that the same nucleotide on the genome can operate multi-functionally, for example as intron and exon of one gene, as exon of another gene, and as transcription factor binding site. Finally, they indicate that annotated genes may have multiple alternative regulatory regions, often beyond what is currently considered to be their annotated 5′ promoters and often overlapping bounds of other genes.
Methods
RACE/array analysis of known protein-coding genes
5′ RACEs were performed on polyA+ RNAs from 12 human tissues and three cell lines using the BD SMART RACE cDNA amplification kit according to the manufacturers’ instructions (BD Clontech). RACE reactions performed with oligonucleotides specific to non-neighboring genes and on the same tissue/cell line cDNA were assembled in pools and hybridized to ENCODE tiling arrays as described in Kapranov et al. (2005). A detailed description of the methods used can be viewed in the Supplement.
Assignment of RACEfrags to the target locus
The hybridization of the 5′ RACE products on the tiling arrays was performed in five pools (each containing ∼80 nonadjacent loci) for each of the tissues/cell lines. The RACEfrags were assigned to a particular locus as follows:
The RACEfrag maps were filtered to remove RACEfrags coming from nonspecific amplicons. RACEfrags that are not specific to any particular pool of oligonucleotides almost certainly represent nonspecific amplicons that are often present in RACE reactions. To remove the products of such amplicons, RACEfrags that did not overlap GENCODE annotations and were not pool-specific were filtered out if they were overlapping RACEfrags from other pools by >50% of their length. In addition, the RACEfrags that overlapped GENCODE exons were subdivided into fragments overlapping and not overlapping exons. The fragmented RACEfrags overlapping exons were kept, whereas the ones not overlapping exons were filtered as above.
A RACE reaction was considered positive if at least one target exon was overlapping a RACEfrag. The target region was defined as the genomic landscape between the index exon in which the original 5′ RACE oligonucleotide was designed and the GENCODE-annotated 5′ terminus of the locus (Harrow et al. 2006). Target exons were defined as annotated exons within the target region. With these criteria we found 73% of positive reactions and 89% of loci positive in at least one of the tissues tested. For the subsequent assignment procedure, only the target loci yielding positive reactions were considered.
The non-assignable RACEfrags, which map 3′ to all target loci belonging to the pool, were discarded. Another group of RACEfrags was classified as ambiguous if they localized 5′ to a pair of target loci mapping on opposite strands. Overall, this resulted in 72% of assignable and 12% of ambiguous RACEfrags of the total number of RACEfrags kept after step 1. The final filter applied to all RACEfrags was to remove the ones overlapping target exons from other pools in order to rule out pooling errors. At the final assignment step, the remaining RACEfrags that were internal to the corresponding target locus were assigned to that target locus. RACEfrags found outside of the bounds of any target loci were assigned to the most proximal 3′ target locus. The ambiguous RACEfrags were assigned to both possible loci, with high or low level of confidence: When the RACEfrag was closer to one locus than to the other (difference of distances >100 kb), the assignment was considered as highly confident for the closest locus (provided that the RACEfrag was <100 kb from the locus); otherwise, the assignments to both loci were considered as not confident. The final set of RACEfrags we describe contains only confidently assigned RACEfrags, representing 75% of all the RACEfrags.
Note that while the RACEfrags were assigned to the 3′ most proximal target locus, we could envision scenarios where the RACEfrags are linked to target loci separated by other target loci. We indeed observed numerous cases of extensions reaching across several loci. However, the verifications based on RT-PCR reactions allowed us to confirm the connectivity between RACEfrags and target loci, suggesting that the assignments were correct in most of the cases (see main text for results and below for procedure).
Furthermore, we were conservative, as non-pool-specific RACEfrags overlapping target exons from genes from other pools were discarded in case some pooling errors had occurred. As described in the main text, the RACE reactions revealed numerous cases of transcription-induced chimeras (Akiva et al. 2006; Parra et al. 2006); thus, some of these discarded RACEfrags could well have come from the correct target locus. Furthermore, as the target exons of other pools (i.e., the exons between the RACE oligonucleotide and the 5′ end of the locus) were discarded, the proportion of RACEfrags overlapping first exons is probably underestimated, and the RACEfrags reaching into 3′ exons of neighboring genes are probably not the most distal ones.
Finally, it is worth mentioning that the hybridization data will be problematic to interpret in clusters of orthologous genes because the amplification products might hybridize to multiple genes from a given cluster, thus creating artifactual chimeras. Aware of this possibility and in order to minimize it, we performed two different analyses: The first includes all chimeras, while the second specifically targets chimeras with no loci that are part of clusters of genes.
RT-PCR of RACEfrags
Five hundred thirty-eight RACEfrags were selected for independent verification of their connectivity with the original annotated gene by RT-PCR on oligo dT-primed and/or gene-specific-primed cDNA as described previously (Kapranov et al. 2002; Reymond et al. 2002a; see Supplemental section for details).
Assignment of RT-PCRfrags
The pooling of RT-PCR reactions for array hybridizations was done such that assignment of RT-PCRfrags to each target locus would be unambiguous; i.e., each pool contained RT-PCR reactions derived from different ENCODE regions. RT-PCRfrags mapping between forward and reverse RT-PCR primers pairs were assigned to the corresponding RT-PCR reaction. An RT-PCR reaction was scored as positive based on the profile of microarray hybridization (see Supplemental section for particulars).
Cloning and sequencing of the RACE/array products
The RT-PCR reactions were either sequenced directly or subcloned and sequenced before submission to GenBank under accession numbers DQ655905-DQ656069 and EF070113–EF070122. A detailed procedure can be viewed in the Supplemental section.
Sequence conservation analysis
Sequence conservation among mammalian species was calculated as follows. For each particular human feature under consideration (exonic sequence or splice site), a subalignment was extracted from the MAVID alignment corresponding to the October 2005 ENCODE data freeze. Gaps with respect to the human sequence and sequences of nonmammalian species were removed. Then, for each column of the alignment, the number of conserved bases and total number of bases aligned to human were tallied. The total number of conserved bases across all columns divided by the total number of aligned bases across all columns gives the conservation score for a feature. Gap characters were ignored in this analysis. Statistical significance for conservation and splice site strength was determined by two-tailed t-tests. Nonparametric tests gave similar results.
Hybridization of RNA samples on tiling arrays
PolyA+ RNA was treated with DNase I, converted into double-stranded cDNA, and hybridized to ENCODE tiling arrays as described (Cheng et al. 2005). We measured intensity signals of the probes overlapping the regions where the RACEfrags mapped. Four sets of RACEfrags were considered: RACEfrags mapping to exons of the RACEd locus (“exonic”), unannotated RACEfrags mapping into introns of the RACEd locus (“intronic”), unannotated RACEfrags mapping externally to the RACEd locus (“external”), and annotated RACEfrags mapping externally to the RACEd locus, i.e., linking the RACEd locus to a locus upstream in a possible chimera (“chimeric”). In the “chimeric” subset, all index genes that were part of clusters of paralogous genes were discarded, as it was impossible to know if the linking between two genes of a cluster is genuine or due to cross-hybridization. The expression levels in the different sets were calculated by averaging the median intensities of positive probes in each RACEfrags/exons among all the exons/RACEfrags in the set.
Overlaps of RACEfrags with other data sets
Four different sets of RACEfrags identified using RNAs from the 12 human tissues more or less enriched for putative 5′ ends of transcripts and HL60 RACEfrags not annotated as first exons were overlapped with 5′-end-related data sets and ChIP-on-chip hits produced by the ENCODE Consortium (The ENCODE Project Consortium 2007), respectively. A complete list of the exploited data sets and a detailed description of the RACEfrags sets can be found in the Supplemental section. The percentages of RACEfrags having 1-bp overlap with the ENCODE data sets (stranded when the dataset included a strand information) were calculated for each RACEfrags set, as well as for random sets (100 random sets mimicking each of the sets) to compare the random overlap to the observed overlap.
Acknowledgments
We thank The ENCODE Project Consortium for making its data publicly available and Urmila Choudhury for comments. This work was funded by National Human Genome Research Institute (NHGRI)/National Institutes of Health (NIH) grants to the GENCODE [#U01HG03150] and Affymetrix, Inc [#U01HG03147], subgroups of the ENCODE project. This work was also supported in part with Federal Funds from the National Cancer Institute, National Institutes of Health, under Contract # N01-CO-12400 (to T.R.G.) and by Affymetrix, Inc. We acknowledge grants from the Swiss National Science Foundation (S.E.A. and A.R.); the Spanish Ministerio de Educación y Ciencia (R.G.), the NCCR Frontiers in Genetics (S.E.A.), the European Union (S.E.A., R.G., and A.R.), and the Jérôme Lejeune (S.E.A. and A.R.), the Childcare (S.E.A.), and the Novartis (A.R.) Foundations.
Footnotes
[Supplemental material is available online at www.genome.org. The sequence data from this study have been submitted to DDBJ/GenBank/EMBL under accession numbers DQ655905-DQ656069 and EF070113-EF070122.]
Article is online at http://www.genome.org/cgi/doi/10.1101/gr.5660607
References
- Adams M.D., Celniker S.E., Holt R.A., Evans C.A., Gocayne J.D., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Celniker S.E., Holt R.A., Evans C.A., Gocayne J.D., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Holt R.A., Evans C.A., Gocayne J.D., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Evans C.A., Gocayne J.D., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Gocayne J.D., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Li P.W., Hoskins R.A., Galle R.F., Hoskins R.A., Galle R.F., Galle R.F., et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Akiva P., Toporik A., Edelheit S., Peretz Y., Diber A., Shemesh R., Novik A., Sorek R., Toporik A., Edelheit S., Peretz Y., Diber A., Shemesh R., Novik A., Sorek R., Edelheit S., Peretz Y., Diber A., Shemesh R., Novik A., Sorek R., Peretz Y., Diber A., Shemesh R., Novik A., Sorek R., Diber A., Shemesh R., Novik A., Sorek R., Shemesh R., Novik A., Sorek R., Novik A., Sorek R., Sorek R. Transcription-mediated gene fusion in the human genome. Genome Res. 2006;16:30–36. doi: 10.1101/gr.4137606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Dolinski K., Dwight S.S., Eppig J.T., Dwight S.S., Eppig J.T., Eppig J.T., et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertone P., Stolc V., Royce T.E., Rozowsky J.S., Urban A.E., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Stolc V., Royce T.E., Rozowsky J.S., Urban A.E., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Royce T.E., Rozowsky J.S., Urban A.E., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Rozowsky J.S., Urban A.E., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Urban A.E., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Tongprasit W., Samanta M., Weissman S., Samanta M., Weissman S., Weissman S., et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–2246. doi: 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- Bray N., Pachter L., Pachter L. MAVID: Constrained ancestral alignment of multiple sequences. Genome Res. 2004;14:693–699. doi: 10.1101/gr.1960404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- C. elegans Sequencing Consortium, Genome sequence of the nematode C. elegans: A platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- Carninci P., Kasukawa T., Katayama S., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., Kasukawa T., Katayama S., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., Katayama S., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., Oyama R., Ravasi T., Lenhard B., Wells C., Ravasi T., Lenhard B., Wells C., Lenhard B., Wells C., Wells C., et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Carninci P., Sandelin A., Lenhard B., Katayama S., Shimokawa K., Ponjavic J., Semple C.A., Taylor M.S., Engstrom P.G., Frith M.C., Sandelin A., Lenhard B., Katayama S., Shimokawa K., Ponjavic J., Semple C.A., Taylor M.S., Engstrom P.G., Frith M.C., Lenhard B., Katayama S., Shimokawa K., Ponjavic J., Semple C.A., Taylor M.S., Engstrom P.G., Frith M.C., Katayama S., Shimokawa K., Ponjavic J., Semple C.A., Taylor M.S., Engstrom P.G., Frith M.C., Shimokawa K., Ponjavic J., Semple C.A., Taylor M.S., Engstrom P.G., Frith M.C., Ponjavic J., Semple C.A., Taylor M.S., Engstrom P.G., Frith M.C., Semple C.A., Taylor M.S., Engstrom P.G., Frith M.C., Taylor M.S., Engstrom P.G., Frith M.C., Engstrom P.G., Frith M.C., Frith M.C., et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 2006;38:626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- Cheng J., Kapranov P., Drenkow J., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Kapranov P., Drenkow J., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Drenkow J., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Patel S., Long J., Stern D., Tammana H., Helt G., Long J., Stern D., Tammana H., Helt G., Stern D., Tammana H., Helt G., Tammana H., Helt G., Helt G., et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium, The ENCODE (ENCyclopedia of DNA Elements) Project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium, Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007 doi: 10.1038/nature05874. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engstrom P.G., Suzuki H., Ninomiya N., Akalin A., Sessa L., Lavorgna G., Brozzi A., Luzi L., Tan S.L., Yang L., Suzuki H., Ninomiya N., Akalin A., Sessa L., Lavorgna G., Brozzi A., Luzi L., Tan S.L., Yang L., Ninomiya N., Akalin A., Sessa L., Lavorgna G., Brozzi A., Luzi L., Tan S.L., Yang L., Akalin A., Sessa L., Lavorgna G., Brozzi A., Luzi L., Tan S.L., Yang L., Sessa L., Lavorgna G., Brozzi A., Luzi L., Tan S.L., Yang L., Lavorgna G., Brozzi A., Luzi L., Tan S.L., Yang L., Brozzi A., Luzi L., Tan S.L., Yang L., Luzi L., Tan S.L., Yang L., Tan S.L., Yang L., Yang L., et al. Complex loci in human and mouse genomes. PLoS Genet. 2006;2:e47. doi: 10.1371/journal.pgen.0020047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffeau A., Barrell B.G., Bussey H., Davis R.W., Dujon B., Feldmann H., Galibert F., Hoheisel J.D., Jacq C., Johnston M., Barrell B.G., Bussey H., Davis R.W., Dujon B., Feldmann H., Galibert F., Hoheisel J.D., Jacq C., Johnston M., Bussey H., Davis R.W., Dujon B., Feldmann H., Galibert F., Hoheisel J.D., Jacq C., Johnston M., Davis R.W., Dujon B., Feldmann H., Galibert F., Hoheisel J.D., Jacq C., Johnston M., Dujon B., Feldmann H., Galibert F., Hoheisel J.D., Jacq C., Johnston M., Feldmann H., Galibert F., Hoheisel J.D., Jacq C., Johnston M., Galibert F., Hoheisel J.D., Jacq C., Johnston M., Hoheisel J.D., Jacq C., Johnston M., Jacq C., Johnston M., Johnston M., et al. Life with 6000 genes. Science. 1996;274:546, 563–567. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- Harrow J., Denoeud F., Frankish A., Reymond A., Chen C.K., Chrast J., Lagarde J., Gilbert J.G., Storey R., Swarbreck D., Denoeud F., Frankish A., Reymond A., Chen C.K., Chrast J., Lagarde J., Gilbert J.G., Storey R., Swarbreck D., Frankish A., Reymond A., Chen C.K., Chrast J., Lagarde J., Gilbert J.G., Storey R., Swarbreck D., Reymond A., Chen C.K., Chrast J., Lagarde J., Gilbert J.G., Storey R., Swarbreck D., Chen C.K., Chrast J., Lagarde J., Gilbert J.G., Storey R., Swarbreck D., Chrast J., Lagarde J., Gilbert J.G., Storey R., Swarbreck D., Lagarde J., Gilbert J.G., Storey R., Swarbreck D., Gilbert J.G., Storey R., Swarbreck D., Storey R., Swarbreck D., Swarbreck D., et al. GENCODE: Producing a reference annotation for ENCODE. Genome Biol. 2006;7(Suppl 1):S4.1–S4.9. doi: 10.1186/gb-2006-7-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi T., Aigaki T., Aigaki T. Alternative trans-splicing: A novel mode of pre-mRNA processing. Biol. Cell. 2006;98:135–140. doi: 10.1042/BC20050002. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium, Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- Jenuwein T., Allis C.D., Allis C.D. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Kampa D., Cheng J., Kapranov P., Yamanaka M., Brubaker S., Cawley S., Drenkow J., Piccolboni A., Bekiranov S., Helt G., Cheng J., Kapranov P., Yamanaka M., Brubaker S., Cawley S., Drenkow J., Piccolboni A., Bekiranov S., Helt G., Kapranov P., Yamanaka M., Brubaker S., Cawley S., Drenkow J., Piccolboni A., Bekiranov S., Helt G., Yamanaka M., Brubaker S., Cawley S., Drenkow J., Piccolboni A., Bekiranov S., Helt G., Brubaker S., Cawley S., Drenkow J., Piccolboni A., Bekiranov S., Helt G., Cawley S., Drenkow J., Piccolboni A., Bekiranov S., Helt G., Drenkow J., Piccolboni A., Bekiranov S., Helt G., Piccolboni A., Bekiranov S., Helt G., Bekiranov S., Helt G., Helt G., et al. Novel RNAs identified from an in-depth analysis of the transcriptome of human chromosomes 21 and 22. Genome Res. 2004;14:331–342. doi: 10.1101/gr.2094104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P., Cawley S.E., Drenkow J., Bekiranov S., Strausberg R.L., Fodor S.P., Gingeras T.R., Cawley S.E., Drenkow J., Bekiranov S., Strausberg R.L., Fodor S.P., Gingeras T.R., Drenkow J., Bekiranov S., Strausberg R.L., Fodor S.P., Gingeras T.R., Bekiranov S., Strausberg R.L., Fodor S.P., Gingeras T.R., Strausberg R.L., Fodor S.P., Gingeras T.R., Fodor S.P., Gingeras T.R., Gingeras T.R. Large-scale transcriptional activity in chromosomes 21 and 22. Science. 2002;296:916–919. doi: 10.1126/science.1068597. [DOI] [PubMed] [Google Scholar]
- Kapranov P., Drenkow J., Cheng J., Long J., Helt G., Dike S., Gingeras T.R., Drenkow J., Cheng J., Long J., Helt G., Dike S., Gingeras T.R., Cheng J., Long J., Helt G., Dike S., Gingeras T.R., Long J., Helt G., Dike S., Gingeras T.R., Helt G., Dike S., Gingeras T.R., Dike S., Gingeras T.R., Gingeras T.R. Examples of the complex architecture of the human transcriptome revealed by RACE and high-density tiling arrays. Genome Res. 2005;15:987–997. doi: 10.1101/gr.3455305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama S., Tomaru Y., Kasukawa T., Waki K., Nakanishi M., Nakamura M., Nishida H., Yap C.C., Suzuki M., Kawai J., Tomaru Y., Kasukawa T., Waki K., Nakanishi M., Nakamura M., Nishida H., Yap C.C., Suzuki M., Kawai J., Kasukawa T., Waki K., Nakanishi M., Nakamura M., Nishida H., Yap C.C., Suzuki M., Kawai J., Waki K., Nakanishi M., Nakamura M., Nishida H., Yap C.C., Suzuki M., Kawai J., Nakanishi M., Nakamura M., Nishida H., Yap C.C., Suzuki M., Kawai J., Nakamura M., Nishida H., Yap C.C., Suzuki M., Kawai J., Nishida H., Yap C.C., Suzuki M., Kawai J., Yap C.C., Suzuki M., Kawai J., Suzuki M., Kawai J., Kawai J., et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- Kimura K., Wakamatsu A., Suzuki Y., Ota T., Nishikawa T., Yamashita R., Yamamoto J., Sekine M., Tsuritani K., Wakaguri H., Wakamatsu A., Suzuki Y., Ota T., Nishikawa T., Yamashita R., Yamamoto J., Sekine M., Tsuritani K., Wakaguri H., Suzuki Y., Ota T., Nishikawa T., Yamashita R., Yamamoto J., Sekine M., Tsuritani K., Wakaguri H., Ota T., Nishikawa T., Yamashita R., Yamamoto J., Sekine M., Tsuritani K., Wakaguri H., Nishikawa T., Yamashita R., Yamamoto J., Sekine M., Tsuritani K., Wakaguri H., Yamashita R., Yamamoto J., Sekine M., Tsuritani K., Wakaguri H., Yamamoto J., Sekine M., Tsuritani K., Wakaguri H., Sekine M., Tsuritani K., Wakaguri H., Tsuritani K., Wakaguri H., Wakaguri H., et al. Diversification of transcriptional modulation: Large-scale identification and characterization of putative alternative promoters of human genes. Genome Res. 2006;16:55–65. doi: 10.1101/gr.4039406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Devon K., Dewar K., Doyle M., FitzHugh W., Dewar K., Doyle M., FitzHugh W., Doyle M., FitzHugh W., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Manak J.R., Dike S., Sementchenko V., Kapranov P., Biemar F., Long J., Cheng J., Bell I., Ghosh S., Piccolboni A., Dike S., Sementchenko V., Kapranov P., Biemar F., Long J., Cheng J., Bell I., Ghosh S., Piccolboni A., Sementchenko V., Kapranov P., Biemar F., Long J., Cheng J., Bell I., Ghosh S., Piccolboni A., Kapranov P., Biemar F., Long J., Cheng J., Bell I., Ghosh S., Piccolboni A., Biemar F., Long J., Cheng J., Bell I., Ghosh S., Piccolboni A., Long J., Cheng J., Bell I., Ghosh S., Piccolboni A., Cheng J., Bell I., Ghosh S., Piccolboni A., Bell I., Ghosh S., Piccolboni A., Ghosh S., Piccolboni A., Piccolboni A., et al. Biological function of unannotated transcription during the early development of Drosophila melanogaster. Nat. Genet. 2006;38:1151–1158. doi: 10.1038/ng1875. [DOI] [PubMed] [Google Scholar]
- Margulies E.H., Cooper G.M., Asimenos G., Thomas D.J., Dewey C.N., Siepel A., Birney E., Keefe D., Schwartz A.S., Hou M., Cooper G.M., Asimenos G., Thomas D.J., Dewey C.N., Siepel A., Birney E., Keefe D., Schwartz A.S., Hou M., Asimenos G., Thomas D.J., Dewey C.N., Siepel A., Birney E., Keefe D., Schwartz A.S., Hou M., Thomas D.J., Dewey C.N., Siepel A., Birney E., Keefe D., Schwartz A.S., Hou M., Dewey C.N., Siepel A., Birney E., Keefe D., Schwartz A.S., Hou M., Siepel A., Birney E., Keefe D., Schwartz A.S., Hou M., Birney E., Keefe D., Schwartz A.S., Hou M., Keefe D., Schwartz A.S., Hou M., Schwartz A.S., Hou M., Hou M., et al. Analyses of deep mammalian sequence alignments and constraint predictions for 1% of the human genome. Genome Res. 2007 doi: 10.1101/gr.6034307. (this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques A.C., Dupanloup I., Vinckenbosch N., Reymond A., Kaessmann H., Dupanloup I., Vinckenbosch N., Reymond A., Kaessmann H., Vinckenbosch N., Reymond A., Kaessmann H., Reymond A., Kaessmann H., Kaessmann H. Emergence of young human genes after a burst of retroposition in primates. PLoS Biol. 2005;3:e357. doi: 10.1371/journal.pbio.0030357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C., Zhang Y., Zhang Y. The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- Misra S., Crosby M.A., Mungall C.J., Matthews B.B., Campbell K.S., Hradecky P., Huang Y., Kaminker J.S., Millburn G.H., Prochnik S.E., Crosby M.A., Mungall C.J., Matthews B.B., Campbell K.S., Hradecky P., Huang Y., Kaminker J.S., Millburn G.H., Prochnik S.E., Mungall C.J., Matthews B.B., Campbell K.S., Hradecky P., Huang Y., Kaminker J.S., Millburn G.H., Prochnik S.E., Matthews B.B., Campbell K.S., Hradecky P., Huang Y., Kaminker J.S., Millburn G.H., Prochnik S.E., Campbell K.S., Hradecky P., Huang Y., Kaminker J.S., Millburn G.H., Prochnik S.E., Hradecky P., Huang Y., Kaminker J.S., Millburn G.H., Prochnik S.E., Huang Y., Kaminker J.S., Millburn G.H., Prochnik S.E., Kaminker J.S., Millburn G.H., Prochnik S.E., Millburn G.H., Prochnik S.E., Prochnik S.E., et al. Annotation of the Drosophila melanogaster euchromatic genome: A systematic review. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-12-research0083. RESEARCH0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng P., Wei C.L., Sung W.K., Chiu K.P., Lipovich L., Ang C.C., Gupta S., Shahab A., Ridwan A., Wong C.H., Wei C.L., Sung W.K., Chiu K.P., Lipovich L., Ang C.C., Gupta S., Shahab A., Ridwan A., Wong C.H., Sung W.K., Chiu K.P., Lipovich L., Ang C.C., Gupta S., Shahab A., Ridwan A., Wong C.H., Chiu K.P., Lipovich L., Ang C.C., Gupta S., Shahab A., Ridwan A., Wong C.H., Lipovich L., Ang C.C., Gupta S., Shahab A., Ridwan A., Wong C.H., Ang C.C., Gupta S., Shahab A., Ridwan A., Wong C.H., Gupta S., Shahab A., Ridwan A., Wong C.H., Shahab A., Ridwan A., Wong C.H., Ridwan A., Wong C.H., Wong C.H., et al. Gene identification signature (GIS) analysis for transcriptome characterization and genome annotation. Nat. Methods. 2005;2:105–111. doi: 10.1038/nmeth733. [DOI] [PubMed] [Google Scholar]
- Pan Q., Saltzman A.L., Kim Y.K., Misquitta C., Shai O., Maquat L.E., Frey B.J., Blencowe B.J., Saltzman A.L., Kim Y.K., Misquitta C., Shai O., Maquat L.E., Frey B.J., Blencowe B.J., Kim Y.K., Misquitta C., Shai O., Maquat L.E., Frey B.J., Blencowe B.J., Misquitta C., Shai O., Maquat L.E., Frey B.J., Blencowe B.J., Shai O., Maquat L.E., Frey B.J., Blencowe B.J., Maquat L.E., Frey B.J., Blencowe B.J., Frey B.J., Blencowe B.J., Blencowe B.J. Quantitative microarray profiling provides evidence against widespread coupling of alternative splicing with nonsense-mediated mRNA decay to control gene expression. Genes & Dev. 2006;20:153–158. doi: 10.1101/gad.1382806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra G., Reymond A., Dabbouseh N., Dermitzakis E.T., Castelo R., Thomson T.M., Antonarakis S.E., Guigo R., Reymond A., Dabbouseh N., Dermitzakis E.T., Castelo R., Thomson T.M., Antonarakis S.E., Guigo R., Dabbouseh N., Dermitzakis E.T., Castelo R., Thomson T.M., Antonarakis S.E., Guigo R., Dermitzakis E.T., Castelo R., Thomson T.M., Antonarakis S.E., Guigo R., Castelo R., Thomson T.M., Antonarakis S.E., Guigo R., Thomson T.M., Antonarakis S.E., Guigo R., Antonarakis S.E., Guigo R., Guigo R. Tandem chimerism as a means to increase protein complexity in the human genome. Genome Res. 2006;16:37–44. doi: 10.1101/gr.4145906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reboul J., Vaglio P., Rual J.F., Lamesch P., Martinez M., Armstrong C.M., Li S., Jacotot L., Bertin N., Janky R., Vaglio P., Rual J.F., Lamesch P., Martinez M., Armstrong C.M., Li S., Jacotot L., Bertin N., Janky R., Rual J.F., Lamesch P., Martinez M., Armstrong C.M., Li S., Jacotot L., Bertin N., Janky R., Lamesch P., Martinez M., Armstrong C.M., Li S., Jacotot L., Bertin N., Janky R., Martinez M., Armstrong C.M., Li S., Jacotot L., Bertin N., Janky R., Armstrong C.M., Li S., Jacotot L., Bertin N., Janky R., Li S., Jacotot L., Bertin N., Janky R., Jacotot L., Bertin N., Janky R., Bertin N., Janky R., Janky R., et al. C. elegans ORFeome version 1.1: Experimental verification of the genome annotation and resource for proteome-scale protein expression. Nat. Genet. 2003;34:35–41. doi: 10.1038/ng1140. [DOI] [PubMed] [Google Scholar]
- Reymond A., Camargo A.A., Deutsch S., Stevenson B.J., Parmigiani R.B., Ucla C., Bettoni F., Rossier C., Lyle R., Guipponi M., Camargo A.A., Deutsch S., Stevenson B.J., Parmigiani R.B., Ucla C., Bettoni F., Rossier C., Lyle R., Guipponi M., Deutsch S., Stevenson B.J., Parmigiani R.B., Ucla C., Bettoni F., Rossier C., Lyle R., Guipponi M., Stevenson B.J., Parmigiani R.B., Ucla C., Bettoni F., Rossier C., Lyle R., Guipponi M., Parmigiani R.B., Ucla C., Bettoni F., Rossier C., Lyle R., Guipponi M., Ucla C., Bettoni F., Rossier C., Lyle R., Guipponi M., Bettoni F., Rossier C., Lyle R., Guipponi M., Rossier C., Lyle R., Guipponi M., Lyle R., Guipponi M., Guipponi M., et al. Nineteen additional unpredicted transcripts from human chromosome 21. Genomics. 2002a;79:824–832. doi: 10.1006/geno.2002.6781. [DOI] [PubMed] [Google Scholar]
- Reymond A., Marigo V., Yaylaoglu M.B., Leoni A., Ucla C., Scamuffa N., Caccioppoli C., Dermitzakis E.T., Lyle R., Banfi S., Marigo V., Yaylaoglu M.B., Leoni A., Ucla C., Scamuffa N., Caccioppoli C., Dermitzakis E.T., Lyle R., Banfi S., Yaylaoglu M.B., Leoni A., Ucla C., Scamuffa N., Caccioppoli C., Dermitzakis E.T., Lyle R., Banfi S., Leoni A., Ucla C., Scamuffa N., Caccioppoli C., Dermitzakis E.T., Lyle R., Banfi S., Ucla C., Scamuffa N., Caccioppoli C., Dermitzakis E.T., Lyle R., Banfi S., Scamuffa N., Caccioppoli C., Dermitzakis E.T., Lyle R., Banfi S., Caccioppoli C., Dermitzakis E.T., Lyle R., Banfi S., Dermitzakis E.T., Lyle R., Banfi S., Lyle R., Banfi S., Banfi S., et al. Human chromosome 21 gene expression atlas in the mouse. Nature. 2002b;420:582–586. doi: 10.1038/nature01178. [DOI] [PubMed] [Google Scholar]
- Venter J.C., Adams M.D., Myers E.W., Li P.W., Mural R.J., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., Adams M.D., Myers E.W., Li P.W., Mural R.J., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., Myers E.W., Li P.W., Mural R.J., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., Li P.W., Mural R.J., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., Mural R.J., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., Smith H.O., Yandell M., Evans C.A., Holt R.A., Yandell M., Evans C.A., Holt R.A., Evans C.A., Holt R.A., Holt R.A., et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Vinckenbosch N., Dupanloup I., Kaessmann H., Dupanloup I., Kaessmann H., Kaessmann H. Evolutionary fate of retroposed gene copies in the human genome. Proc. Natl. Acad. Sci. 2006;103:3220–3225. doi: 10.1073/pnas.0511307103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washietl S., Pedersen J.S., Korbel J.O., Stocsits C., Gruber A.R., Hackermüller J., Hertel J., Lindemeyer M., Reiche K., Tanzer A., Pedersen J.S., Korbel J.O., Stocsits C., Gruber A.R., Hackermüller J., Hertel J., Lindemeyer M., Reiche K., Tanzer A., Korbel J.O., Stocsits C., Gruber A.R., Hackermüller J., Hertel J., Lindemeyer M., Reiche K., Tanzer A., Stocsits C., Gruber A.R., Hackermüller J., Hertel J., Lindemeyer M., Reiche K., Tanzer A., Gruber A.R., Hackermüller J., Hertel J., Lindemeyer M., Reiche K., Tanzer A., Hackermüller J., Hertel J., Lindemeyer M., Reiche K., Tanzer A., Hertel J., Lindemeyer M., Reiche K., Tanzer A., Lindemeyer M., Reiche K., Tanzer A., Reiche K., Tanzer A., Tanzer A., et al. Structured RNAs in the ENCODE selected regions of the human genome. Genome Res. 2006 doi: 10.1101/gr.5650707. (this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D., Frankish A., Baertsch R., Kapranov P., Reymond A., Choo S.W., Lu Y., Denoeud F., Antonarakis S.E., Snyder M., Frankish A., Baertsch R., Kapranov P., Reymond A., Choo S.W., Lu Y., Denoeud F., Antonarakis S.E., Snyder M., Baertsch R., Kapranov P., Reymond A., Choo S.W., Lu Y., Denoeud F., Antonarakis S.E., Snyder M., Kapranov P., Reymond A., Choo S.W., Lu Y., Denoeud F., Antonarakis S.E., Snyder M., Reymond A., Choo S.W., Lu Y., Denoeud F., Antonarakis S.E., Snyder M., Choo S.W., Lu Y., Denoeud F., Antonarakis S.E., Snyder M., Lu Y., Denoeud F., Antonarakis S.E., Snyder M., Denoeud F., Antonarakis S.E., Snyder M., Antonarakis S.E., Snyder M., Snyder M., et al. Pseudogenes in the ENCODE regions: Consensus annotation, analysis of transcription, and evolution. Genome Res. 2006 doi: 10.1101/gr.5586307. (this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]