Abstract
A key component of the ongoing ENCODE project involves rigorous comparative sequence analyses for the initially targeted 1% of the human genome. Here, we present orthologous sequence generation, alignment, and evolutionary constraint analyses of 23 mammalian species for all ENCODE targets. Alignments were generated using four different methods; comparisons of these methods reveal large-scale consistency but substantial differences in terms of small genomic rearrangements, sensitivity (sequence coverage), and specificity (alignment accuracy). We describe the quantitative and qualitative trade-offs concomitant with alignment method choice and the levels of technical error that need to be accounted for in applications that require multisequence alignments. Using the generated alignments, we identified constrained regions using three different methods. While the different constraint-detecting methods are in general agreement, there are important discrepancies relating to both the underlying alignments and the specific algorithms. However, by integrating the results across the alignments and constraint-detecting methods, we produced constraint annotations that were found to be robust based on multiple independent measures. Analyses of these annotations illustrate that most classes of experimentally annotated functional elements are enriched for constrained sequences; however, large portions of each class (with the exception of protein-coding sequences) do not overlap constrained regions. The latter elements might not be under primary sequence constraint, might not be constrained across all mammals, or might have expendable molecular functions. Conversely, 40% of the constrained sequences do not overlap any of the functional elements that have been experimentally identified. Together, these findings demonstrate and quantify how many genomic functional elements await basic molecular characterization.
The identification of sequences under evolutionary constraint is a powerful approach for inferring the locations of functional elements in a genome; mutations that affect bases with sequence-specific functionality will often be deleterious to the organism and be eliminated by purifying selection (Kimura 1983). This paradigm can be leveraged to identify both protein-coding and noncoding functions, and represents one of the best computational methods available for annotating genomic sites that are likely to be of functional, phenotypic importance (Nobrega and Pennacchio 2004). Indeed, leveraging evolutionary constraints is a cornerstone approach of modern genomics, motivating many vertebrate genome-sequencing efforts (Collins et al. 2003; Margulies et al. 2005b) as well as similar projects involving model organism taxa (Cliften et al. 2003; Kellis et al. 2003; Stein et al. 2003; Davis and White 2004).
The ENCODE Project Consortium set an ambitious goal of identifying all functional elements in the human genome, including regulators of gene expression, chromatin structural components, and sites of protein–DNA interaction (The ENCODE Project Consortium 2004). In its pilot phase, ENCODE targeted 44 individual genomic regions (see http://genome.ucsc.edu/ENCODE/regions.html for details on the target selection process) that total roughly 30 Mb (∼1% of the human genome) for functional annotation. A major component of this effort has been to generate a large resource of multispecies sequence data orthologous to these human genomic regions. The rationale for a major comparative genomics component of ENCODE includes the following:
Comparative sequence analyses reveal evolutionary constraint, and this is complementary to experimental assays because it is agnostic to any specific function. Furthermore, the experimental assays used to date by ENCODE only investigate a subset of potential functions and mostly emphasize the use of cell culture systems, which are limited in their ability to detect functional processes unique to the development, physiology, and anatomy of an organism.
Significant technical challenges regarding the alignment and analysis of deep mammalian genome sequence data sets remain unsolved and reduce the efficacy of comparative analyses. Systematic evaluation and comparison of the best computational tools, which requires such a large comparative genomics data set, would be a valuable contribution to future efforts.
Until now, no synchronized effort between evolutionarily deep comparative sequence analyses and comprehensive identification of broad classes of functional elements has been pursued. The selected ENCODE regions of the human genome provide such a test bed for exploring the relationship between evolutionary sequence constraint and sequence function in a systematic way.
Results and Discussion
Comparative sequence data
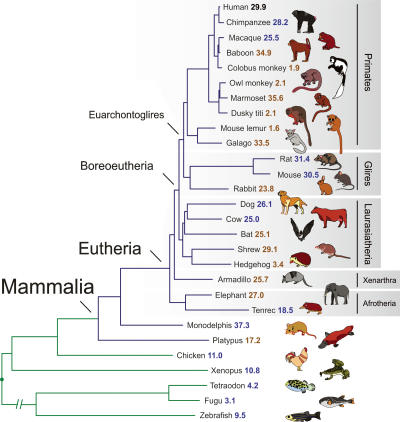
We generated and/or obtained sequences orthologous to the 44 ENCODE regions (The ENCODE Project Consortium 2004) from 28 vertebrates (Fig. 1; Supplemental Table S1). For 14 mammals, a total of 206 Mb of sequence was obtained from mapped bacterial artificial chromosomes (BACs) and finished to “comparative-grade” standards (Blakesley et al. 2004) specifically for these studies; for another 14 species, a total of 340 Mb of sequence was obtained from genome-wide sequencing efforts at varying levels of completeness and quality (Aparicio et al. 2002; International Mouse Genome Sequencing Consortium 2002; International Chicken Genome Sequencing Consortium 2004; International Human Genome Sequencing Consortium 2004; Jaillon et al. 2004; Rat Genome Sequencing Project Consortium 2004; Chimpanzee Sequencing and Analysis Consortium 2005; Lindblad-Toh et al. 2005; Margulies et al. 2005b) (see also Methods and Supplemental Material).
Figure 1.
Phylogenetic tree relating the set of analyzed species. The depicted topology and branch lengths illustrate the relationships among the analyzed species’ sequences. Analytical support for the represented tree is provided elsewhere (Nikolaev et al. 2007). The numbers next to each species name indicate the amount of sequence (in Mb) examined in this study (some species have >30 Mb of sequence either as a result of lineage-specific expansions of these regions or the resolution with which orthologous sequences can be identified before alignment) (see Supplemental Material for additional details); (red numbers) BAC-derived sequence sequenced to “comparative grade” (see Methods); (blue numbers) sequence obtained from whole-genome sequencing efforts; and (black numbers) finished human sequence. Blue and green branches distinguish mammalian from non-mammalian sequences, respectively.
Generation of multisequence alignments
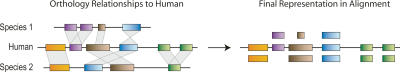
For each human base in the ENCODE regions, we aimed to identify an orthologous genomic position in every other species. Toward that end, we generated four sets of multisequence alignments, and refer to each by the name of the principal program used—namely, MAVID (Bray and Pachter 2004), MLAGAN (Brudno et al. 2003), TBA (Blanchette et al. 2004), and the recently developed PECAN (B. Paten and B.E. Pecan, in prep.). The multisequence alignments are represented using the human sequence as a reference coordinate system in which non-human sequences are manipulated to be in a “humanized” order and orientation; as such, two nucleotides in a non-human sequence need not be in the same orientation in which they natively reside (Fig. 2). All human bases are present in the resulting alignments, and have at most one aligned nucleotide from each other species. Thus, duplications in non-human lineages were resolved so that a single orthologous copy is aligned; in contrast, non-human bases may be aligned more than once if they are orthologous to multiple human positions as a result of a duplication in the human genome (note that MAVID alignments enforced a strict one-to-one orthology; see below). Although all human bases are present in the final alignments, positions in the non-human sequences may have been omitted. For example, sequence corresponding to large species-specific insertions or human deletions might have been removed due to a lack of orthology with the human sequence. It is thus important to keep in mind that these alignments were built in a “pipeline” fashion, in which nucleotide-level global alignment is only one step.
Figure 2.
“Human-centric” approach for constructing multisequence alignments. The human sequence (middle) is aligned to two other species’ sequences (top and bottom). In the final alignment (right), nucleotides from the other species need not have retained their original order and orientation; they may, for example, have been subjected to inversions (top blue) or duplications (bottom green). Non-human duplications need to be resolved (top magenta), so that each position in the human sequence is aligned to at most one position in any other species’ sequence.
Equally important as generating alignments is defining metrics for alignment quality. Unlike protein alignments, where structural information can be used to generate reference alignments (Van Walle et al. 2005), or the prediction of transcription factor-binding sites, where experimental data can be used to define bound and unbound sites (Tompa et al. 2005), no such “gold standards” exist for genomic sequence alignments. The challenge is to define measurements for alignment specificity (i.e., fraction of orthology predictions that are correct) and sensitivity (i.e., fraction of all truly orthologous relationships that are correctly predicted). Since multisequence alignments are used to generate and test evolutionary hypotheses, measurements of alignment quality should be tied to the quality of the evolutionary inferences derived from them; subsequently, we compare the sets of alignments in this manner. It is worth noting that in many instances, the “true” evolutionary history of a particular nucleotide or region is unknown (and perhaps unknowable), and in many of the concomitant comparisons no definitive assessment of “better” or “worse” can be generated. Whenever such assertions can be made (such as with respect to alignment coverage of protein-coding sequences as a measure of sensitivity), however, we attempt to do so.
Alignment comparison—Region level
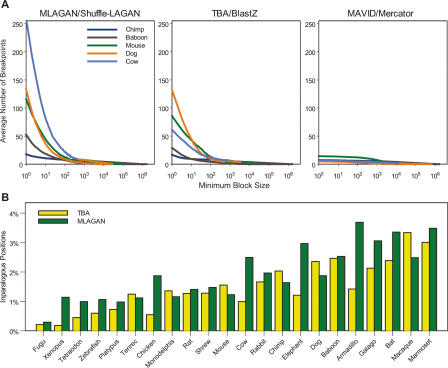
The alignments allow inferences to be made about large-scale evolutionary events that have shaped the ENCODE loci in these mammalian genomes. For 59.2% of ENCODE region–species pairs, a single segment in the query species genome was predicted to contain sequence orthologous to sequence in the human region, indicating that these regions have been largely undisturbed throughout mammalian evolution. However, many small-scale rearrangements were detected (“conserved synteny” of a large genomic region does not imply colinearity of all nucleotides within that region). The number of rearrangement breakpoints within a given region was highly dependent on the size of alignment blocks considered. Figure 3A summarizes the number of rearrangement breakpoints determined by MLAGAN/Shuffle-LAGAN, TBA/BlastZ, and MAVID/Mercator as the minimum block size was varied for five species (see Methods). Blocks of length <100 base pairs (bp) were found to cause the vast majority of the breakpoints, consistent with both higher probabilities of occurrence and an increase in the probability of spurious alignments. Mercator/MAVID predicted very few small-scale rearrangements, while MLAGAN predicted the largest number, particularly with respect to cow. However, the three methods were largely in agreement for rearranged blocks longer than 100 bp. Blocks of at least 1 kb numbered from 70 in marmoset to 101 in rat, as determined in the MLAGAN/Shuffle-LAGAN alignments. For these blocks, the median block lengths were roughly 300 kb and 14 kb, respectively.
Figure 3.
Rearrangements and duplications inferred by the alignments. (A) Number of rearrangement breakpoints in the ENCODE regions as a function of minimum block size, determined by three alignment methods. For each species, the average number of breakpoints over all regions (Y-axis) was calculated for all minimum block sizes (in base pairs; X-axis). The species shown are chimp (dark blue), baboon (brown), mouse (green), dog (orange), and cow (light blue). For each minimum block size, the number of breakpoints in a given region was determined after removing blocks in order of increasing size and joining consistent blocks until no block had size less than the minimum (see Methods). (B) Duplicated human nucleotide positions in the ENCODE regions. The fraction of ENCODE positions that are inparalogous to one another relative to a given species is plotted for each species, as determined by TBA (yellow) and MLAGAN (green). Colobus Monkey, Dusky Titi, Mouse Lemur, and Owl Monkey are not shown because sequence from these species was only obtained for one region (ENm001).
The TBA and MLAGAN alignments allowed multiple human positions to be aligned to a single position in a query species. In such cases, the alignment states that both human positions are orthologous to the query position, and are paralogous to each other as a result of a duplication event in the human lineage since its last common ancestor with the query species. These positions are said to be “inparalogous,” a relationship that depends on a query species (Sonnhammer and Koonin 2002). Figure 3B shows the fraction of ENCODE human positions that were determined to be inparalogous relative to each query species. For 16 of 22 query species that had sequence for all ENCODE regions, MLAGAN predicted more such positions than TBA. For six species, MLAGAN predicted more than twice as many positions as TBA. The fraction of inparalogous positions varied greatly over the different ENCODE regions. For example, >30% of positions in region ENr233 were predicted to be human-specific duplicates relative to marmoset by both aligners, compared to <5% of positions in region ENm004 relative to all species.
Alignment comparison—Nucleotide level
We also sought to compare our alignments at the nucleotide level, as this is the level at which many downstream applications operate. We find that the level of agreement between alignments varies significantly between species, with agreement much higher when comparing alignments of primates versus those of more distant species (Supplemental Table S2). In general, agreement between the different alignments is influenced significantly by the total coverage; for example, MAVID aligns 27.4% of human bases to an armadillo nucleotide, versus 42.4%, 41.2%, and 40.1% for MLAGAN, PECAN, and TBA, respectively; and thus the maximum possible agreement between all the alignments is 27.4%. We find that 17.5% of all human nucleotides are aligned to the same armadillo nucleotide by all four alignments, and 66.1% of all human bases are identically aligned if we consider gapped columns (i.e., columns in which a human nucleotide is predicted not to have an orthologous nucleotide in the armadillo sequence). We conclude that there are substantial variations between the nucleotide-level orthology predictions made by the four alignments, although a significant majority of all human nucleotides are aligned identically between human and a given non-human sequence.
An important use of multisequence alignments is to characterize rates of nucleotide substitution in predominantly neutral DNA. Such estimates are not only important to understand genome evolution, but may also illustrate differences between alignments at the nucleotide level. We therefore estimated rates of evolution in ancestral repeats (AR) in our alignments (Supplemental Table S3; also see Methods). Eutherian ARs are fragments of mobile elements believed to have inserted into the common ancestor of all placental mammals and been retained since then. Assuming these elements are largely not functional, they are free to evolve in the absence of selection (with notable exceptions: Nekrutenko and Li 2001; Jordan et al. 2003; Silva et al. 2003; Cooper et al. 2005; Kamal et al. 2006) and thus constitute a good model for neutral evolution in mammalian genomics (International Mouse Genome Sequencing Consortium 2002; Ellegren et al. 2003; Hardison et al. 2003; Rat Genome Sequencing Project Consortium 2004; Yang et al. 2004). First, we note that rates of evolution in ARs are similar to, but higher than, rates estimated from fourfold degenerate sites within proteins (average increases of 2%–13%, depending on the alignment and region). This may indicate weak purifying selection on synonymous sites (e.g., Kimchi-Sarfaty et al. 2007; Komar 2007), but may also result from an increased proportion of errors in alignment of ARs, which are more difficult to align. We observe considerable variation both between genomic regions and between alignments. Regional rate variation has been well documented for mammalian genomes (International Mouse Genome Sequencing Consortium 2002; Rat Genome Sequencing Project Consortium 2004), and we find similar results here, with a standard deviation (averaged over the four alignments) of 0.15 substitutions per site (∼3.7% of the neutral rate) among the 44 ENCODE loci. Furthermore, while this regional variation is highly correlated among the alignment sets (average pairwise R2 of ∼0.62), we find that the standard deviation between the four alignments in a given locus is 0.2 substitutions per site, roughly similar to the level resulting from regional variation. Thus, while relative rate fluctuations between regions are correlated with legitimate fluctuations in local rates of nucleotide substitution, interpretation of absolute rates of nucleotide substitution for any given region must be done cautiously, with appropriate accommodation of technical error for any downstream application that requires such estimates. The “true” neutral rate for any given region of the human genome is thus only estimable given some nontrivial technical uncertainty.
Assessing alignment coverage
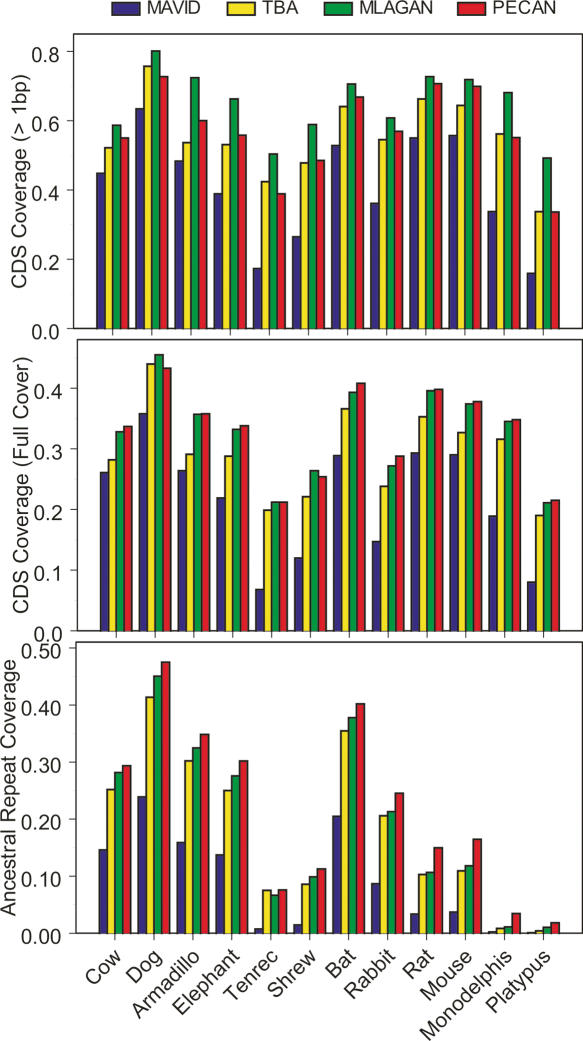
As a surrogate for sensitivity, we determined the coverage of annotated protein-coding sequences in each of our alignments. Since coding exons are regions of the human genome that are largely ancient and likely to be shared among all of the lineages analyzed here, they represent a set of nucleotides heavily enriched for “true positive” (i.e., actually orthologous) positions. We expect that alignment “coverage,” defined by the number of human coding bases aligning to a given non-human species, will be highly correlated with alignment sensitivity. Note that the simple existence of an alignment does not imply that an alignment is correct (“correctness” is addressed below), but we assume that sensitivity will be proportional to the total amount of aligned sequence. We find that coverage of coding exons varies considerably among the different alignments, especially when analyzing alignments between humans and more distant species (i.e., non-primates). When counting the number of coding exons with at least one base pair aligned to a base in the mouse genome, for example, coverage ranges from 55% in MAVID to 72% in MLAGAN (Fig. 4, top panel), with TBA and PECAN showing intermediate values. Alternatively, when looking at only those coding exons that are fully covered (i.e., no gaps), these values range from 29% in MAVID to 38% in PECAN (Fig. 4, middle panel). PECAN and MLAGAN exhibit the highest values by these measures and are similar for most species.
Figure 4.
Alignment coverage of coding exons and ancestral repeats. For a representative group of mammalian species (X-axis), the fraction of human coding exons covered by at least 1 base (top panel) or completely covered (i.e., no gaps, middle panel) are shown for the MAVID (blue), TBA (yellow), MLAGAN (green), and PECAN (red) alignments. For the same set of species, we also show the percentage of all human “ancestral repeat” bases (out of a total of ∼5.8 million) that are aligned to a nucleotide within a mobile element of the same class and family. Note that absolute coverage levels should be interpreted cautiously, as they reflect both phylogenetic signal (i.e., insertions and deletions of DNA between human and the query species) and sequence completeness.
However, quantifying rates of evolution in neutral DNA is dependent on our ability to align orthologous regions that are more dissimilar than typical coding exons. ARs provide a more realistic measure in this regard. To develop a sensitivity measure on the basis of AR alignments, we first independently identified repeats in each aligned species’ sequence using RepeatMasker (http://www.repeatmasker.org). Then, for each alignment, we quantified the number of human AR bases (filtered from the RepeatMasker output as previously described, Margulies et al. 2003) that are aligned to a base within an element of the same class and family in each of the non-human sequences. As above for coding sequence, in principle, these alignments are not necessarily correct. However, it is reasonable to assume that the total amount of aligned mobile element fragments (classified as “ancestral” within humans and independently identified to be of the same class and family in the non-human sequence) is proportional to actual sensitivity. As for coding exons, we find considerable variability between the alignments. In this case, however, PECAN alignments are clearly the most sensitive. For example, >47% of the ∼5.8 million AR bases in the human are aligned to a dog nucleotide by PECAN, while only 24% are aligned by MAVID (Fig. 4, bottom panel). PECAN has an average coverage increase of 2.4%, 3.8%, and 12.5% over MLAGAN, TBA, and MAVID, respectively. Keeping in mind that there are ∼5.8 million AR bases in the human ENCODE regions, we find that there are substantial differences in sensitivity to neutrally evolving DNA among these alignments.
Assessing alignment correctness
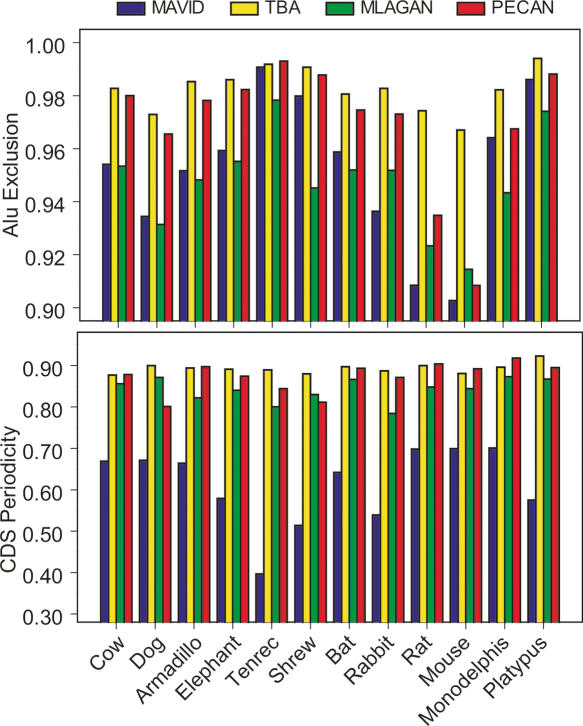
We also sought to estimate the specificity of our alignments, since the simple presence of an alignment does not imply correctness. Because we do not know with certainty what should and should not align, we used two alternate measures as surrogates for alignment specificity. The first approach uses our knowledge of mobile element fragments to measure “false-positive” alignments; since Alu element activity is phylogenetically restricted to primates, alignment of human Alu elements to any non-primate mammalian sequence is a false orthology prediction. Furthermore, since Alus are abundant in the human genome and are also SINEs, they can potentially generate many similar matches between human and even distantly related mammalian species. In this regard, they are a direct and stringent measurement of incorrect orthology predictions. On the basis of the ∼3.8 million Alu bases in the ENCODE targets, we observe that TBA is the most “specific” aligner (Fig. 5, top panel), followed by PECAN, MAVID, and MLAGAN, with an average decrease in Alu exclusion rate of 1.3%, 3.0%, and 3.5%, respectively. As above for the AR analysis, we note that while these numbers appear small, they are substantial, with a 1% difference amounting to nearly 40,000 human nucleotides that are differentially (and incorrectly in this case) aligned.
Figure 5.
Alignment “correctness” as measured by Alu exclusion and periodicity of substitutions in coding exons. For a group of non-primate mammals, the fraction of human Alu bases (out of a total of ∼3.8 million) that are not aligned (i.e., gapped) is shown (top panel). A score of 1 would correlate with complete exclusion of all Alus, as would be the case in alignments with no false-positive orthology predictions. We also show the fraction of human coding exons that show a triplet periodicity in substitutions in the pairwise alignment between human and each query species (see Methods). Note that this is purely a relative measure, since we exclude exons that are completely gapped in at least one alignment, or fail to show periodicity in at least one alignment.
The second measure exploits our knowledge of coding sequence, where we expect that correct alignments will exhibit periodicity in the pattern of inferred nucleotide substitutions due to the enrichment of synonymous sites at codon third positions. Thus, we quantified the levels of periodicity in the coding exon alignments as a proxy measure for their nucleotide-level specificity (see Methods). Furthermore, to eliminate those coding exons that are missing in a particular species or not periodic (i.e., due to a false prediction or too few synonymous changes, as often occurs between human and chimp sequence), we only include those coding exons that exhibit periodicity in at least one alignment and some level of nucleotide coverage in all alignments. These specificity measures are therefore not confounded by differences in coverage (see above) or false coding exons. TBA and PECAN exhibit the highest levels of codon periodicity (and thus inferred specificity), with TBA being on average slightly higher (1.4%) than PECAN (Fig. 5, bottom panel). In contrast, MLAGAN is moderately weaker than TBA (average decrease of 4.4%), while the MAVID alignments have the lowest levels of periodicity (average decrease from TBA of ∼21.3%).
Explaining alignment discrepancies
We observed substantial differences between the four alignments; determining the sources for these differences is difficult, but a few conclusions can be drawn. For example, MAVID’s lower coverage estimates likely result from the strict one-to-one orthology requirement, which eliminates human-specific duplications. The discrepancy in coverage between MAVID and the other aligners that is due to this restriction can be upper-bounded by the amount of inparalogous human sequence, as predicted by the other aligners. Up to 4% of human bases in the ENCODE regions were predicted to be inparalogous, depending on the query species (Fig. 3B). These bases represent up to roughly 10% of those covered by the aligners. Furthermore, some of the randomly picked ENCODE regions have very low gene content, which may affect the sensitivity of Mercator’s (the region-level orthology prediction algorithm used by MAVID; see Methods) primarily exon-based orthology detection process. On the other hand, MLAGAN and PECAN coverage estimates generally appear higher. The Shuffle-LAGAN “humanization” step is somewhat lenient and rearranges the original sequences with a rather coarse resolution; nonorthologous pieces may be kept if they fall between long stretches of orthologous sequences, for example, and rearrangement boundaries are generally approximate. The reduced specificity seen for MLAGAN may result from this leniency in combination with the fact that MLAGAN preserves all of the input sequence in its output, resulting in alignments that aggressively span nonorthologous regions. Conversely, PECAN, which uses the same Shuffle-LAGAN humanization step but showed higher specificity levels than the MLAGAN alignments, does not force an alignment along the entire input. In addition, PECAN uses “consistency,” which has been shown to give marked improvements in protein alignments (Notredame et al. 2000; Do et al. 2005) but is a novel addition to genomic sequence alignments. The TBA alignments generally have the highest specificity, and are the most effective at ignoring highly similar, but nonorthologous, alignments resulting from Alu elements. Since the blocks produced by TBA emerge from local alignments, they usually have tight boundaries and are fairly compact, and can avoid the long insertions that are harder to dismiss by the three global alignment techniques.
Identification and measurement of constraint in the ENCODE regions
Our multisequence alignments covered more of the human genome at greater evolutionary depth than previous studies, which have either used whole-genome sequences from only a few species (International Mouse Genome Sequencing Consortium 2002; Cooper et al. 2004; Rat Genome Sequencing Project Consortium 2004; Lindblad-Toh et al. 2005; Siepel et al. 2005) or included many species’ sequences but were limited to single loci <2 Mb in size (Boffelli et al. 2003; Margulies et al. 2003; Cooper et al. 2005). These alignments thus provided a unique opportunity to systematically identify constrained sequences for a large segment of the human genome. Evolutionary constraint was detected using three distinct methods: phastCons, which uses a phylo-HMM (Siepel et al. 2005); GERP, which exploits single-site maximum likelihood rate estimation (Cooper et al. 2005); and binCons, which quantifies pairwise similarities using a binomial distribution from sliding windows (Margulies et al. 2003). Details for each of these methods are provided in their respective citations, and additional details about the use of each algorithm are also available at the UCSC Genome Browser (http://genome.ucsc.edu; Kent et al. 2002; Karolchik et al. 2003) and in the Methods section. Each method analyzed the same human-referenced multisequence alignments (performed separately with three of the alignments; note that PECAN alignments were not included because of its recent development) to generate scores and element predictions across all ENCODE regions. We further equalized constraint detection thresholds using an empirically generated, standardized “null” alignment for each ENCODE-region alignment. In all cases, sequences deemed as constrained are significant at a “relative” false-positive rate of <5% (see Methods).
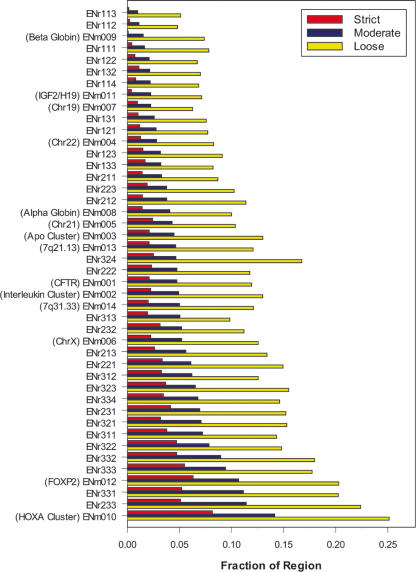
In addition to nine independent sets of constrained sequences (three methods analyzing three alignments), we also generated constraint annotations that integrate these data. Three annotation sets emerged from this integration: a “loose” set, defined by the union of all bases predicted as constrained for any method on any alignment; a “moderate” set, defined by the union of all bases predicted as constrained for at least two methods on at least two alignments; and a “strict” set, defined by the intersection of all three methods on all three alignments. Overall, the loose, moderate, and strict data sets identify 11.8%, 4.9%, and 2.4% of the ENCODE regions, respectively. We observe considerable variation among the different regions in the total fraction of constrained sequence, likely reflecting the genomic diversity of the 44 ENCODE regions and the biology encoded therein (Fig. 6). We find that both sensitivity and the level of error rise, as expected, from the strict to loose sets. For example, treating coding exons as a set of true positives, the strict set has a sensitivity of 44%, which increases to 69% and 88% (measured per nucleotide) in the moderate and loose sets, respectively. Conversely, using mammalian ancestral repeats as a surrogate of neutrally evolving sequences (i.e., “true negatives”) (International Mouse Genome Sequencing Consortium 2002; Margulies et al. 2003), we estimate that the false-discovery rate increases from the strict to moderate to loose sets as 0.1%, 0.5%, and 4.8% (measured per nucleotide) of constrained bases, respectively. This likely indicates a decrease in specificity concomitant with the increase in sensitivity in the three sets.
Figure 6.
Constrained bases in each ENCODE region. For each ENCODE region (Y-axis), the percentage of nucleotides found to be under evolutionary constraint in the strict (red), moderate (blue), and loose sets (yellow) is shown (X-axis). The 44 regions are ranked from top to bottom by the fraction of bases in the moderate (green) annotations. For all the manually picked regions, their biological significance is noted in parentheses.
Explaining constraint prediction discrepancies
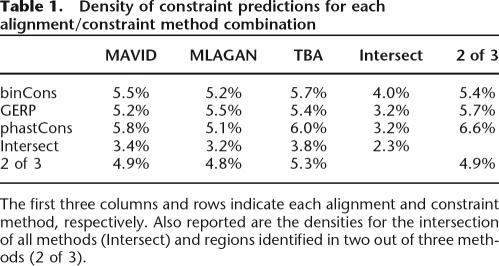
While our false-discovery rate standardization accounts for a significant fraction of aligner-specific behaviors in neutrally evolving DNA (see Methods), alignment discrepancies are clearly contributors to differences in constraint predictions. Even within an alignment, however, we observe that the methods used for inference of constraint make distinct predictions, with approximately one-third of the predicted constrained bases being discrepantly predicted by at least one method (Table 1). Manual analyses reveal that one of the most informative classes of such differences reveals a dichotomy between the high-resolution, phylogenetic methods (phastCons, GERP) and the more heuristic binCons approach, which uses a 25-bp sliding window. While binCons is incapable of detecting many of the smaller elements identified by the phylogenetic approaches (phastCons and GERP elements have median sizes of 15–20 bases), we found that binCons is less sensitive to spurious alignments resulting from short regions of high similarity between distant species. Another important difference arises from the handling of regions of the alignment that exhibit low neutral diversity, such as might be seen in an alignment of only a handful of primate sequences. While GERP explicitly ignored columns with <0.5 substitutions per neutral site, phastCons and binCons did not and may occasionally annotate constrained sequences within these regions (which of statistical necessity are generally long elements that therefore inflate the level of disagreement between methods). We also note that a major fraction of the discrepancies among the nine annotation sets results from the precise definition of constrained sequence boundaries rather than the presence of constraint per se; ∼80% of constrained sequence regions (as opposed to nucleotides) overlap by at least one base in the intersection of all nine annotations, in contrast with 60%–70% of all nucleotides (analogous to the distinction in element overlaps made in Supplemental Fig. S1).
Table 1.
Density of constraint predictions for each alignment/constraint method combination
The first three columns and rows indicate each alignment and constraint method, respectively. Also reported are the densities for the intersection of all methods (Intersect) and regions identified in two out of three methods (2 of 3).
Comparative analyses of ENCODE experimental annotations
Elsewhere we report on the extent of correlation between the moderate set of constrained sequences and each class of experimentally annotated element (The ENCODE Project Consortium 2007). We noted that 40% of the moderate constrained sequence represents protein-coding exons and their associated untranslated regions, and an additional 20% of the constrained sequence overlaps other experimentally identified functional regions, leaving 40% of the constrained sequence without any ENCODE-generated experimental annotation.
Constrained sequences not overlapping experimental annotations
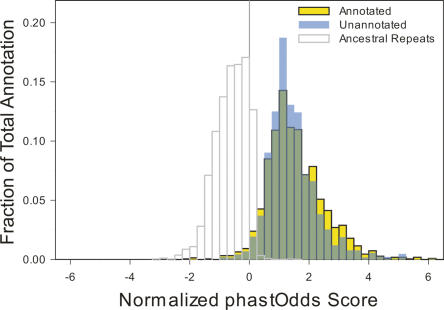
Two independent lines of evidence suggest a functional role for these remaining constrained sequences, despite a lack of experimental annotation. First, these sequences are not enriched for weakly constrained bases (Fig. 7), as would be expected if our analyses yielded too many false-positive results (i.e., neutral sequence falsely identified as constrained). In fact, the region of greatest evolutionary constraint (based on length and per-position alignment score, residing within an intron of FOXP2) as well as 16 of the top 50 constrained sequences do not overlap an experimental annotation (Supplemental Table S2). Second, analyses of human polymorphisms show that constrained sequences (both the annotations specifically described here and others in general) correlate with reduced heterozygosity and derived allele frequencies, indicative of recent purifying selection in humans (Drake et al. 2006; The ENCODE Project Consortium 2007). Thus, constrained sequences are neither mutational cold spots nor do they appear to have lost function recently in human evolution.
Figure 7.
Annotated versus unannotated constrained sequences. For each block of constrained sequence, a score based on the log-likelihood of observing such a sequence under a model of constrained versus neutral evolution was computed using the phastOdds program (Siepel et al. 2005). These values were divided by the length of each block to compute a normalized per-base log-likelihood that reflects constraint intensity (X-axis). These values were plotted as a frequency histogram (Y-axis) for the blocks of constrained sequences that do (yellow) or do not (blue) overlap an experimental annotation. The distributions largely overlap (green), even at the extreme positive end in which highly constrained sequences reside. For comparison, the distribution for ancestral repeat sequences is shown as a representation of largely neutral DNA.
It is also unlikely that the unannotated constrained sequences primarily encode unknown proteins, as we observe little overlap with predictions of coding potential analyzed from multisequence alignments (Siepel and Haussler 2004a; see Supplemental Material). These sequences therefore likely reflect functional elements that were not detected by the assays used to date by the ENCODE project. For example, functional elements involved in embryonic development might have escaped detection due to an emphasis on using cells grown in tissue culture. Indeed, recent experiments show that many highly constrained pan-vertebrate sequences are developmental enhancers (Nobrega et al. 2003; Woolfe et al. 2005; Pennacchio et al. 2006), with functions that are perhaps only detectable in the context of the developing organism. In addition, while the array of functions examined by ENCODE is broad, certain known classes (e.g., enhancer and silencer elements) have only been assayed indirectly (e.g., by DNase I hypersensitivity or DNA–protein binding) or not at all. Finally, it is almost certain that as-yet-unknown types of function are conferred by some of the unannotated constrained sequences.
We thus conclude that many (at least 40%; this number is likely to be larger given the limited resolution for some experimental assays) constrained sequences have received no purported functional annotation to date, despite considerable experimental effort by the ENCODE project. Indeed, we show that there are many regions of the human genome that likely have functions critical to mammalian biology but that have not been detected by the experimental assays employed thus far.
Assessing evolutionary constraints on experimentally annotated sequences
While the association of constrained sequence and genome function is well established (Hardison 2000), the converse relationship—i.e., the extent to which the sequences of functional elements are under evolutionary constraint—has not been explored in detail. Elsewhere, we examine the overlap between constrained sequences and each class of experimental annotation (The ENCODE Project Consortium 2007). We noted that most experimentally identified elements showed a significant level of overlap with constrained sequences, but there was a wide variation in the amount of that overlap. While coding exons appeared to have the majority of their bases constrained, noncoding functional elements overlap considerably less (although still statistically significant), with some subclasses failing to exhibit a nonrandom level of overlap. Since the experimental assays employed by the ENCODE project to date appear to be generally reliable and have tolerably low false-positive rates (The ENCODE Project Consortium 2007), we explored a number of explanations for the relative paucity of constraint within experimentally annotated noncoding elements. We note that these are not mutually exclusive.
First, some fraction of bases within experimentally annotated sequence is unlikely to be part of the corresponding functional elements because of resolution limitations of the experimental assay. An experimentally annotated element may therefore be a mixture of functional and nonfunctional sequence, and thus contain significant amounts of unconstrained sequence. Elsewhere, we showed that most such annotated elements have several “islands” of constrained sequences within them, with many experimentally annotated elements overlapping constrained sequence more significantly at the annotation level than at the base level (The ENCODE Project Consortium 2007; also see Supplemental Fig. S1). For example, while non-protein-coding transcripts of unknown function (TUFs) (see Supplemental Box S1) exhibit relatively weak evidence for evolutionary constraint on average over all of their bases (Supplemental Fig. S1, yellow bars, column 4), they are significantly enriched for annotations that overlap at least some amount of constrained sequence (Supplemental Fig. S1, blue bars, column 4).
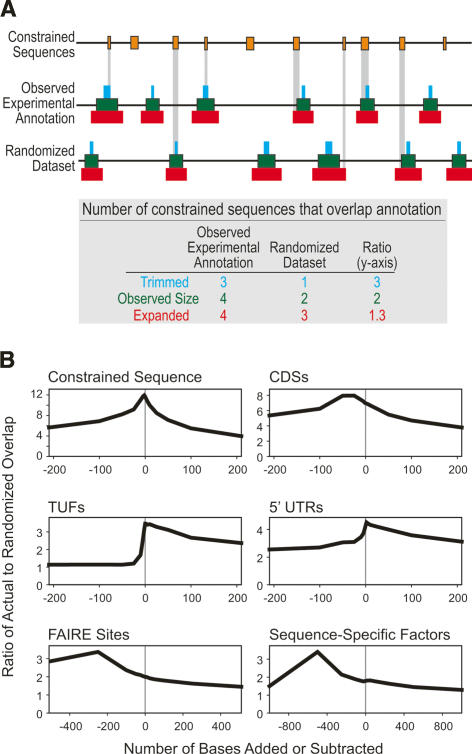
To test the possibility that this “island effect” could result from the relatively low resolution of the experimental methods used to establish these annotations, we asked whether the overlap between constrained sequences and experimentally annotated elements could be improved by “trimming” the latter from either end, leveraging the hypothesis that the functional subregions would, on average, be toward the center of these annotated regions. We find that this is, indeed, the case for certain experimental annotations (Fig. 8; Supplemental Fig. S2), particularly so for assays that detect protein–DNA binding of sequence-specific transcription factors. Thus, it is plausible that the functional portion of these experimentally annotated elements may be only a handful of bases long and correspond more closely to the constrained sequence than the extent of the experimental annotation suggests. This is in contrast to annotations with precise borders (such as UTRs), where it is clear that only portions of the functional element are evolutionarily constrained (Fig. 8).
Figure 8.
Significance of constrained sequence overlapping various experimental annotations. We quantified the ratio of “observed” to “randomized” overlaps between constrained sequences and experimental annotations (see Supplemental Box S1), after adding and subtracting a given number of bases to the ends of each experimentally identified annotation. Randomized data sets were generated by randomizing the start positions of features within each ENCODE target, preserving the length distribution of each feature set and any target-specific regional effects. (A) This analysis is illustrated for a hypothetical set of annotations. (Orange bars) The positions of constrained sequences; trimmed (blue bars), observed (green bars), and expanded (red) experimental annotations. (Vertical gray bars) Regions of overlap between constrained sequences and experimental annotations. A table summarizing the overlaps among the different scenarios is provided below the diagram. For this hypothetical example, note how the ratio of overlap between the observed and randomized data sets increases as the experimental annotations are trimmed, indicating an enrichment of constrained sequence in the trimmed annotations. (B) This analysis for several experimentally identified elements is plotted, where the X-axis indicates the amount of trimmed (negative) or expanded (positive) sequence on each element, and the Y-axis indicates the ratio of observed-to-randomized overlap (scale varies between plots). Note that CDSs exhibit a slight enrichment after deletion of a small number of bases at either end, but are very similar to what is expected given the theoretically optimal self–self overlap (“Constrained Sequence”), where we know that trimming should not increase specificity. For many annotations (e.g., “TUFs” and “5′-UTRs”) (see Supplemental Box S1), such enrichment quickly drops off as the annotations are expanded or trimmed. However, some annotations, such as “FAIRE Sites” and “Sequence-Specific Factors,” exhibit a clear improvement in overlap after trimming substantial amounts of sequence from either end (250 and 500 bases for “FAIRE Sites” and “Sequence-Specific Factors,” respectively). Similar plots for all experimental annotations are available as Supplemental Figure S4.
Second, analyses of evolutionary constraint fail to detect functional constraint that is not reflected in primary sequence conservation (e.g., Ludwig et al. 2000). For example, we note that 60% of the detected transcriptional promoters fail to overlap any constrained sequence whatsoever. Promoters can be detected with several orthogonal and highly reliable assays, and their locations are often conserved between humans and mice (Trinklein et al. 2004). At the very least, the core promoter of ∼50 bases within the majority of these annotations must be functional sequence, yet in many cases it is not under detectable evolutionary constraint, suggesting that characteristics other than primary sequence are important for conferring function.
A third possibility that could explain these unconstrained experimental annotations is that they are only functional within a subset of the mammalian phylogeny, such as primates. This explanation is consistent with the identification of purifying selection against human polymorphisms even after excluding pan-mammalian constrained sequences (The ENCODE Project Consortium 2007). By definition, these elements are either completely absent or have evolved swiftly in some lineages, significantly reducing the chance that we would identify them as being under constraint (Stone et al. 2005). To address the possibility of primate-specific constraint (other patterns of constraint gains and losses are also possible, see Supplemental Material), we used a novel algorithm to detect lineage-specific constrained sequences (Siepel et al. 2006). Although our power to detect primate-specific constrained sequences is relatively weak, especially if they are short or have become constrained very recently, we found 94 such sequences (median length 164 bases; range 69–615 bases), some of which are quite striking (Supplemental Figs. S3 and S4). These results suggest that, while most constrained sequences are shared among mammals, there are some that are specific to primates, and these sequences account for a small portion of the apparently unconstrained experimental annotations. As more primate sequence data become available, our power to detect such regions in the genome will improve.
Fourth, it is conceivable that there are genomic regions that reproducibly appear to be “functional” by an experimental assay (e.g., transcription-factor-binding sites or RNA transcription units) that are of no consequence to the organism, and thus are “invisible” to natural selection. Such elements might exist in the genome at a steady-state frequency dictated by the sequence specificity of the function and the rate of neutral turnover of genomic sequence throughout evolution. Short and degenerate elements, for example, could emerge often in a large genome and be quite abundant, while larger and more complex elements would be rare. This is consistent with our observation that many annotated sites of protein–DNA interaction, in many cases thought to be dictated (at least in part) by short and degenerate motifs, do not overlap any constrained sequence, while nearly all coding exons, which would emerge at random extremely rarely, are under constraint. Thus, it is plausible that many biochemically functional but biologically inert elements exist in the human genome and provide evolutionary potential from which new functions may arise.
It is interesting to note that a sizable fraction of each class of experimental annotation is not evolutionarily constrained by the methods used here. If the corresponding elements are, indeed, important for human biology, then it becomes important to establish how their function is encoded in the absence of evolutionary constraint at the primary sequence level (The ENCODE Project Consortium 2007). Alternatively, if some of the annotations reflect functional elements that are of no consequence to the organism, then our definition of biological function will require refinement not unlike the expansion of our understanding of evolution that came about with the development of the neutral theory (Kimura 1983).
Summary
Comparative analyses necessitating accurate alignments of multiple, large genomic sequences are now crucial parts of many biological analyses. Here, we describe one of the largest comparative genomic challenges documented, generating and analyzing alignments of 30 Mb of human sequence to 27 other vertebrate species. This field remains an active area of research and development, as the four prominent alignment tools that we have used show significant levels of discrepancies. It is impossible at the moment to make definitive qualitative statements concerning the alignment tools, as there are distinct trade-offs in their behaviors; for example, alignments produced by MLAGAN exhibit global increases in alignment coverage when compared to TBA and MAVID, but this includes increases in incorrect alignments (Figs. 4, 5). PECAN may be achieving a better compromise in this regard, with better specificity than MLAGAN but similar levels of sensitivity. Thus, these alignments offer distinct specificity/sensitivity trade-offs that are reflected in changes in the inferred rates of both indel and substitution events (Supplemental Table S3). Other factors may also influence alignment choice, such as the basic modeling assumption used concerning the types of orthology that are to be predicted (Dewey and Pachter 2005). Additionally, PECAN is at the moment only a global aligner and therefore incapable of handling rearranged sequences. Thus, choice of alignment method and goals depends on many factors and ultimately should be dictated by the downstream application employed. Furthermore, all downstream applications should be cognizant of such technical discrepancies, and account for uncertainty whenever resulting parameters, such as rates of nucleotide substitution in neutral sites, are utilized. Similar qualitative caveats can be made with respect to inferring the locations of evolutionarily constrained sequences in the human genome. For example, one trade-off that we identified is that the sliding-window approach employed by binCons, while being less sensitive to many of the smaller elements that phastCons and GERP identify with confidence, is less prone to annotate alignment artifacts resulting from isolated and short but highly similar sequence matches between humans and distantly related species. We find that there is significant room for improvement in the computational analyses of diverse mammalian sequences. A particularly pertinent area will be the standardization of benchmarks and, perhaps more importantly, concepts and definitions for both multisequence alignments and analyses of constrained sequences.
However, despite this uncertainty, we show that comparative sequence analyses are a critical component of efforts to systematically identify and characterize functional elements in the human genome. Our observation that 40% of all constrained sequences fail to overlap any ENCODE experimental annotation suggests that future efforts aimed at the comprehensive identification of genomic functional elements require a more diverse array of experimental approaches, and also lends support to incorporating medium-to-high-throughput model-organism experimentation. We also demonstrate that constraint analyses can be used to refine experimental annotations made with relatively low-resolution methods, and that such efforts can likely guide future experimental and computational analyses of these experimental data. Our studies have thus yielded both an important resource for comparative genomics and biological insights to guide future functional analyses of the entire human genome.
Methods
Data availability
Alignments and other annotations generated and used for the studies reported here are available at http://genome.ucsc.edu/ENCODE (click on the “Downloads” link in the blue column along the left side of the page). They are also displayed in the UCSC Genome Browser under the “ENCODE Comparative Genomics” set of tracks. PECAN alignments are available at http://www.ebi.ac.uk/~bjp/pecan/encode_sept_pecan_mfas_proj.tar.bz2. All experimental annotations were obtained from publicly available ENCODE project data (The ENCODE Project Consortium 2007); a bulk download of these data is available at http://www.nisc.nih.gov/data.
ENCODE genomic sequence data
The ENCODE regions represent a mix of manually and randomly selected targets, with details available at http://genome.ucsc.edu/ENCODE/regions.html (Thomas et al. 2006). In addition to the NISC BAC-based comparative grade sequence data generated specifically for this project, orthologous regions of the following whole-genome assemblies were used: chicken (CGSC_Feb._2004, galGal2); chimpanzee (NCBI_Build_1_v1, panTro1); dog (Broad_Institute_v._1.0, canFam1); Fugu (IMCB/JGI, fr1); macaque (BCM, rheMac1); monodelphis (Broad_Institute, monDom1); mouse (NCBI_Build_33, mm6); rat (Baylor_HGSC_v3.1, rn3); tetraodon (Genoscope_V7, tetNig1); Xenopus (JGI, xenTro1); and zebrafish (Sanger_Zv4, danRer2). For non-human vertebrate species with genome-wide assemblies, the identification of orthologous regions (i.e., large genomic intervals in each non-human sequence that are orthologous to each ENCODE target) was done with the liftOver program (Kent et al. 2003) and the Mercator program (Dewey 2006). These predictions were merged to produce a comprehensive sequence data set, which was then RepeatMasked. All analyses presented here use a sequence freeze dated September 2005 (labeled as SEP-2005).
Alignments
TBA/BLASTZ
The Threaded Blockset Aligner (TBA) (Blanchette et al. 2004) was used to generate multisequence alignments as follows. First, combinatorial pairwise alignments were generated with BLASTZ (Schwartz et al. 2003) using the following command-line parameters: Y=3400 H=2000. For mammalian-sequence comparisons, we additionally added B=2 C=0. For all other comparisons (except tetraodon and Fugu, which were treated as a mammalian comparison), we instead used the HoxD55 alternate scoring matrix (Margulies et al. 2005a). Pairwise alignments that included the human reference sequence were permitted to include sequence from the other species to align to more than one position. The pairwise sequence alignments, along with a generally accepted tree topology (Murphy et al. 2001; Thomas et al. 2003; Margulies et al. 2005a), were used to generate the multisequence alignment, which was then projected onto the human sequence to remove alignment blocks that did not contain the human reference sequence.
MLAGAN/Shuffle-LAGAN
MLAGAN alignments were produced by a pipeline specifically designed for ENCODE. First, WU-BLAST (W. Gish, 1996–2004; http://blast.wustl.edu) was used to find local similarities (anchors) between the human sequence and the sequence of every other species. Then, Shuffle-LAGAN was used to calculate the highest-scoring human-monotonic chain of these local similarities (according to a scoring scheme that penalized evolutionary rearrangements) and (with the help of a utility called SuperMap) produce a map of orthologous segments in increasing human coordinates. This map was used to “undo” the genomic rearrangements of the other sequence and convert it to a form that was directly alignable to the human sequence. The new humanized sequences, together with the human sequence, were then multiply aligned using MLAGAN. The resulting alignments were subsequently refined using MUSCLE (Edgar 2004), which processed small nonoverlapping alignment windows and realigned them in an iterative fashion, keeping the refined alignment if it had a better sum-of-pairs score than the original. Finally, a pairwise refinement round was performed, during which the pieces that had very low identity (in the induced pairwise alignments between human and each species) were removed from the alignment.
MAVID/Mercator
One set of alignments was created by a combination of Mercator (Dewey 2006), an orthology mapping program, and MAVID (Bray and Pachter 2004), a multiple global alignment program. For each ENCODE region, Mercator was first used to determine a small-scale collinear orthology map: sets of orthologous and collinear segments within the sequences given for that region. These sets of segments were determined in a symmetric fashion, without the use of the human sequence as a reference, and included sets that contained segments from only a subset of the input species. The orthology maps determined by Mercator were one to one, and thus had the property that a sequence position in any species was present in at most one segment set. Given the orthology maps, MAVID was then used to produce nucleotide-level multisequence alignments of each segment set. Only those segment sets that contained human sequence were retained for downstream analyses. Several programs were used to generate the input for Mercator. First, GENSCAN (Burge and Karlin 1997) was used to predict coding exons in all of the input sequences. The amino acid sequences corresponding to the coding exons were then compared to each other in an all-versus-all fashion with BLAT (Kent 2002). In order to detect noncoding rearrangements in the input sequences, MUMmer (Kurtz et al. 2004) was run to detect exact matches of length at least 20 bases between all pairs of genomes. The output of MUMmer was processed to produce a set of noncoding and nonoverlapping landmarks in each of the genomes. Mercator was then run with both coding and noncoding landmarks to determine an orthology map for each ENCODE region, as well as a set of alignment constraints within the segment sets based on matched landmarks. Nucleotide-level multisequence alignments of each segment set that obeyed the alignment constraints were constructed by MAVID. As part of its progressive multisequence alignment strategy, MAVID utilized a phylogenetic tree of the species with branch lengths determined from fourfold degenerate sites in all ENCODE regions.
PECAN
PECAN is a global alignment algorithm that has similarities with the Probcons (Do et al. 2005) and T-Coffee (Notredame et al. 2000) programs, but is adapted to deal with arbitrarily long sequences by a process of “sequence progressive” iteration (B. Paten and B.E. Pecan, in prep.). Sequences were first reordered in reference to the human sequence using Shuffle-LAGAN (see above). PECAN alignments were generated by running the program in three stages. First, the primate sequences were aligned, followed by the alignment of the placental mammals, and finally the more distant species were added. As PECAN can currently only align sequences, it was necessary to convert the intermediate products of the alignments (first the primate, then the placental mammal) to consensus ancestral sequences, for which we used Felsenstein’s algorithm (Felsenstein 1981). We avoided the issue of ancestral insertions and deletions by computing the consensus sequence based on the human sequence. Thus, all and only the bases present in the human sequence were included. This human-centric approach is sensible in light of ENCODE’s overall goals, the problems of partial sequence coverage in non-human species (which may be incorrectly inferred as gaps), and the general limited availability of algorithm implementations for accurately computing insertion and deletion histories. Prior to alignment, some training of PECAN’s pair hidden Markov models was performed using rearranged sequences from a subset of the ENCODE regions. Alignments have not been post-processed and largely represent the default parameters of the program (v0.6).
Inferring rearrangement events
For all ENCODE alignments, a pairwise alignment between human and each other species was extracted. The pairwise alignments were converted into a “threaded block set” format (Blanchette et al. 2004), where each block was required to be ungapped. Blocks that were species-specific or duplicated in human were removed, and neighboring collinear blocks were merged. For a given minimum block size, blocks were removed from the block set in order of increasing size, with adjacent collinear blocks merged after each removal stage, until all blocks had size greater than or equal to the minimum. The number of breakpoints was simply the number blocks remaining minus one. The number and length of blocks in a given alignment were calculated based on the blocks removed from the alignment in the process described, and not on all blocks present in the initial alignment.
Estimating rates of evolution at neutral sites
We first generated a tree on the basis of aligned fourfold degenerate sites within coding exons (taken from the longest transcript if there was more than one at a given locus). For any given non-human sequence, sites that fell within gaps or that were no longer synonymous (because of changes in the first two bases) were treated as missing data. Substitution rates were estimated by maximum likelihood with the PHAST package (Siepel and Haussler 2004b) and the XRATE package (Klosterman et al. 2006). A generally accepted tree topology for the analyzed species was used. The most general reversible substitution model (REV) was used, and no molecular clock was assumed. Further details are available as Supplemental Material.
Assessment of periodicity in coding exons
The periodicity assessment considers the mutation pattern between human and each non-human, “informant” species. We expect the pattern of mutations to be 3-periodic as a result of degeneracy in the third base of many codons. The assessment determines, for each CDS in the test set and for each species in the alignment, whether the alignment for the species, when paired with human, exhibits evidence of a 3-periodic substitution pattern either over the whole length of the CDS or in at least one 48-bp window. Evidence of periodicity is taken to be a “hi_spi” value of 3 or above. The “hi_spi” value is calculated as the ratio of the number of substitutions in frame “2,” divided by the number of substitutions in frame “0,” where frame “2” is identified as the frame with the highest number of substitutions. If the denominator is zero, it is changed to 1. The analyses are referenced to human annotations, thus gaps in the human sequence were removed from both species before the substitution counts were made. Because closely related species and some highly conserved genes have low levels of synonymous substitution, it is not possible to detect periodicity in all exons, and this will vary from species to species. Therefore, for each species, we count how many of the exons exhibit periodicity in at least one alignment method (n) and divide the raw counts by n to give the percent figures displayed in Figure 5.
Identification of constrained sequence
PhastCons
PhastCons parses a multisequence alignment into constrained and nonconstrained regions using a phylo-HMM. The phylo-HMM has two states, one for constrained regions and one for nonconstrained regions, and these states are associated with identical phylogenetic models, except that the branch lengths of the constrained phylogeny are scaled by a factor ρ (0 < ρ < 1). Constrained elements are predicted using the Viterbi algorithm. The predictions depend on several parameters, including the scaling parameter ρ, two parameters γ and ω that define the state-transition probabilities, and the parameters of the shared phylogenetic model (branch lengths and substitution rate matrix). We used a parameter estimation procedure slightly different from the one described in Siepel et al. (2005). Briefly, the nonconstrained model was estimated separately, from fourfold degenerate sites in coding regions (using the REV substitution model and the phyloFit program) (Siepel and Haussler 2004b), and other parameter estimates were conditioned on this model. We allowed phastCons to estimate the scaling parameter ρ by maximum likelihood, and adjusted the tuning parameters γ and ω to achieve the desired false discovery rate (see below).
GERP
Genomic evolutionary rate profiling (GERP) was run as described (Cooper et al. 2005). Briefly, each position of the human-projected multisequence alignment was evaluated independently, with a resulting estimate of both the observed (obtained with maximum likelihood under an HKY 85 model of nucleotide substitution) and expected (on the basis of a neutral tree; see above) substitution count obtained. All gapped species were eliminated from consideration at each column. Subsequently, each group of consecutive columns (with each column corresponding to one human nucleotide) in which the observed counts are smaller than the expected counts were identified as candidate constrained elements, with a merging tolerance of one unconstrained base. These candidates are summed according to the total deviation between observed and expected counts, with those meeting a certain threshold (using the target/alignment null model defined below) retained as legitimately constrained sequences.
BinCons
The binomial-based conservation approach was used essentially as described (Margulies et al. 2003, 2004). Briefly, the amount of sequence conservation is calculated for each overlapping 25-base window, where each species’ contribution is weighted by its corresponding neutral rate (as calculated above). In this fashion, more diverged sequences contribute more to the overall conservation score than do less diverged sequences. This is computed with a cumulative binomial distribution, with the neutral rate of each species representing the null distribution. For the calculations reported here, the exact amount of constrained sequence predicted by this method was tuned to the mean amount of predicted sequence by GERP and phastCons.
Standardizing false-discovery rates
Given the diversity of methodologies employed, we sought to simplify and standardize parameter choices among the methods as much as possible. The most crucial parameter is a threshold for differentiating regions that are truly constrained (i.e., subject to purifying selection) from those that appear constrained by chance. While ideally such a measure would use a set of true positives and true negatives, such elements are unavailable. Coding exons are generally true positives, for example, but are well known to be a nonrepresentative minority of the total space of constrained sequence. On the other hand, ancestral repeats are generally thought to evolve neutrally, but have been previously shown to include a nontrivial amount of constrained DNA (Silva et al. 2003; Cooper et al. 2005; Kamal et al. 2006). We therefore adopted an empirical approach to measure and standardize false-discovery rates that can also effectively cope with both region and alignment variation in the underlying neutral rates, similar to a previously described method (Cooper et al. 2005). For each ENCODE-region alignment, we generated a bootstrapped null or “neutral” alignment of 1 Mb in length. Specificity thresholds were then defined on the basis of the number of “constrained” bases identified in these bootstrapped alignments (false positives). Thresholds were set such that the number of false positives amounted to 5% of the total number of constrained bases identified in the true alignment (for example, if 50,000 bases are annotated to be constrained in the real alignment, 2500 would be annotated in the bootstrapped alignment).
Statistical evaluation of overlaps
We quantified the overlap between constrained sequences and each class of experimentally identified element at both the nucleotide and regional levels (Fig. 6); this same method was used elsewhere (The ENCODE Project Consortium 2007). We used an implementation of the Block Bootstrap (Künsch 1989) to model the variance in randomly expected levels of overlap. This empirical method agrees well with analytical variance computations (achievable for the nucleotide-level overlaps, but not for region-level overlaps), and also accounts for the intrinsic biases against repetitive sequence observed in both the constraint and experimental annotations (see the Supplemental Material). All confidence intervals shown for the overlap statistics are at 99.8% (Fig. 6).
Note added in proof
Recent reports resolve an early node (Murphy et al. 2007) and an internal node (Nishihara et al. 2006) of the boreoeutherian tree differently than shown in Figure 1. However, it is unlikely that these differences in tree topology will have a significant impact on the conclusions drawn here.
Acknowledgments
We thank F. Collins for critical review of the manuscript; all other ENCODE analysis subgroups for their camaraderie and collaboration; P. Good, E. Feingold, and L. Liefer for ENCODE Consortium guidance and administrative assistance; the Wellcome Trust Sanger Institute, the Max Planck Institute for Developmental Biology, and The Netherlands Institute for Developmental Biology for providing a draft zebrafish genome sequence prior to publication; the DOE Joint Genome Institute for providing a draft Xenopus sequence prior to publication; G. Schuler for making ENCODE comparative sequence data available at NCBI; D. Church for coordinating the identification of finished mouse sequence orthologous to ENCODE regions; and the anonymous reviewers of this manuscript for their constructive comments on previous drafts. This research was supported in part by the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health (E.H.M., J.C.M., and E.D.G.). G.M.C. was a Howard Hughes Medical Institute pre-doctoral Fellow. G.A. is a Bio-X Graduate Student Fellow. D.J.T. is supported by NIH 1 P41 HG02371-05. C.N.D. is supported in part by NIH HG003150. M.H., J.T., and W.M. are supported in part by R01:HG002238. T.M. was supported by BBSRC grant 721/BEP17055. I.H. was funded in part by NIH/NHGRI grant 1R01GM076705-01. S.E.A., S.N., and J.I.M. thank the “Vital IT” computational platform and are supported by grants from NIH ENCODE, Swiss National Science Foundation, European Union, and the ChildCare Foundation. L.P. is supported in part by R01:HG02632 and U01:HG003150. N.G. was supported in part by the Wellcome Trust. D.H. and A. Sidow are supported by funds from NHGRI. A. Siepel was supported by the UCBREP GREAT fellowship (University of California Biotechnology Research and Education Program Graduate Research and Education in Adaptive Biotechnology).
Footnotes
[Supplemental material is available online at www.genome.org.]
Article is online at http://www.genome.org/cgi/doi/10.1101/gr.6034307
References
- Aparicio S., Chapman J., Stupka E., Putnam N., Chia J.-M., Dehal P., Christoffels A., Rash S., Hoon S., Smit A., Chapman J., Stupka E., Putnam N., Chia J.-M., Dehal P., Christoffels A., Rash S., Hoon S., Smit A., Stupka E., Putnam N., Chia J.-M., Dehal P., Christoffels A., Rash S., Hoon S., Smit A., Putnam N., Chia J.-M., Dehal P., Christoffels A., Rash S., Hoon S., Smit A., Chia J.-M., Dehal P., Christoffels A., Rash S., Hoon S., Smit A., Dehal P., Christoffels A., Rash S., Hoon S., Smit A., Christoffels A., Rash S., Hoon S., Smit A., Rash S., Hoon S., Smit A., Hoon S., Smit A., Smit A., et al. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science. 2002;297:1301–1310. doi: 10.1126/science.1072104. [DOI] [PubMed] [Google Scholar]
- Blakesley R.W., Hansen N.F., Mullikin J.C., Thomas P.J., McDowell J.C., Maskeri B., Young A.C., Benjamin B., Brooks S.Y., Coleman B.I., Hansen N.F., Mullikin J.C., Thomas P.J., McDowell J.C., Maskeri B., Young A.C., Benjamin B., Brooks S.Y., Coleman B.I., Mullikin J.C., Thomas P.J., McDowell J.C., Maskeri B., Young A.C., Benjamin B., Brooks S.Y., Coleman B.I., Thomas P.J., McDowell J.C., Maskeri B., Young A.C., Benjamin B., Brooks S.Y., Coleman B.I., McDowell J.C., Maskeri B., Young A.C., Benjamin B., Brooks S.Y., Coleman B.I., Maskeri B., Young A.C., Benjamin B., Brooks S.Y., Coleman B.I., Young A.C., Benjamin B., Brooks S.Y., Coleman B.I., Benjamin B., Brooks S.Y., Coleman B.I., Brooks S.Y., Coleman B.I., Coleman B.I., et al. An intermediate grade of finished genomic sequence suitable for comparative analyses. Genome Res. 2004;14:2235–2244. doi: 10.1101/gr.2648404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette M., Kent W.J., Riemer C., Elnitski L., Smit A.F.A., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Kent W.J., Riemer C., Elnitski L., Smit A.F.A., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Riemer C., Elnitski L., Smit A.F.A., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Elnitski L., Smit A.F.A., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Smit A.F.A., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Rosenbloom K., Clawson H., Green E.D., Clawson H., Green E.D., Green E.D., et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boffelli D., McAuliffe J., Ovcharenko D., Lewis K.D., Ovcharenko I., Pachter L., Rubin E.M., McAuliffe J., Ovcharenko D., Lewis K.D., Ovcharenko I., Pachter L., Rubin E.M., Ovcharenko D., Lewis K.D., Ovcharenko I., Pachter L., Rubin E.M., Lewis K.D., Ovcharenko I., Pachter L., Rubin E.M., Ovcharenko I., Pachter L., Rubin E.M., Pachter L., Rubin E.M., Rubin E.M. Phylogenetic shadowing of primate sequences to find functional regions of the human genome. Science. 2003;299:1391–1394. doi: 10.1126/science.1081331. [DOI] [PubMed] [Google Scholar]
- Bray N., Pachter L., Pachter L. MAVID: Constrained ancestral alignment of multiple sequences. Genome Res. 2004;14:693–699. doi: 10.1101/gr.1960404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brudno M., Do C.B., Cooper G.M., Kim M.F., Davydov E., Green E.D., Sidow A., Batzoglou S., Do C.B., Cooper G.M., Kim M.F., Davydov E., Green E.D., Sidow A., Batzoglou S., Cooper G.M., Kim M.F., Davydov E., Green E.D., Sidow A., Batzoglou S., Kim M.F., Davydov E., Green E.D., Sidow A., Batzoglou S., Davydov E., Green E.D., Sidow A., Batzoglou S., NISC Comparative Sequencing Program, Green E.D., Sidow A., Batzoglou S., Sidow A., Batzoglou S., Batzoglou S. LAGAN and Multi-LAGAN: Efficient tools for large-scale multiple alignment of genomic DNA. Genome Res. 2003;13:721–731. doi: 10.1101/gr.926603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge C., Karlin S., Karlin S. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- Chimpanzee Sequencing and Analysis Consortium, Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- Cliften P., Sudarsanam P., Desikan A., Fulton L., Fulton B., Majors J., Waterston R., Cohen B.A., Johnston M., Sudarsanam P., Desikan A., Fulton L., Fulton B., Majors J., Waterston R., Cohen B.A., Johnston M., Desikan A., Fulton L., Fulton B., Majors J., Waterston R., Cohen B.A., Johnston M., Fulton L., Fulton B., Majors J., Waterston R., Cohen B.A., Johnston M., Fulton B., Majors J., Waterston R., Cohen B.A., Johnston M., Majors J., Waterston R., Cohen B.A., Johnston M., Waterston R., Cohen B.A., Johnston M., Cohen B.A., Johnston M., Johnston M. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science. 2003;301:71–76. doi: 10.1126/science.1084337. [DOI] [PubMed] [Google Scholar]
- Collins F.S., Green E.D., Guttmacher A.E., Guyer M.S., Green E.D., Guttmacher A.E., Guyer M.S., Guttmacher A.E., Guyer M.S., Guyer M.S. A vision for the future of genomics research: A blueprint for the genomic era. Nature. 2003;422:835–847. doi: 10.1038/nature01626. [DOI] [PubMed] [Google Scholar]
- Cooper G.M., Sidow A., Sidow A. Genomic regulatory regions: Insights from comparative sequence analysis. Curr. Opin. Genet. Dev. 2003;13:604–610. doi: 10.1016/j.gde.2003.10.001. [DOI] [PubMed] [Google Scholar]
- Cooper G.M., Brudno M., Stone E.A., Dubchak I., Batzoglou S., Sidow A., Brudno M., Stone E.A., Dubchak I., Batzoglou S., Sidow A., Stone E.A., Dubchak I., Batzoglou S., Sidow A., Dubchak I., Batzoglou S., Sidow A., Batzoglou S., Sidow A., Sidow A. Characterization of evolutionary rates and constraints in three mammalian genomes. Genome Res. 2004;14:539–548. doi: 10.1101/gr.2034704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper G.M., Stone E.A., Asimenos G., Green E.D., Batzoglou S., Sidow A., Stone E.A., Asimenos G., Green E.D., Batzoglou S., Sidow A., Asimenos G., Green E.D., Batzoglou S., Sidow A., Green E.D., Batzoglou S., Sidow A., Batzoglou S., Sidow A., Sidow A. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15:901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M.B., White K.P., White K.P. Recent advances in Drosophila genomics. Genome Biol. 2004;5:339. doi: 10.1186/gb-2004-5-8-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewey C. “Whole-genome alignments and polytopes for comparative genomics” Ph.D. thesis. 2006. University of California, Berkeley. [Google Scholar]
- Do C.B., Mahabhashyam M.S., Brudno M., Batzoglou S., Mahabhashyam M.S., Brudno M., Batzoglou S., Brudno M., Batzoglou S., Batzoglou S. ProbCons: Probabilistic consistency-based multiple sequence alignment. Genome Res. 2005;15:330–340. doi: 10.1101/gr.2821705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake J.A., Bird C., Nemesh J., Thomas D.J., Newton-Cheh C., Reymond A., Excoffier L., Attar H., Antonarakis S.E., Dermitzakis E.T., Bird C., Nemesh J., Thomas D.J., Newton-Cheh C., Reymond A., Excoffier L., Attar H., Antonarakis S.E., Dermitzakis E.T., Nemesh J., Thomas D.J., Newton-Cheh C., Reymond A., Excoffier L., Attar H., Antonarakis S.E., Dermitzakis E.T., Thomas D.J., Newton-Cheh C., Reymond A., Excoffier L., Attar H., Antonarakis S.E., Dermitzakis E.T., Newton-Cheh C., Reymond A., Excoffier L., Attar H., Antonarakis S.E., Dermitzakis E.T., Reymond A., Excoffier L., Attar H., Antonarakis S.E., Dermitzakis E.T., Excoffier L., Attar H., Antonarakis S.E., Dermitzakis E.T., Attar H., Antonarakis S.E., Dermitzakis E.T., Antonarakis S.E., Dermitzakis E.T., Dermitzakis E.T., et al. Conserved noncoding sequences are selectively constrained and not mutation cold spots. Nat. Genet. 2006;38:223–227. doi: 10.1038/ng1710. [DOI] [PubMed] [Google Scholar]
- Edgar R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H., Smith N.G., Webster M.T., Smith N.G., Webster M.T., Webster M.T. Mutation rate variation in the mammalian genome. Curr. Opin. Genet. Dev. 2003;13:562–568. doi: 10.1016/j.gde.2003.10.008. [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium, The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium, Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007 doi: 10.1038/nature05874. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Evolutionary trees from DNA sequences: A maximum likelihood approach. J. Mol. Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- Hardison R.C. Conserved noncoding sequences are reliable guides to regulatory elements. Trends Genet. 2000;16:369–372. doi: 10.1016/s0168-9525(00)02081-3. [DOI] [PubMed] [Google Scholar]
- Hardison R.C., Roskin K.M., Yang S., Diekhans M., Kent W.J., Weber R., Elnitski L., Li J., O’Connor M., Kolbe D., Roskin K.M., Yang S., Diekhans M., Kent W.J., Weber R., Elnitski L., Li J., O’Connor M., Kolbe D., Yang S., Diekhans M., Kent W.J., Weber R., Elnitski L., Li J., O’Connor M., Kolbe D., Diekhans M., Kent W.J., Weber R., Elnitski L., Li J., O’Connor M., Kolbe D., Kent W.J., Weber R., Elnitski L., Li J., O’Connor M., Kolbe D., Weber R., Elnitski L., Li J., O’Connor M., Kolbe D., Elnitski L., Li J., O’Connor M., Kolbe D., Li J., O’Connor M., Kolbe D., O’Connor M., Kolbe D., Kolbe D., et al. Covariation in frequencies of substitution, deletion, transposition, and recombination during eutherian evolution. Genome Res. 2003;13:13–26. doi: 10.1101/gr.844103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Chicken Genome Sequencing Consortium, Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium, Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- International Mouse Genome Sequencing Consortium, Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Jaillon O., Aury J.-M., Brunet F., Petit J.-L., Stange-Thomann N., Mauceli E., Bouneau L., Fischer C., Ozouf-Costaz C., Bernot A., Aury J.-M., Brunet F., Petit J.-L., Stange-Thomann N., Mauceli E., Bouneau L., Fischer C., Ozouf-Costaz C., Bernot A., Brunet F., Petit J.-L., Stange-Thomann N., Mauceli E., Bouneau L., Fischer C., Ozouf-Costaz C., Bernot A., Petit J.-L., Stange-Thomann N., Mauceli E., Bouneau L., Fischer C., Ozouf-Costaz C., Bernot A., Stange-Thomann N., Mauceli E., Bouneau L., Fischer C., Ozouf-Costaz C., Bernot A., Mauceli E., Bouneau L., Fischer C., Ozouf-Costaz C., Bernot A., Bouneau L., Fischer C., Ozouf-Costaz C., Bernot A., Fischer C., Ozouf-Costaz C., Bernot A., Ozouf-Costaz C., Bernot A., Bernot A., et al. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature. 2004;431:946–957. doi: 10.1038/nature03025. [DOI] [PubMed] [Google Scholar]
- Jordan I.K., Rogozin I.B., Glazko G.V., Koonin E.V., Rogozin I.B., Glazko G.V., Koonin E.V., Glazko G.V., Koonin E.V., Koonin E.V. Origin of a substantial fraction of human regulatory sequences from transposable elements. Trends Genet. 2003;19:68–72. doi: 10.1016/s0168-9525(02)00006-9. [DOI] [PubMed] [Google Scholar]
- Kamal M., Xie X., Lander E.S., Xie X., Lander E.S., Lander E.S. A large family of ancient repeat elements in the human genome is under strong selection. Proc. Natl. Acad. Sci. 2006;103:2740–2745. doi: 10.1073/pnas.0511238103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D., Baertsch R., Diekhans M., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Baertsch R., Diekhans M., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Diekhans M., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Schwartz M., Sugnet C.W., Thomas D.J., Sugnet C.W., Thomas D.J., Thomas D.J., et al. The USCS genome browser database ( http://nar.oupjournals.org/cgi/content/abstract/31/1/51) Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis M., Patterson N., Endrizzi M., Birren B., Lander E.S., Patterson N., Endrizzi M., Birren B., Lander E.S., Endrizzi M., Birren B., Lander E.S., Birren B., Lander E.S., Lander E.S. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- Kent W.J. BLAT—The BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Pringle T.H., Zahler A.M., Haussler D., Zahler A.M., Haussler D., Haussler D. The human genome browser at UCSC ( http://www.genome.org/cgi/content/abstract/12/6/996) Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W.J., Baertsch R., Hinrichs A., Miller W., Haussler D., Baertsch R., Hinrichs A., Miller W., Haussler D., Hinrichs A., Miller W., Haussler D., Miller W., Haussler D., Haussler D. Evolution’s cauldron: Duplication, deletion, and rearrangement in the mouse and human genomes. Proc. Natl. Acad. Sci. 2003;100:11484–11489. doi: 10.1073/pnas.1932072100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimchi-Sarfaty C., Oh J.M., Kim I.W., Sauna Z.E., Calcagno A.M., Ambudkar S.V., Gottesman M.M., Oh J.M., Kim I.W., Sauna Z.E., Calcagno A.M., Ambudkar S.V., Gottesman M.M., Kim I.W., Sauna Z.E., Calcagno A.M., Ambudkar S.V., Gottesman M.M., Sauna Z.E., Calcagno A.M., Ambudkar S.V., Gottesman M.M., Calcagno A.M., Ambudkar S.V., Gottesman M.M., Ambudkar S.V., Gottesman M.M., Gottesman M.M. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- Kimura M. The neutral theory of molecular evolution. Cambridge University Press; Cambridge, UK: 1983. [Google Scholar]
- Klosterman P.S., Uzilov A.V., Bendana Y.R., Bradley R.K., Chao S., Kosiol C., Goldman N., Holmes I., Uzilov A.V., Bendana Y.R., Bradley R.K., Chao S., Kosiol C., Goldman N., Holmes I., Bendana Y.R., Bradley R.K., Chao S., Kosiol C., Goldman N., Holmes I., Bradley R.K., Chao S., Kosiol C., Goldman N., Holmes I., Chao S., Kosiol C., Goldman N., Holmes I., Kosiol C., Goldman N., Holmes I., Goldman N., Holmes I., Holmes I. XRate: A fast prototyping, training and annotation tool for phylo-grammars. BMC Bioinformatics. 2006;7:428. doi: 10.1186/1471-2105-7-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komar A.A. Genetics. SNPs, silent but not invisible. Science. 2007;315:466–467. doi: 10.1126/science.1138239. [DOI] [PubMed] [Google Scholar]
- Künsch H.R. The jackknife and the bootstrap for general stationary observations. Ann. Statist. 1989;17:1217–1241. [Google Scholar]
- Kurtz S., Phillippy A., Delcher A.L., Smoot M., Shumway M., Antonescu C., Salzberg S.L., Phillippy A., Delcher A.L., Smoot M., Shumway M., Antonescu C., Salzberg S.L., Delcher A.L., Smoot M., Shumway M., Antonescu C., Salzberg S.L., Smoot M., Shumway M., Antonescu C., Salzberg S.L., Shumway M., Antonescu C., Salzberg S.L., Antonescu C., Salzberg S.L., Salzberg S.L. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblad-Toh K., Wade C.M., Mikkelsen T.S., Karlsson E.K., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., Zody M.C., Wade C.M., Mikkelsen T.S., Karlsson E.K., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., Zody M.C., Mikkelsen T.S., Karlsson E.K., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., Zody M.C., Karlsson E.K., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., Zody M.C., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., Zody M.C., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., Zody M.C., Clamp M., Chang J.L., Kulbokas E.J., Zody M.C., Chang J.L., Kulbokas E.J., Zody M.C., Kulbokas E.J., Zody M.C., Zody M.C., et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- Ludwig M.Z., Bergman C., Patel N.H., Kreitman M., Bergman C., Patel N.H., Kreitman M., Patel N.H., Kreitman M., Kreitman M. Evidence for stabilizing selection in a eukaryotic enhancer element. Nature. 2000;403:564–567. doi: 10.1038/35000615. [DOI] [PubMed] [Google Scholar]
- Margulies E.H., Blanchette M., Haussler D., Green E.D., Blanchette M., Haussler D., Green E.D., NISC Comparative Sequencing Program, Haussler D., Green E.D., Green E.D. Identification and characterization of multi-species conserved sequences. Genome Res. 2003;13:2507–2518. doi: 10.1101/gr.1602203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies E.H., Green E.D., NISC Comparative Sequencing Program, Green E.D. Detecting highly conserved regions of the human genome by multispecies sequence comparisons. Cold Spring Harb. Symp. Quant. Biol. 2004;68:255–263. doi: 10.1101/sqb.2003.68.255. [DOI] [PubMed] [Google Scholar]
- Margulies E.H., Maduro V.V.B., Thomas P.J., Tomkins J.P., Amemiya C.T., Luo M., Green E.D., NISC Comparative Sequencing Program, Maduro V.V.B., Thomas P.J., Tomkins J.P., Amemiya C.T., Luo M., Green E.D., Thomas P.J., Tomkins J.P., Amemiya C.T., Luo M., Green E.D., Tomkins J.P., Amemiya C.T., Luo M., Green E.D., Amemiya C.T., Luo M., Green E.D., Luo M., Green E.D., Green E.D. Comparative sequencing provides insights about the structure and conservation of marsupial and monotreme genomes. Proc. Natl. Acad. Sci. 2005a;102:3354–3359. doi: 10.1073/pnas.0408539102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies E.H., Vinson J.P., Miller W., Jaffe D.B., Lindblad-Toh K., Chang J.L., Green E.D., Lander E.S., Mullikin J.C., Vinson J.P., Miller W., Jaffe D.B., Lindblad-Toh K., Chang J.L., Green E.D., Lander E.S., Mullikin J.C., NISC Comparative Sequencing Program, Miller W., Jaffe D.B., Lindblad-Toh K., Chang J.L., Green E.D., Lander E.S., Mullikin J.C., Jaffe D.B., Lindblad-Toh K., Chang J.L., Green E.D., Lander E.S., Mullikin J.C., Lindblad-Toh K., Chang J.L., Green E.D., Lander E.S., Mullikin J.C., Chang J.L., Green E.D., Lander E.S., Mullikin J.C., Green E.D., Lander E.S., Mullikin J.C., Lander E.S., Mullikin J.C., Mullikin J.C., et al. An initial strategy for the systematic identification of functional elements in the human genome by low-redundancy comparative sequencing. Proc. Natl. Acad. Sci. 2005b;102:4795–4800. doi: 10.1073/pnas.0409882102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy W.J., Eizirik E., O’Brien S.J., Madsen O., Scally M., Douady C.J., Teeling E., Ryder O.A., Stanhope M.J., de Jong W.W., Eizirik E., O’Brien S.J., Madsen O., Scally M., Douady C.J., Teeling E., Ryder O.A., Stanhope M.J., de Jong W.W., O’Brien S.J., Madsen O., Scally M., Douady C.J., Teeling E., Ryder O.A., Stanhope M.J., de Jong W.W., Madsen O., Scally M., Douady C.J., Teeling E., Ryder O.A., Stanhope M.J., de Jong W.W., Scally M., Douady C.J., Teeling E., Ryder O.A., Stanhope M.J., de Jong W.W., Douady C.J., Teeling E., Ryder O.A., Stanhope M.J., de Jong W.W., Teeling E., Ryder O.A., Stanhope M.J., de Jong W.W., Ryder O.A., Stanhope M.J., de Jong W.W., Stanhope M.J., de Jong W.W., de Jong W.W., et al. Resolution of the early placental mammal radiation using Bayesian phylogenetics. Science. 2001;294:2348–2351. doi: 10.1126/science.1067179. [DOI] [PubMed] [Google Scholar]
- Murphy W.J., Pringle T.H., Crider T.A., Springer M.S., Miller W., Pringle T.H., Crider T.A., Springer M.S., Miller W., Crider T.A., Springer M.S., Miller W., Springer M.S., Miller W., Miller W. Using genomic data to unravel the root of the placental mammal phylogeny. Genome Res. 2007;17:413–421. doi: 10.1101/gr.5918807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrutenko A., Li W.H., Li W.H. Transposable elements are found in a large number of human protein-coding genes. Trends Genet. 2001;17:619–621. doi: 10.1016/s0168-9525(01)02445-3. [DOI] [PubMed] [Google Scholar]
- Nikolaev S., Montoya-Burgos J.I., Margulies E.H., Rougemont J., Nyffeler B., Antonarakis S.E., Montoya-Burgos J.I., Margulies E.H., Rougemont J., Nyffeler B., Antonarakis S.E., Margulies E.H., Rougemont J., Nyffeler B., Antonarakis S.E., NISC Comparative Sequencing Program, Rougemont J., Nyffeler B., Antonarakis S.E., Nyffeler B., Antonarakis S.E., Antonarakis S.E. Early history of mammalian evolution is elucidated with the ENCODE multiple species sequencing data. PLoS Genet. 2007;3:e2. doi: 10.1371/journal.pgen.0030002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihara H., Hasegawa M., Okada N., Hasegawa M., Okada N., Okada N. Pegasoferae, an unexpected mammalian clade revealed by tracking ancient retroposon insertions. Proc. Natl. Acad. Sci. 2006;103:9929–9934. doi: 10.1073/pnas.0603797103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobrega M.A., Pennacchio L.A., Pennacchio L.A. Comparative genomic analysis as a tool for biological discovery. J. Physiol. 2004;554:31–39. doi: 10.1113/jphysiol.2003.050948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobrega M.A., Ovcharenko I., Afzal V., Rubin E.M., Ovcharenko I., Afzal V., Rubin E.M., Afzal V., Rubin E.M., Rubin E.M. Scanning human gene deserts for long-range enhancers. Science. 2003;302:413. doi: 10.1126/science.1088328. [DOI] [PubMed] [Google Scholar]
- Notredame C., Higgins D.G., Heringa J., Higgins D.G., Heringa J., Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- Pennacchio L.A., Ahituv N., Moses A.M., Prabhakar S., Nobrega M.A., Shoukry M., Minovitsky S., Dubchak I., Holt A., Lewis K.D., Ahituv N., Moses A.M., Prabhakar S., Nobrega M.A., Shoukry M., Minovitsky S., Dubchak I., Holt A., Lewis K.D., Moses A.M., Prabhakar S., Nobrega M.A., Shoukry M., Minovitsky S., Dubchak I., Holt A., Lewis K.D., Prabhakar S., Nobrega M.A., Shoukry M., Minovitsky S., Dubchak I., Holt A., Lewis K.D., Nobrega M.A., Shoukry M., Minovitsky S., Dubchak I., Holt A., Lewis K.D., Shoukry M., Minovitsky S., Dubchak I., Holt A., Lewis K.D., Minovitsky S., Dubchak I., Holt A., Lewis K.D., Dubchak I., Holt A., Lewis K.D., Holt A., Lewis K.D., Lewis K.D., et al. In vivo enhancer analysis of human conserved noncoding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- Rat Genome Sequencing Project Consortium, Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- Schwartz S., Kent W.J., Smit A., Zhang Z., Baertsch R., Hardison R.C., Haussler D., Miller W., Kent W.J., Smit A., Zhang Z., Baertsch R., Hardison R.C., Haussler D., Miller W., Smit A., Zhang Z., Baertsch R., Hardison R.C., Haussler D., Miller W., Zhang Z., Baertsch R., Hardison R.C., Haussler D., Miller W., Baertsch R., Hardison R.C., Haussler D., Miller W., Hardison R.C., Haussler D., Miller W., Haussler D., Miller W., Miller W. Human–mouse alignments with BLASTZ. Genome Res. 2003;13:103–107. doi: 10.1101/gr.809403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A., Haussler D., Haussler D. Proceedings of the 8th Annual International Conference on Computational Biology. 2004a. Computational identification of evolutionarily conserved exons; pp. 177–186. (RECOMB’04) [Google Scholar]
- Siepel A., Haussler D., Haussler D. Phylogenetic estimation of context-dependent substitution rates by maximum likelihood. Mol. Biol. Evol. 2004b;21:468–488. doi: 10.1093/molbev/msh039. [DOI] [PubMed] [Google Scholar]
- Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Clawson H., Spieth J., Hillier L.W., Richards S., Spieth J., Hillier L.W., Richards S., Hillier L.W., Richards S., Richards S., et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A., Pollard K., Haussler D., Pollard K., Haussler D., Haussler D. Proceedings of the 10th Annual International Conference on Research in Computational Biology. 2006. New methods for detecting lineage-specific selection. [Google Scholar]
- Silva J.C., Shabalina S.A., Harris D.G., Spouge J.L., Kondrashovi A.S., Shabalina S.A., Harris D.G., Spouge J.L., Kondrashovi A.S., Harris D.G., Spouge J.L., Kondrashovi A.S., Spouge J.L., Kondrashovi A.S., Kondrashovi A.S. Conserved fragments of transposable elements in intergenic regions: Evidence for widespread recruitment of MIR- and L2-derived sequences within the mouse and human genomes. Genet. Res. 2003;82:1–18. doi: 10.1017/s0016672303006268. [DOI] [PubMed] [Google Scholar]
- Sonnhammer E.L., Koonin E.V., Koonin E.V. Orthology, paralogy and proposed classification for paralog subtypes. Trends Genet. 2002;18:619–620. doi: 10.1016/s0168-9525(02)02793-2. [DOI] [PubMed] [Google Scholar]
- Stein L.D., Bao Z., Blasiar D., Blumenthal T., Brent M.R., Chen N., Chinwalla A., Clarke L., Clee C., Coghlan A., Bao Z., Blasiar D., Blumenthal T., Brent M.R., Chen N., Chinwalla A., Clarke L., Clee C., Coghlan A., Blasiar D., Blumenthal T., Brent M.R., Chen N., Chinwalla A., Clarke L., Clee C., Coghlan A., Blumenthal T., Brent M.R., Chen N., Chinwalla A., Clarke L., Clee C., Coghlan A., Brent M.R., Chen N., Chinwalla A., Clarke L., Clee C., Coghlan A., Chen N., Chinwalla A., Clarke L., Clee C., Coghlan A., Chinwalla A., Clarke L., Clee C., Coghlan A., Clarke L., Clee C., Coghlan A., Clee C., Coghlan A., Coghlan A., et al. The genome sequence of Caenorhabditis briggsae: A platform for comparative genomics. PLoS Biol. 2003;1:e45. doi: 10.1371/journal.pbio.0000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone E.A., Cooper G.M., Sidow A., Cooper G.M., Sidow A., Sidow A. Trade-offs in detecting evolutionarily constrained sequence by comparative genomics. Annu. Rev. Genomics Hum. Genet. 2005;6:143–164. doi: 10.1146/annurev.genom.6.080604.162146. [DOI] [PubMed] [Google Scholar]
- Thomas J.W., Touchman J.W., Blakesley R.W., Bouffard G.G., Beckstrom-Sternberg S.M., Margulies E.H., Blanchette M., Siepel A.C., Thomas P.J., McDowell J.C., Touchman J.W., Blakesley R.W., Bouffard G.G., Beckstrom-Sternberg S.M., Margulies E.H., Blanchette M., Siepel A.C., Thomas P.J., McDowell J.C., Blakesley R.W., Bouffard G.G., Beckstrom-Sternberg S.M., Margulies E.H., Blanchette M., Siepel A.C., Thomas P.J., McDowell J.C., Bouffard G.G., Beckstrom-Sternberg S.M., Margulies E.H., Blanchette M., Siepel A.C., Thomas P.J., McDowell J.C., Beckstrom-Sternberg S.M., Margulies E.H., Blanchette M., Siepel A.C., Thomas P.J., McDowell J.C., Margulies E.H., Blanchette M., Siepel A.C., Thomas P.J., McDowell J.C., Blanchette M., Siepel A.C., Thomas P.J., McDowell J.C., Siepel A.C., Thomas P.J., McDowell J.C., Thomas P.J., McDowell J.C., McDowell J.C., et al. Comparative analyses of multispecies sequences from targeted genomic regions. Nature. 2003;424:788–793. doi: 10.1038/nature01858. [DOI] [PubMed] [Google Scholar]
- Thomas D.J., Rosenbloom K.R., Clawson H., Hinrichs A.S., Trumbower H., Raney B.J., Karolchik D., Barber G.P., Harte R.A., Hillman-Jackson J., Rosenbloom K.R., Clawson H., Hinrichs A.S., Trumbower H., Raney B.J., Karolchik D., Barber G.P., Harte R.A., Hillman-Jackson J., Clawson H., Hinrichs A.S., Trumbower H., Raney B.J., Karolchik D., Barber G.P., Harte R.A., Hillman-Jackson J., Hinrichs A.S., Trumbower H., Raney B.J., Karolchik D., Barber G.P., Harte R.A., Hillman-Jackson J., Trumbower H., Raney B.J., Karolchik D., Barber G.P., Harte R.A., Hillman-Jackson J., Raney B.J., Karolchik D., Barber G.P., Harte R.A., Hillman-Jackson J., Karolchik D., Barber G.P., Harte R.A., Hillman-Jackson J., Barber G.P., Harte R.A., Hillman-Jackson J., Harte R.A., Hillman-Jackson J., Hillman-Jackson J., et al. The ENCODE Project at UC Santa Cruz. Nucleic Acids Res. 2006;35:D663–D667. doi: 10.1093/nar/gkl1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompa M., Li N., Bailey T.L., Church G.M., Moor B.D., Eskin E., Favorov A.V., Frith M.C., Fu Y., Kent W.J., Li N., Bailey T.L., Church G.M., Moor B.D., Eskin E., Favorov A.V., Frith M.C., Fu Y., Kent W.J., Bailey T.L., Church G.M., Moor B.D., Eskin E., Favorov A.V., Frith M.C., Fu Y., Kent W.J., Church G.M., Moor B.D., Eskin E., Favorov A.V., Frith M.C., Fu Y., Kent W.J., Moor B.D., Eskin E., Favorov A.V., Frith M.C., Fu Y., Kent W.J., Eskin E., Favorov A.V., Frith M.C., Fu Y., Kent W.J., Favorov A.V., Frith M.C., Fu Y., Kent W.J., Frith M.C., Fu Y., Kent W.J., Fu Y., Kent W.J., Kent W.J., et al. Assessing computational tools for the discovery of transcription factor binding sites. Nat. Biotechnol. 2005;23:137–144. doi: 10.1038/nbt1053. [DOI] [PubMed] [Google Scholar]
- Trinklein N.D., Aldred S.F., Hartman S.J., Schroeder D.I., Otillar R.P., Myers R.M., Aldred S.F., Hartman S.J., Schroeder D.I., Otillar R.P., Myers R.M., Hartman S.J., Schroeder D.I., Otillar R.P., Myers R.M., Schroeder D.I., Otillar R.P., Myers R.M., Otillar R.P., Myers R.M., Myers R.M. An abundance of bidirectional promoters in the human genome. Genome Res. 2004;14:62–66. doi: 10.1101/gr.1982804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Walle I., Lasters I., Wyns L., Lasters I., Wyns L., Wyns L. SABmark—A benchmark for sequence alignment that covers the entire known fold space. Bioinformatics. 2005;21:1267–1268. doi: 10.1093/bioinformatics/bth493. [DOI] [PubMed] [Google Scholar]
- Woolfe A., Goodson M., Goode D.K., Snell P., McEwen G.K., Vavouri T., Smith S.F., North P., Callaway H., Kelly K., Goodson M., Goode D.K., Snell P., McEwen G.K., Vavouri T., Smith S.F., North P., Callaway H., Kelly K., Goode D.K., Snell P., McEwen G.K., Vavouri T., Smith S.F., North P., Callaway H., Kelly K., Snell P., McEwen G.K., Vavouri T., Smith S.F., North P., Callaway H., Kelly K., McEwen G.K., Vavouri T., Smith S.F., North P., Callaway H., Kelly K., Vavouri T., Smith S.F., North P., Callaway H., Kelly K., Smith S.F., North P., Callaway H., Kelly K., North P., Callaway H., Kelly K., Callaway H., Kelly K., Kelly K., et al. Highly conserved noncoding sequences are associated with vertebrate development. PLoS Biol. 2005;3:e7. doi: 10.1371/journal.pbio.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Smit A.F., Schwartz S., Chiaromonte F., Roskin K.M., Haussler D., Miller W., Hardison R.C., Smit A.F., Schwartz S., Chiaromonte F., Roskin K.M., Haussler D., Miller W., Hardison R.C., Schwartz S., Chiaromonte F., Roskin K.M., Haussler D., Miller W., Hardison R.C., Chiaromonte F., Roskin K.M., Haussler D., Miller W., Hardison R.C., Roskin K.M., Haussler D., Miller W., Hardison R.C., Haussler D., Miller W., Hardison R.C., Miller W., Hardison R.C., Hardison R.C. Patterns of insertions and their covariation with substitutions in the rat, mouse, and human genomes. Genome Res. 2004;14:517–527. doi: 10.1101/gr.1984404. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Alignments and other annotations generated and used for the studies reported here are available at http://genome.ucsc.edu/ENCODE (click on the “Downloads” link in the blue column along the left side of the page). They are also displayed in the UCSC Genome Browser under the “ENCODE Comparative Genomics” set of tracks. PECAN alignments are available at http://www.ebi.ac.uk/~bjp/pecan/encode_sept_pecan_mfas_proj.tar.bz2. All experimental annotations were obtained from publicly available ENCODE project data (The ENCODE Project Consortium 2007); a bulk download of these data is available at http://www.nisc.nih.gov/data.