Abstract
ChIP-chip studies have revealed that many in vivo binding sites have a weak match to the consensus sequence for the transcription factor being analyzed. Possible explanations for these observations include (1) the in vitro-derived consensus site does not represent the in vivo binding site and/or (2) the factor is recruited to a weak binding site via interaction with another protein. To address these possibilities, we developed an approach (ChIPMotifs) that incorporates a bootstrap resampling method to statistically infer the optimal cutoff threshold for a position weight matrix (PWM) of a motif identified from ChIP-chip data by ab initio motif discovery programs. Using OCT4 ChIP-chip data and the ChIPMotifs approach, we first developed a refined OCT4 PWM. We then used the refined PWM and a ChIPModules approach to identify transcription factors colocalizing with OCT4 in Ntera2 testicular embryonal carcinoma cells. We found that the consensus binding site for SRY, a transcription factor critical for testis development, colocalizes with the OCT4 PWM. To further characterize the relationship between OCT4 and SRY, we performed ChIP-chip experiments with human promoter microarrays, and found that 49% of the top ∼1000 OCT4 target promoters were also bound by SRY. This analysis represents the first identification of SRY target promoters. Interestingly, we determined that promoters bound by OCT4 and SRY, but not those bound by SRY alone, were also bound by the transcriptional repressor KAP1. Our studies not only validate the ChIPMotifs and ChIPModules combinatorial approach but also identify a possible new regulatory partner of OCT4.
During the past decade, several computational approaches have been developed to study large and complex data sets generated from high-throughput technologies such as mRNA expression profiling (Schena et al. 1995; Lockhart et al. 1996), ChIP-chip (Ren et al. 2000; Iyer et al. 2001), DamID (van Steensel and Henikoff 2000), DNase-chip (Crawford et al. 2006), and ChIP-PET (Loh et al. 2006). Many approaches (such as ModuleSearcher, ModuleScanner, CRÈME, CONFAC, ROVER, and oPOSSUM) have been applied to problems such as identifying binding sites and putative cis-regulatory modules in the promoters of coexpressed genes (Wasserman and Fickett 1998; Krivan and Wasserman 2001; Aerts et al. 2003; Sharan et al. 2003; Haverty et al. 2004; Karanam and Moreno 2004; Ho-Sui et al. 2005). Other approaches (Zhou and Wong 2004; Gupta and Liu 2005; Hong et al. 2005a; Smith et al. 2005; Wang et al. 2005; Cheng et al. 2006; Jin et al. 2006; Li et al. 2006) have been used to identify motifs derived from ChIP-chip data. The computational algorithms behind the approaches listed above include (1) statistically driven ab initio motif discovery methods such as hidden Markov models (Pedersen and Moult 1996), Gibbs sampling (Lawrence et al. 1993), greedy alignment algorithms (CONSENSUS) (Hertz and Stormo 1999), expectation-maximization (MEME) (Bailey and Elkan 1995), probabilistic mixture modeling (NestedMica) (Down and Hubbard 2005), exhaustive enumeration (Weeder) (Pavesi et al. 2004), and words enumeration with a positional weight matrix updating (Liu et al. 2002); and (2) prior-compiled PWMs library-based motifs detection methods such as MATCH (Kel et al. 2003) combined with the TRANSFAC database (Wingender et al. 2000) and MSCAN (Alkema et al. 2004) combined with the JASPAR database (Sandelin et al. 2004a).
All of the abovementioned methods have proven to be useful in detecting novel motifs and deciphering the logics of transcription regulatory networks. However, there are still several major challenges facing these de novo methods. First, because a transcription factor binding site (TFBS) is a short (10–20 base pairs [bp]) and degenerate sequence, it is difficult to detect among the noise of much longer background sequences. Second, the issue of binding-site variability for each given transcription factor is not well understood, making it difficult to accurately predict sites using only computational approaches. Third, the consensus sites used by many of the programs have been derived from a small number of in vitro interactions. Some of these challenges in identifying motifs can be minimized by using ChIP-chip data to derive a consensus binding site to which a factor is bound in vivo. Also, some of the issues concerned with background (control) sequences can be eliminated using a bootstrap resampling of the data. Bootstrap resampling is a methodology that creates many repeated data sets from a single set of experimental data and makes inference statistically from those samples without knowing the theoretical distribution of the data samples (see Efron 1979; Felsenstein 1985).
In this study, we have incorporated a modified bootstrap resampling into our de novo motif discovery approach (named ChIPMotifs) to statistically infer the optimal cutoff threshold for the PWM of an OCT4 motif initially identified from ChIP-chip data using ab initio motif discovery programs. We then used the refined OCT4 PWM (OCT4H_PWM) and a set of high-confidence data obtained from ChIP-chip experiments to identify five cis-regulatory modules (using a previously described ChIPModules approach). Finally, we experimentally validated one of the computationally identified modules (see Fig. 1B for an overview of our approach). Our results suggest that SRY is a potential new regulatory partner of OCT4.
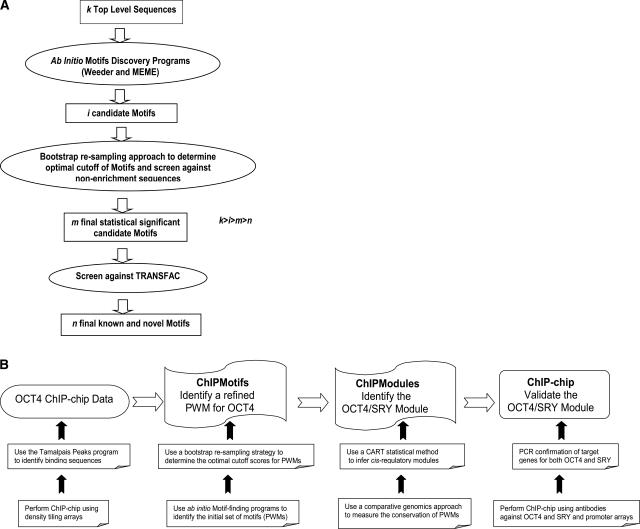
Figure 1.
(A) A diagram of the integrated ChIPMotifs approach. Beginning with a set of k high-confidence level ChIP-chip sequences, ab initio motifs discovery programs such as Weeder and MEME are used to identify i candidate motifs. Then, a bootstrap resampling approach is used to determine cutoffs for the motifs and to screen against nonenriched sequences to obtain m final statistically significant candidate motifs. Finally, the motifs are screened against TRANSFAC to retrieve n known and novel motifs. (B) A strategy diagram showing how our ChIPMotifs and ChIPModules approaches work in concert to efficiently mine OCT4 ChIP-chip data, to allow the development of de novo OCT4 motifs, to identify new cis-regulatory modules of OCT4 and SRY, and finally to develop an experimentally confirmed set of OCT4 and SRY targets.
Results
De novo OCT4-binding-site motif discovery
We have previously reported the identification of a set of OCT4-binding sites identified by ChIP-chip analysis using Ntera2 testicular embryonal carcinoma cells (O’Geen et al. 2006). These binding sites were identified using ENCODE arrays, which are high-density oligonucleotide arrays on which 44 regions of the human genome (The ENCODE Project Consortium 2004) are tiled at a density of one 50 mer every 38 bp. There are a total of ∼380,000 probes on the array, representing the nonrepetitive portion of ∼30 Mb (1%) of the human genome. The OCT4-binding sites (defined as Data set 1; see Methods) that serve as the basis for the analyses in this study represent a very high confidence set of binding sites for three reasons. First, the set of 154 binding sites was identified using the L1 criteria of the Tamalpais peak calling program (Bieda et al. 2006; see also http://chipanalysis.genomecenter.ucdavis.edu/cgi-bin/tamalpais.cgi), which is defined as peaks in the top 2% of the array data that have a P-value <0.0001. Second, these sites were identified in both data sets from two different biologically independent experiments (i.e., the cells were grown and cross-linked on separate days). Third, for hybridization to the arrays, we used pooled ChIP samples (10 ChIPs were pooled for each experiment) and thus eliminated any potential artifacts that might arise during amplification of a ChIP sample. However, although these sites are high-confidence OCT4-binding sites, further characterization indicates that only 13.6% (21) of them contain the conventional OCT4 consensus motif, ATGC(A/T)AAT (Pesce and Scholer 2001).
Because the OCT4 motif was originally defined using in vitro studies, it was possible that OCT4 might bind to a different consensus under physiologically normal conditions in a chromatin environment. Therefore, as our first step in the characterization of OCT4-binding sites, we developed a de novo motif-finding approach (termed ChIPMotifs) as shown in Figure 1A, which uses the ab initio motif-finding programs such as Weeder (Pavesi et al. 2004) and MEME (Bailey and Gribskov 1997), followed by a modified bootstrap resampling statistical inference method to identify significantly overrepresented motifs from ChIP-chip data.
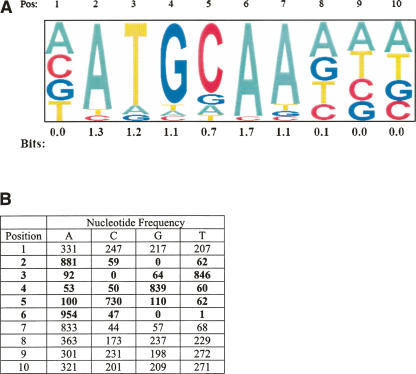
The ChIPMotifs approach (Fig. 1A) began with inputting a set of 154 in vivo OCT4-binding sequences into the Weeder and MEME programs. Using these programs, we identified 10 candidate motifs, each having a length of 8–12 bp. We then constructed 10 positional weight matrices (PWMs) for each candidate motif. We randomized the sequences of each of the 154 OCT4-binding sites 100 times to generate a set of 15,400 randomized sequences. These randomized sequences no longer correspond to binding sites, but have the same nucleotide frequencies as the original binding sites and are therefore used as a negative control set for motif finding. We then scanned these randomized sequences for each candidate motif (using the PWMs derived from Weeder and MEME) starting at a minimal core score of 0.5 and a minimal PWM score of 0.5. Then, we retrieved core scores and PWM scores at the top 0.1% percentile (one-tailed P-value is <0.001), the top 0.5% percentile (one-tailed P-value is <0.005), and the top 1% percentile (one-tailed P-value is <0.01), respectively. Using these scores, we tested the 154 OCT4-binding regions (Data set 1) and 499 regions that were not bound by OCT4 (defined as Data set 2; see Methods). A Fisher test was applied, and the P-value was used to define the significant cutoff for these scores. Only those motifs that were found in the OCT4-binding sites, but not in the control Data set 2, were considered to be overrepresented motifs; these motifs have a confidence level at the top 0.1% percentile and a Fisher test P-value <0.001. Thus, a P-value of 0.00026 for the OCT4H_PWM at the top 0.1% percentile with a core score of 0.88 and PWM score of 0.85 is considered to be significant. As such, the motif NATG CAAANN, which resembles the OCT4 consensus site of ATG CAAAT (Fig. 2A), was identified.
Figure 2.
Refinement of the OCT4 consensus binding site. (A) A sequence log of the OCT4 consensus site that was derived from OCT4 ChIP-chip data from Ntera2 cells. (B) The positional weight matrix (OCT4H_PWM) built by the ChIPMotifs approach, with a core score computed from position 2 to 6 (6 bases) (bold) and a PWM score from 1 to 10 (10 bases).
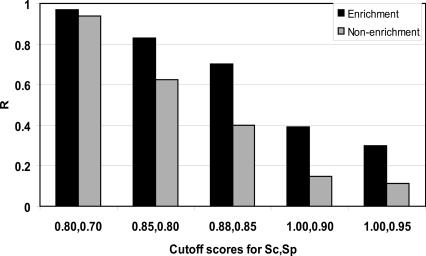
Importantly, our ChIPMotifs analysis provided not only a consensus site, but also a position weight matrix (OCT4H_PWM) for in vivo OCT4 binding (Fig. 2B). We then used the OCT4-binding regions (Data set 1) and the control regions (Data set 2) to determine cutoff thresholds for this newly constructed OCT4H_PWM (Fig. 3). Allowing too many changes from the consensus motif results in the identification of OCT4-binding sites in the great majority of both data sets, whereas requiring a complete match to the consensus eliminates the majority of the true binding sites. We found that a 0.88 match to the core sequences (Sc) and a 0.85 match to the PWM (Sp) clearly distinguish the OCT4 data set from the control set (with a P-value at 0.00026) and demonstrate high specificity (eliminating 60% of the fragments in the negative control set) and high sensitivity (capturing ∼70% of the binding sites). However, when using 0.88 (Sc) and 0.85 (Sp) criteria, 28.6% of the experimentally determined Oct4-binding regions still lack a match to the OCT4H_PWM.
Figure 3.
A histogram plot showing the prediction rate (R) of OCT4 target promoters versus non-OCT4 target promoters at several combinations of the match to the core sequences (Sc_h) and to the PWMs (Sp_h). The values of 0.88 for Sc_h, 0.85 for Sc_p were chosen as cutoff thresholds for further analyses; these scores had a P-value of 0.00026.
OCT4-binding sites are predominantly found within transcribed sequences
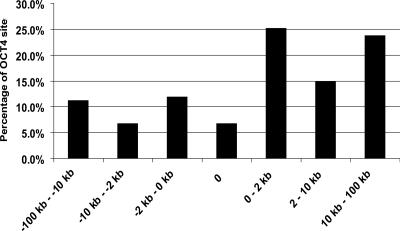
We were curious as to whether the ∼70% of the OCT4-binding regions that contained good matches to the OCT4H_PWM (having a core score ≥0.88 and a PWM score ≥0.85) had different characteristics from the ∼30% of the sites that had only low matches. One characteristic that we analyzed was the location of the OCT4-binding site relative to the start site of transcription. We annotated the 154 human OCT4-binding sequences based on the GENCODE Database (Harrow et al. 2006). First, we defined an OCT4-binding region that is >100 kb upstream of or downstream from a transcription start site as being in a gene desert. Interestingly, 13 (8.4%) of the OCT4-binding sites are 100 kb away from any known gene and thus in the gene desert category (Supplemental Fig. 3B). We then categorized the remaining binding sites into groups based on two different classification schemes: gene structure and distance relative to the transcription start site. For the gene structure classification, we defined the regions as upstream of a gene (between −100 kb and +1), overlapping with the transcription start site, within a transcribed region (without distinguishing between exons and introns), and within 100 kb downstream from the 3′-UTR (Supplemental Fig. 3A). If a site fell in between two genes (but is not in a gene desert), it was assigned to the gene for which the transcription start site is closest to the binding site. Of the 141 binding sites that were not in gene deserts, 40 (28.4%) were located within the upstream promoter/enhancer region, nine (5.8%) were overlapping with a known transcription start site, 70 (49.6%) mapped within transcribed regions, and 22 (15.6%) were within 100 kb downstream from a gene (Supplemental Fig. 3B). For the second classification scheme, distance relative to a transcription start site, we defined the following categories: overlapping with a transcription start site and 0–2 kb, 2–10 kb, or 10–100 kb either upstream of or downstream from the start site. As shown in Figure 4, ∼19% of the OCT4-binding sites that are not in gene deserts are located in a proximal region upstream of the start site. Interestingly, >25% of the sites are located in a proximal region downstream from the start of transcription.
Figure 4.
Distribution of OCT4-binding sites on the basis of distance relative to a start site. Shown is the percentage of OCT4-binding sites in the different regions. The categories include from 10 to 100 kb upstream of a start site, from 2 to 10 kb upstream of a start site, from 10 bp to 2 kb upstream of a start site, overlapping a start site, between 10 bp and 2 kb downstream from a start site, between 2 and 10 kb downstream from a start site, between 10 and 100 kb downstream from a start site, and those sites that are not within 100 kb upstream or downstream of a start site. The OCT4-binding regions were defined as peaks detected on duplicate ENCODE arrays using a peak calling program developed for ChIP-chip experiments (Bieda et al. 2006); the gene list was based on GENCODE Genes (Harrow et al. 2006).
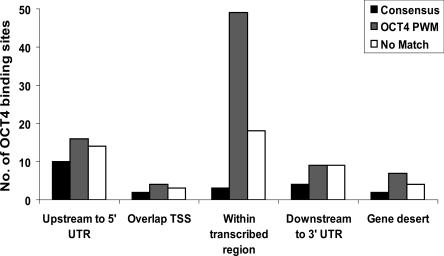
Having determined the location of all the OCT4-binding sites, we then examined whether those sites having a close match to the consensus showed a different localization from those sites having no match to the consensus (Fig. 5). We found that the OCT4-binding sites located within the transcribed region of a gene mostly represent the sites that contain a good match (but not the actual consensus sequence) to the OCT4H_PWM. Interestingly, the consensus site is very rarely represented at this location. The other locations (upstream of, overlapping the start site, and downstream from the gene) have very similar percentages of consensus, match to the PWM, and no match sites. In particular, the sites located upstream of the start site are not more enriched for consensus sites or close matches to the PWM than those sites located downstream from the transcribed region. Our findings regarding the location analysis of OCT4 are similar to those previously reported for sites discovered in mouse embryonic stem cells using ChIP-PET, a non-array-based method of identifying the location of binding sites (Loh et al. 2006). We identified 154–318 OCT4-binding sites (depending on the stringency selected), using arrays that contain only 1% of the genome. This suggests that there may be as many as 15,000–30,000 OCT4-binding sites in the entire human genome. Although the ENCODE regions may not be completely representative of the entire genome, these numbers are far larger than the 1083 Oct4 targets in mouse embryonic stem cells reported in Loh et al. (2006) and the 628 OCT4 targets in human embryonic stem cells reported in Boyer et al. (2005). This suggests that there are likely many OCT4-binding sites that have not yet been identified.
Figure 5.
A comparison of the distribution of binding sites that (1) contain a conventional OCT consensus (ATGC[A/T]AAT); (2) lack the consensus but contain a match to the OCT PWM identified by the ChIPMotifs approach with 1.0 > Sc_h ≥ 0.88 and 0.95 > Sp_h ≥ 0.85 (the consensus sites would have also been identified by the OCT PWM but have not been included in this set for comparison purposes); and (3) contain no match to the OCT PWM above Sc_h 0.88, Sp_h 0.85. The OCT4-binding regions were defined as peaks detected on duplicate ENCODE arrays using a peak calling program developed for ChIP-chip experiments (Bieda et al. 2006); the gene list was based on GENCODE Genes (Harrow et al. 2006).
Identifying cis-regulatory modules in OCT4 target promoters
Because only 13.6% of the OCT4-binding sites contained consensus motifs, it seemed likely that other transcription factors could be cooperating with OCT4 to stabilize its binding to low-affinity sites (i.e., sites that were fairly good matches to the PWM but were not consensus sites). Others have reported that SOX2 and NANOG bind to a large percentage of the OCT4 target promoters in embryonic stem cells (Boyer et al. 2005; Loh et al. 2006). We did not identify the SOX2 or NANOG motifs in the ENCODE OCT4 data sets using our de novo ChIPMotifs approach. However, when we used the PWMs of Sox2 and Nanog derived in mouse from a previous study (Loh et al. 2006), we found that 60 (39%) and 19 (12%) of the 154 OCT4 sequences did have SOX2- and NANOG-binding sites, respectively, within a distance of 270 bp of an OCT4-binding site. This suggested that perhaps other motifs may colocalize with the OCT4 motif to enhance binding at the other sites. Our next series of experiments was then focused on the application of several different bioinformatics approaches for the identification of other transcription factors that colocalize with OCT4.
The first method that we used to identify cis-regulatory modules was the ChIPModules approach (Jin et al. 2006). In brief, our ChIPModules approach begins with a set of experimentally identified binding sites for a given factor of interest. Then, PWMs and evolutionary conservation are used to refine the set of binding sites. Finally, binding sites for other factors are identified that occur within a short distance of the first factor. The predicted ChIPModules are then confirmed experimentally using ChIP-chip assays and arrays that contain tens of thousands of human promoters. Two important aspects of the ChIPModules approach are (1) the human target promoters are compared to the homologous promoters in mouse so that only those modules that occur in both species are identified; and (2) experimental confirmation of the identified modules is performed using follow-up ChIP-chip experiments with promoter arrays (see Fig. 1B).
To obtain a larger number of OCT4-binding sites to use in our search for colocalizing factors, we performed duplicate OCT4 ChIP-chip analyses using arrays that contain ∼24,000 human promoters, with the region from −1300 to +200 of the promoter being represented by 15 different oligonucleotides. We chose 187 promoters that were in the top 500 ranked OCT4-binding sites on both of the arrays and that had homologous mouse counterparts in the OMGProm database to begin our ChIPModules approach (Data set 3; see Methods and Supplemental Methods for more details) as a training data set. As a negative set of promoters, we chose 94 promoters that were not bound by OCT4 but were bound by POLR2A in Ntera2 cells and that had homologous mouse counterparts in the OMGProm database (Data set 4; see Methods). We chose to use promoters bound by POLR2A, but not by OCT4, as the negative control set for several reasons. Many regions on promoter arrays are misidentified and do not actually correspond to promoters. Such regions may have a different overall nucleotide frequency than promoter regions (which tend to be GC rich). However, because the regions in our negative set are bound by POLR2A in Ntera2 cells, they have been confirmed to be bona fide promoters. Second, these promoters were identified in ChIP experiments (using an antibody to POLR2A instead of to OCT4). Therefore, if there are characteristics of certain promoters that allow them to be easily immunoprecipitated or easily identified on arrays, this negative set would contain those same characteristics.
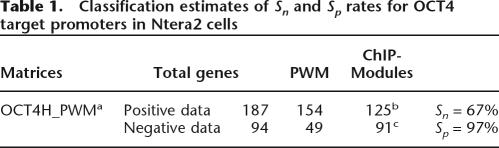
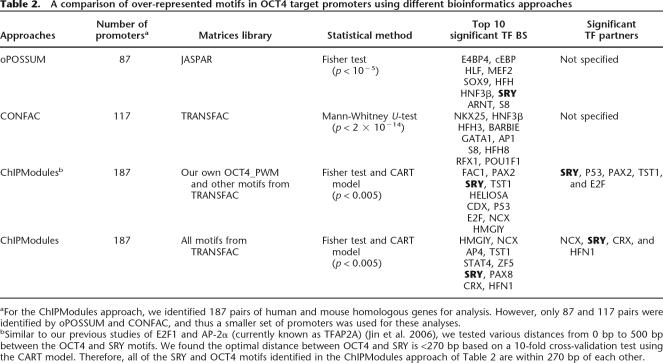
After identifying a conserved (human and mouse) set of positive and negative control promoters, the ChIPModules approach next requires that a set of cutoff scores for the PWM be defined that discriminate the positive 187 OCT4 targets (Data set 3) from the negative 94 promoters (Data set 4) that were not bound by OCT4 (Jin et al. 2006). After testing a number of cutoff scores (data not shown), we selected cutoff thresholds of 0.88 for the core score and of 0.85 for the refined PWM score for the human promoters and cutoff thresholds of 0.80 for the core score and 0.70 for the refined PWM score for the mouse promoters. Of the 187 OCT4 target promoters, 154 (82%) have an OCT4 motif above the cutoff thresholds, whereas only 49 (52%) of the 94 promoters in the negative control set have an OCT4 motif above the cutoff thresholds, which means 45 (48%) of them have no OCT4 Motifs (Table 1). The 154 OCT4 targets and 49 non-OCT4 targets that had matches to the refined OCT4 motif were modeled by the CART method (Breiman et al. 1984) to identify colocalizing motifs (see Jin et al. 2006). For the PWMs of other transcription factors, we used default thresholds defined in “minFN_good83.prf” profile (profile of cutoff values with minimum number of false-negative predictions) from the TRANSFAC database, and 60% identity was used as a conservation cutoff value for ClustalW-aligned human and mouse orthologous pairs. We identified 43 motifs within a distance of 270 bp of the OCT4-binding site (270 bp was chosen on the basis of our previous study) (Jin et al. 2006), and 17 of the 43 motifs had a P-value <0.005. However, only five of the 17 motifs (SRY, P53, TST1, E2F, and PAX2) were identified as significant classifiers with a sensitivity (Sn) of 81% (125 of the 154 OCT4 target promoters contained one of the five motifs) and a specificity (Sp) of 94% (46 of the 49 nontargets lacked the motifs). Thus, as indicated in Table 1, the identified ChIPModules captured 67% (125 of 187) of the OCT4 targets and excluded 97% (91 of 94) of the promoters not bound by OCT4 (including those promoters that both contained and lacked a conserved match to the refined OCT4 PWM). Specifically, 75 (49%) of the OCT4 targets contained an SRY motif, 18 (12%) contained a P53 motif, 14 (9%) contained a TST1 motif, 12 (8%) contained an E2F motif, and four (3%) contained a PAX2 motif. For comparison, we performed the same analyses using the original OCT_PWM (Supplemental Fig. 2) from the TRANSFAC database (with cutoff thresholds of human core score 0.95, human PWM score 0.90, mouse core score 0.8, and mouse PWM score 0.7). Four motifs—NCX, SRY, CRX, and HFN1—were identified as significant transcription factor partners using the TRANSFAC OCT_PWM (Table 2).
Table 1.
Classification estimates of Sn and Sp rates for OCT4 target promoters in Ntera2 cells
aThe OCT4H_PWM was built from our ChIPMotifs approach using an optimal cutoff threshold of 0.88 for Sc_h and 0.85 for Sp_h to identify the OCT4-binding sites.
bThe number of promoters that contained one of the five identified modules.
cThe number of promoters that lacked one of the five identified modules.
Table 2.
A comparison of over-represented motifs in OCT4 target promoters using different bioinformatics approaches
aFor the ChIPModules approach, we identified 187 pairs of human and mouse homologous genes for analysis. However, only 87 and 117 pairs were identified by oPOSSUM and CONFAC, and thus a smaller set of promoters was used for these analyses.
bSimilar to our previous studies of E2F1 and AP-2α (currently known as TFAP2A) (Jin et al. 2006), we tested various distances from 0 bp to 500 bp between the OCT4 and SRY motifs. We found the optimal distance between OCT4 and SRY is <270 bp based on a 10-fold cross-validation test using the CART model. Therefore, all of the SRY and OCT4 motifs identified in the ChIPModules approach of Table 2 are within 270 bp of each other.
We next applied other bioinformatics approaches to our data sets to compare their performances to our ChIPModules approach. The Web-based programs oPOSSUM (Ho-Sui et al. 2005) and CONFAC (Karanam and Moreno 2004) were chosen since both programs apply comparative genomics, using human and mouse homologous conservation information, to identify overrepresented binding sites. A detailed comparison of the results obtained using all of the different approaches is shown in Table 2. Different sets of motifs were identified using the different approaches, most likely due to the different statistical methods used in identifying overrepresented motifs and to the different background sequences chosen for control data sets. Also, our ChIPModules approach uses an advanced CART model in addition to the traditional statistical test and narrows down the number of motifs to only those within a short distance (270 bp) of an OCT4-binding site.
Experimental validation for the ChIPModule of OCT4 and SRY
Although the different bioinformatics approaches identified different sets of colocalizing motifs, the SRY motif was identified in three of the four analyses. SRY is a transcription factor that shows very restricted expression patterns; it is expressed at high levels mainly in the testis. Because Ntera2 cells are derived from a testicular germ cell tumor, they express SRY. Therefore, we chose to test whether SRY did, in fact, bind to promoters within 270 bp of the OCT4-binding sites in Ntera2 cells. For these experiments, we performed ChIP-chip experiments with antibodies to OCT4 and SRY; each immunoprecipitation was performed in duplicate using two independent cultures of Ntera2 cells. Amplicons prepared from each of the four samples were then analyzed in ChIP-chip experiments. We used arrays that correspond to −1300 to +200 of ∼24,100 different regions that correspond to ∼18,000 annotated human genes. Using the peakCalling program developed in our previous study (Jin et al. 2006), we identified a set of 1104 OCT4 targets and a set of 1344 SRY targets (each target was identified by the particular antibody in both of the ChIP-chip experiments). We note that a relatively small number of OCT4 target promoters is detected using the 1.5-kb arrays; only 1000–3000 targets were identified, which is much less than the 15,400 predicted from analysis of 1% of the genome on ENCODE arrays. This is due to the fact that ∼16% of OCT4 targets bind in proximal upstream promoter regions (see Fig. 4). Extrapolation of the promoter array data again suggests there could be as many as 15,000 OCT4-binding sites in the entire human genome. When the OCT4 and SRY targets were compared, we found that 538 (49%) of the 1104 OCT4 target promoters were also bound by SRY. A Monte Carlo simulation revealed that only 54 common targets were found using randomly generated data sets obtained from the OCT4 and SRY ChIP-chip data (P < 10−6). A list of OCT4 and SRY targets as well as the commonly bound targets is shown in Supplemental Table S1; the entire data set for the four ChIP-chip experiments including the enrichment values for all 24,000 promoters is shown in Supplemental Table S2. We also compared our list of OCT4 targets with the list reported by Boyer et al. (2005). We first determined that 290 of the 603 binding sites identified in the previous study are represented on the NimbleGen 1.5-kb promoter arrays. Of these 290 promoters, 97 (33%) are also in our list of OCT4 targets (see Supplemental Table S3). This low overlap may be due to the fact that Boyer et al. (2005) used embryonic stem cells, whereas the cell line used in this study was a testicular embryonal carcinoma.
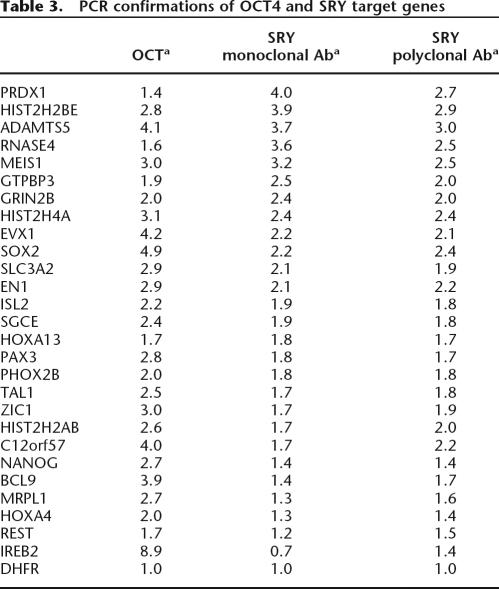
To confirm the array data, we randomly chose a set of 29 promoters identified by the arrays as being OCT4 and SRY target genes and performed PCR reactions using amplicons prepared from OCT4 and SRY ChIP samples that were distinct from the samples used in the duplicate array experiments; a region of the DHFR gene was used as a negative control. The results of the PCR assays are shown in Table 3. Because these are the first identified SRY target genes, we felt that it was critical that care be taken to ensure that the target promoters identified by our ChIP-chip experiments were, in fact, bound by SRY. Therefore, for the confirmation experiments, we performed SRY ChIP assays using both the same SRY antibody as was used for the ChIP-chip experiments as well as an SRY antibody distinct from the one used for the array experiments (a goat polyclonal antibody was used for the ChIP-chip experiments, and a mouse monoclonal antibody was used for the confirmation ChIP experiments). As can be seen in Table 3, promoters identified by ChIP-chip using the goat polyclonal SRY antibody were confirmed to be bound by SRY in independent ChIP assays using both the goat polyclonal and the mouse monoclonal SRY antibodies. Also, of the 1344 SRY targets identified by our ChIP-chip assays, 84% have a match to the SRY_PWM from TRANSFAC, with a core score >1.0 and a PWM score >0.95. (Note: these scores are recommended by the TRANSFAC database.)
Table 3.
PCR confirmations of OCT4 and SRY target genes
Two different antibodies were used to prepare SRY ChIP samples for confirmation PCR assays; the targets in this list were identified originally by ChIP-chip using the SRY polyclonal antibody.
aThe values in the table represent the fold enrichment, which was compared to the enrichment of the total input and normalized to the negative control. The signals were within the linear range of the assay, providing a semiquantitative analysis. For each of these experiments, IgG ChIP samples were performed; the promoters did not show enrichment in the IgG samples.
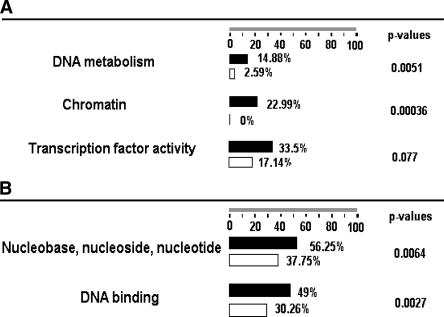
To better understand the biological functions for these identified OCT4 and SRY target genes, we applied the FatiGO program (Al-Shahrour et al. 2005), which is publicly available online at http://fatigo.bioinfo.cipf.es/, to characterize the promoters that are bound by both OCT4 and SRY versus a group of promoters bound only by OCT4 or only by SRY (Fig. 6). The FatiGO program is designed to compare (according to GO annotations) two sets of genes identified from large-scale experiments and to identify categories of genes that are significantly overrepresented in one set versus the other set. Using the FatiGO program, we found that 277 of the 538 common targets of OCT4 and SRY, 229 of 566 OCT4-only targets, and 415 of the 806 SRY-only targets have GO annotations. We first compared the OCT4 + SRY target set to the set of promoters bound by OCT4 but not by SRY. As shown in Figure 6A, genes in the categories of DNA metabolism, chromatin, and transcriptional activity are significantly overrepresented in the common targets group than they are in OCT4-only targets. We next compared the OCT4 + SRY set to the SRY-only set. We found that genes with nucleobase, nucleoside, nucleotide, and DNA binding are more significant in the common targets compared to SRY-only targets (Fig. 6B).
Figure 6.
(A) The comparison of GO annotations for the 538 common targets of OCT4 and SRY (black bars) versus 566 OCT4 targets only (white bars). (B) The comparison of GO annotations for the 538 common targets of OCT4 and SRY (black bars) versus 806 SRY (white bars) targets only.
Target genes bound by OCT4 and SRY are also bound by the transcriptional repressor KAP1
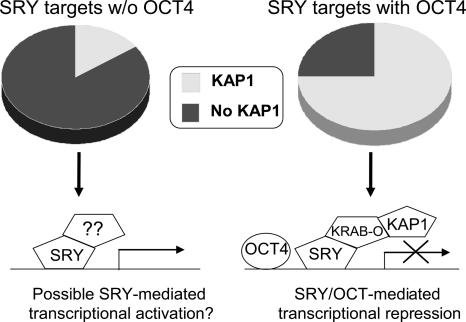
There is very little known about the mechanisms by which SRY may regulate transcription. However, two proteins implicated in nuclear import function, calmodulin and importin beta, have been identified as interacting with SRY (Sweitzer and Hanover 1996; Forwood et al. 2001). Also, previous studies (Poulat et al. 1997; Oh et al. 2005) have used human or mouse SRY as bait in yeast two-hybrid screens to identify SRY-interacting protein (SIP-1) and KRAB only (KRAB-O). KRAB-O is encoded by an alternatively spliced transcript of ZNF208, a zinc-finger-containing gene (Oh and Lau 2005; Oh et al. 2005). However, the KRAB-O transcript does not contain the zinc fingers normally found in ZNF208, suggesting that recruitment of the KRAB-O protein to the DNA may require interaction with a site-specific DNA-binding protein (such as SRY). KRAB-O encodes a protein that contains a KRAB domain, a highly conserved protein domain found in about one-third of Kruppel-type (C2H2) zinc-finger domain proteins. The KRAB domain binds the KRAB-associating protein KAP1 (Friedman et al. 1996; Kim and Shapiro 1996; Looman et al. 2002). KAP1 is thought to act as a scaffolding protein to recruit chromatin-modifying enzymes and the transcriptional repressor HP1. It has been hypothesized that SRY functions as a transcriptional repressor via interaction with KRAB-O and KAP1. However, it is also possible that SRY can, in some cases, function as a transcriptional activator. The mechanism(s) by which SRY regulates transcription have not been elucidated due to the lack of known target genes. Having identified a large set of SRY target genes, we could test various models of SRY function. In particular, we were interested in determining (1) if SRY target genes are bound by KAP1; and (2) if the set of target promoters bound by both SRY and OCT4 has different characteristics from the set of promoters bound only by SRY.
To determine if SRY target promoters are also bound by KAP1, we used an antibody to KAP1 and performed two independent ChIP-chip assays using NimbleGen 1.5-kb human promoter arrays. We compared the overlap of the top KAP1 targets with the top SRY targets in each of the two experiments and found that, in both cases, 48% of the target genes were the same. Thus, our experiments confirm the immunofluorescence colocalization studies of SRY and KAP1 (Oh et al. 2005). We then separated the SRY target promoters into those also bound by OCT4 versus those bound only by SRY. Examining the KAP1 target list, we found that 75% of the SRY targets also bound by OCT4 were bound by KAP1, whereas only 15% of the SRY targets not bound by OCT4 were bound by KAP1. This analysis suggests that SRY might function in two different ways, as a transcriptional repressor in a complex containing OCT4 and KAP1, and in a complex that lacks both OCT4 and KAP1.
Discussion
OCT4 is a key regulator in maintaining the pluripotency and self-renewal of human embryonic stem cells, germ cells, and tumor cells. Recent studies have identified a set of target genes for OCT4 in different human or mouse embryonic stem cells (Boyer et al. 2005; Loh et al. 2006; Player et al. 2006; Squazzo et al. 2006). Based on this previous work, several factors have been identified that regulate a common set of genes as does OCT4. For example, SOX2 and NANOG have been implicated as positive coregulatory factors with OCT4 (Boyer et al. 2005; Loh et al. 2006) in embryonic stem cells. This study, using an integration of high-throughput experimental techniques and computational approaches, suggests that OCT4 may also cooperate with SRY to regulate a set of genes in Ntera2 cells.
Integration of experimental data and computational analyses is becoming increasingly important as more and more data sets are generated from high-throughput technologies. As discussed in a recent review (Elnitski et al. 2006), the analysis of ChIP-chip data using various computational approaches is providing new opportunities to understand transcription networks. Several recent studies have used ChIP-chip data to systematically identify colocalizing transcription factors (Blanchette et al. 2006; Das et al. 2006; Jin et al. 2006). Although these approaches have proven to be quite useful in identifying cis-regulatory modules, the PWMs used in the previous studies have come either from the TRANSFAC database (Jin et al. 2006) or from a few binding sites from previously known target promoters (Cheng et al. 2006). Having available a collection of PWMs derived from in vivo binding sites would be very useful for understanding transcription networks. Toward this goal, a few previous studies (Liu et al. 2002; Harbison et al. 2004; Carroll et al. 2005; Johnson et al. 2006) have derived motifs from sets of in vivo binding sites. In this present study, we have independently developed a de novo motif discovery approach (ChIPMotifs) that can derive in vivo PWMs using ChIP-chip data obtained from arrays that represent 1% of the human genome (The ENCODE Project Consortium 2004). ChIPMotifs (Fig. 1) first detects motifs using ab initio motif programs such as Weeder or MEME and then selects the best motifs using cutoff thresholds defined by a robust, unbiased nonparametric bootstrap resampling method. Most ab initio motif programs identify motifs using only one set of background sequences. When compared to other discriminative motifs approaches (Robison et al. 1998; Workman and Stormo 2000; Benitez-Bellon et al. 2002; Djordjevic et al. 2003; Sinha 2003; Harbison et al. 2004; MacIsaac et al. 2006), the major difference in our approach is that a modified bootstrap resampling strategy is incorporated into our approach so that we can statistically determine the level of stringency of identified motifs using both positive (enriched) and negative (nonenriched) ChIP-chip data sets.
We applied our ChIPMotifs approach to a set of 154 OCT4 target promoters identified by ChIP-chip using ENCODE arrays. We were able to recover an OCT4-binding site motif, NATG CAAANN (p = 0.00026), which has 7-bp matching with the conventional canonical OCT4 motif. We then used the new OCT4 PWM derived from the ChIPMotifs approach to identify regulatory modules using our previously described ChIPModules protocol (Jin et al. 2006), identifying five motifs that colocalize with the OCT4H_PWM (SRY, E2F, P53, TST1, and PAX2). SRY was also identified as one of the top 10 motifs when we applied the program oPOSSUM to our OCT4 ChIP-chip data. Therefore, we chose the OCT4 + SRY regulatory module for further validation, performing ChIP-chip experiments using antibodies to both OCT4 and SRY. We found that the overlap of the top ranked OCT4 and SRY targets (49%) is similar to the 50% overlap of OCT4 and SOX2 targets in human H9 ES cells (Boyer et al. 2005) and the 45% overlap of Oct4 and Nanog targets in E14 mouse ES cells (Loh et al. 2006). Interestingly, both SRY and SOX2 belong to the Sox family (Sry-type high-mobility group [HMG] box) of transcription factors, suggesting that OCT4 may interact with a variety of HMG-box proteins.
SRY is a key regulator of the development of the male gonads and is critical for normal male sex determination (Koopman et al. 2001; Nikolova and Vilain 2006). However, no direct targets of SRY had been identified before this study. The expression of several genes has been shown to be influenced by SRY activity, such as Sox9 (Harley et al. 2003) and Wilm’s Tumor suppressor (Hossain and Saunders 2001). However, direct binding of SRY to the promoter regions of these genes has not been demonstrated. Thus, the ∼1000 SRY target genes that we have identified represent a major advance in the field of testis differentiation and will aid in future studies aimed at dissecting the function of SRY in male sex determination. Using our ChIP-chip analyses, we also tested a previously proposed model for SRY function by demonstrating that a large percentage of SRY target genes are also bound by the KAP1 transcriptional repressor. Interestingly, if we divide the SRY target promoters into subsets that are bound versus not bound by OCT4, we found that the three proteins (OCT4, SRY, and KAP1) often colocalize (see Fig. 7). This suggests that the SRY + OCT4 cis-module that we describe in this study helps to identify a specific set of promoters that may be repressed by SRY. A preliminary analysis, using Illumina Sentrix Beadchips, of the expression levels of the set of genes whose promoters are bound by OCT4 and SRY plus KAP1 indicates that the majority of the genes have very low expression levels in Ntera2 cells (Krig et al. 2007).
Figure 7.
OCT4, SRY, and KAP1 bind to a common set of target promoters. Using OCT4 and SRY ChIP-chip experiments from the same set of cross-linked Ntera2 cells, 880 of the SRY targets were classified as SRY+ and OCT4+ because of their presence in the top 2000 ranked promoters from both lists. To identify the SRY+ and OCT4− promoters, the top 2000 SRY targets were ranked according to their OCT4 enrichment values, and the bottom 389 promoters were chosen. Then, these two sets of promoters (SRY+ OCT4+ and SRY+ OCT4−) were compared to the top 1000 KAP1 targets.
In summary, the computational identification and experimental confirmation of a common set of OCT4 and SRY targets demonstrate that our ChIPMotifs and ChIPModules approaches can work in concert to efficiently mine ChIP-chip data, allowing the development of de novo motifs and the identification of new cis-regulatory modules. Importantly, these approaches allow investigators to develop a PWM for a given factor and search for colocalizing motifs using experimentally identified in vivo binding sites from a specific cell type. This will greatly enable investigations into possible cell-type specificity in the set of target genes and interaction partners for various mammalian transcription factors.
Methods
ChIP-chip data used for modeling
Data set 1—ENCODE regions bound by OCT4
A set of 154 human OCT4 enrichment binding sequences in Ntera2 cells was identified from human ENCODE arrays at the L1 level (p < 0.001) from three biological replicates and used for finding the de novo motifs.
Data set 2—ENCODE regions not bound by OCT4
A set of 499 sequences from the ENCODE regions, each having an average length of 500 bp, that were not bound by OCT4 in Ntera2 cells was selected; each selected sequence was within 10 kb of a promoter region of a known gene.
Data set 3—core promoters bound by OCT4
A set of 293 human OCT4 target promoters in Ntera2 cells was identified from human minipromoter arrays at a rank of the top 500 overlapped targets based on median values from two biological replicate ChIP-chip experiments (see the Supplemental Material for the design of the minipromoter array and processing the data set). Of these 293 targets, 187 have human and mouse orthologous pairs and were further considered to be a positive data set of OCT4 target promoters for training by our ChIPModules approach.
Data set 4—core promoters not bound by OCT4
A set of 3323 human promoters not bound by OCT4 in Ntera2 cells was retrieved from human minipromoter arrays at a rank of bottom 5000 overlapped targets based on median values from two biological replicates. Of these 3323 promoters, 200 show intensities >1.0 in both replicates of the POLR2A ChIP sample and were therefore used as a negative control set. Of these 200 promoters, 94 have human and mouse orthologous pairs and were considered to be a negative data set of non-OCT4 target promoters.
Promoter sequence retrieval
Orthologous promoter sequences, corresponding attributes, and annotation data were retrieved from an integrated information resource (Palaniswamy et al. 2005; http://bioinformatics.med.ohio-state.edu/OMGProm). For the regions identified from the ENCODE arrays, flanking sequences of 1 kb upstream of to 1 kb downstream from each binding region were analyzed. For the target genes identified from the promoter arrays, sequences from 1.3 kb upstream of to 200 bp downstream from each target were analyzed. The sequences were then aligned to mouse orthologous promoter sequences, examining from 10 kb upstream to 10 kb downstream of the transcriptional start site for the orthologous mouse gene using the program ClustalW (Thompson et al. 1994).
Bootstrap resampling strategy
The bootstrap resampling approach is used as a statistical method to determine the optimal cutoffs for the PWM of its motif. Let us consider a set of PWM scores Y = (y1, y2, . . . , yn) predicted from a set of training sequences S = (s1, s2, . . . , sn) for a given transcription factor, TF; F(Y) is the distribution of Y. B is the number of independent random data sets to be generated from the training data set S1*, S2*, . . . , SB*, where S* = (s1*, s2*, . . . , sn*), and each random set has a new set of PWM scores: Y1*, Y2*, . . . , YB*, where Y* = (y1*, y2*, . . . , yn*). Since the F(Y) is an unknown distribution for our PWM scores, therefore we first form the empirical distribution function (EDF) Fn(.| Y). p* is a statistical parameter for F(Y*). The details of the bootstrap resampling process are: Given a set of training sequences S = (s1, s2, . . . , sn) for a given transcription factor TF, generate a set of PWM scores Y = (y1, y2, . . . , yn) from S. For i = 1 to B, for j = 1 to n, first generate sij and then predict yij from sij. Next j. Form the EDF Fn(.|Yi*). Next i. Calculate p(Y*) for F(Y*).
Other approaches have also been used to identify motifs. These approaches all start with a seed matrix (or several seed matrices) identified by other ab initio motif programs such as MDScan, Weeder, MEME, and AlignACE from ChIP-chip data or use known binding profiles (such as from the TRANSFAC database), then further incorporate some statistical methods or machine learning algorithms to discriminate the positive set from the negative set. As described above, our approach uses a modified bootstrap resampling procedure and the Fisher test; DMOTIFs (Sinha 2003) uses a probabilistic analysis with a computed P-value or score; MotifBooster (Hong et al. 2005b) uses a modified confidence-rated boosting (CRB); and THEME (MacIsaac et al. 2006) uses a support vector machine (SVM). We believe that each of these approaches works well on its specific test data sets and is very suitable for a particular array platform. However, besides discriminating the positive data from negative data, our ChIPMotifs approach determines PWM cutoffs, which can be further used to train other ChIP-chip data sets and to identify cis-regulatory modules (see next section).
Identification of cis-regulatory modules
The OCT4 PWM discovered from our ChIPMotifs approach was used for identifying the best OCT4-binding site in each target region. A sliding-window method similar to the method used in Sandelin et al. (2004b) was then used to measure the degree of conservation of a located OCT4-binding site in a pair of orthologous sequences. A site (M) is considered to be conserved if there is at least one site for a given factor in the orthologous sequences within a given window size (e) and the scores are greater than a threshold (T), where T is a user-defined parameter. Binding sites for other transcription factors were identified by the MATCH (Kel et al. 2003) program using the PWMs from the TRANSFAC database (Wingender et al. 2000). For each pair of human and mouse orthologous promoters, we searched for ∼500 PWMs corresponding to ∼300 known human transcription factors using the “minFN_good83.prf” profile (profile of cutoff values with minimum number of false-negative predictions) of MATCH. Each predicted binding site was determined by four parameters: (1) the human core score (Sc_h); (2) the human PWM score (Sp_h); (3) the mouse core score (Sc_m); and (4) the mouse PWM core score (Sp_m). The core and PWM scores, ranging from 0 (worst) to 1 (best), reflect the similarity of predicted sites to the core of the consensus and to the full consensus sequence. The conservation of other binding sites was determined by the percentage of identical base pairs from the ClustalW-aligned sequences.
Calculation of Sn and Sp
A true positive rate termed as sensitivity (Sn), and a true negative rate termed as specificity (Sp) were calculated by the following formulas:
and
where both TP (a true positive) and TN (a true negative) are correct classifications, and both FP (a false positive) and FN (a false negative) are incorrect classifications.
ChIP-chip assays
Ntera2 cells were grown in Dulbecco’s Modified Eagle’s Medium supplemented with 2 mM glutamine, 100 units/mL penicillin and streptomycin, and 10% fetal bovine serum. All cells were incubated at 37°C in a humidified 5% CO2 incubator. ChIP assays (1 × 107 cells/assay) were performed following the protocol provided at http://genomics.ucdavis.edu/farnham/ and http://genomecenter.ucdavis.edu/expression_analysis/. Amplicons were prepared using the whole-genome amplification method (see O’Geen et al. 2006; http://www.genomecenter.ucdavis.edu/farnham/protocol.html; and the Supplemental Material). The OCT4 antibody used in this study was purchased from Santa Cruz Biotechnology (cat# sc-8628X). Two different SRY antibodies were used in this study: ChIP samples that were hybridized on the arrays were obtained using an antibody from Santa Cruz Biotechnology (cat# sc-8232X), while ChIP samples used for PCR validation of the target genes were obtained using both the original antibody and an antibody purchased from Abcam (cat# ab22166). Amplicons were hybridized by the NimbleGen Array Service onto 1.5-kb human promoter arrays created by NimbleGen Systems. For details concerning the generation of amplicons from ChIP samples, see http://genomics.ucdavis.edu/farnham/. For PCR analysis of the ChIP samples prior to amplicon generation, QIA quick-purified immunoprecipitates were dissolved in 50 μL of water, except for input samples, which were dissolved in 100 μL. Standard PCR reactions using 2 μL of the immunoprecipitated DNA were performed. PCR products were separated by electrophoresis through 1.5% agarose gels and visualized using ethidium bromide.
Acknowledgments
This work was supported in part by Public Health Service grants CA45250, HG003129, and DK067889. We also thank the members of the Farnham laboratory for helpful discussions.
Footnotes
[Supplemental material is available online at www.genome.org. The OCT4, SRY, and KAP1 ChIP-chip data has been deposited in GSE6409.]
Article is online at http://www.genome.org/cgi/doi/10.1101/gr.6006107
References
- Aerts S., Van Loo P., Thijs G., Moreau Y., De Moor B., Van Loo P., Thijs G., Moreau Y., De Moor B., Thijs G., Moreau Y., De Moor B., Moreau Y., De Moor B., De Moor B. Computational detection of cis-regulatory modules. Bioinformatics. 2003;19:ii5–ii14. doi: 10.1093/bioinformatics/btg1052. [DOI] [PubMed] [Google Scholar]
- Alkema W.B., Johansson O., Lagergren J., Wasserman W.W., Johansson O., Lagergren J., Wasserman W.W., Lagergren J., Wasserman W.W., Wasserman W.W. MSCAN: Identification of functional clusters of transcription factor binding sites. Nucleic Acids Res. 2004;32:W195–W198. doi: 10.1093/nar/gkh387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Shahrour F., Minguez P., Vaquerizas J.M., Conde L., Dopazo J., Minguez P., Vaquerizas J.M., Conde L., Dopazo J., Vaquerizas J.M., Conde L., Dopazo J., Conde L., Dopazo J., Dopazo J. Babelomics: A suite of Web tools for functional annotation and analysis of groups of genes in high-throughput experiments. Nucleic Acids Res. 2005;33:W460–W464. doi: 10.1093/nar/gki456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T.L., Elkan C., Elkan C. The value of prior knowledge in discovering motifs with MEME. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1995;3:21–29. [PubMed] [Google Scholar]
- Bailey T.L., Gribskov M., Gribskov M. Score distributions for simultaneous matching to multiple motifs. J. Comput. Biol. 1997;4:45–59. doi: 10.1089/cmb.1997.4.45. [DOI] [PubMed] [Google Scholar]
- Benitez-Bellon E., Moreno-Hagelsieb G., Collado-Vides J., Moreno-Hagelsieb G., Collado-Vides J., Collado-Vides J. Evaluation of thresholds for the detection of binding sites for regulatory proteins in Escherichia coli K12 DNA. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-3-research0013. RESEARCH0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieda M., Xu X., Singer M., Green R., Farnham P.J., Xu X., Singer M., Green R., Farnham P.J., Singer M., Green R., Farnham P.J., Green R., Farnham P.J., Farnham P.J. Unbiased location analysis of E2F1 binding sites suggests a widespread role for E2F1 in the human genome. Genome Res. 2006;16:595–605. doi: 10.1101/gr.4887606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette M., Bataille A.R., Chen X., Poitras C., Laganière J., Lefèbvre C., Deblois G., Giguère V., Ferretti V., Bergeron D., Bataille A.R., Chen X., Poitras C., Laganière J., Lefèbvre C., Deblois G., Giguère V., Ferretti V., Bergeron D., Chen X., Poitras C., Laganière J., Lefèbvre C., Deblois G., Giguère V., Ferretti V., Bergeron D., Poitras C., Laganière J., Lefèbvre C., Deblois G., Giguère V., Ferretti V., Bergeron D., Laganière J., Lefèbvre C., Deblois G., Giguère V., Ferretti V., Bergeron D., Lefèbvre C., Deblois G., Giguère V., Ferretti V., Bergeron D., Deblois G., Giguère V., Ferretti V., Bergeron D., Giguère V., Ferretti V., Bergeron D., Ferretti V., Bergeron D., Bergeron D., et al. Genome-wide computational prediction of transcriptional regulatory modules reveals new insights into human gene expression. Genome Res. 2006;16:656–668. doi: 10.1101/gr.4866006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer L.A., Lee T.I., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Lee T.I., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Kumar R.M., Murray H.L., Jenner R.G., Murray H.L., Jenner R.G., Jenner R.G., et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman L., Friedman J.H., Olshen R.A., Stone C.J., Friedman J.H., Olshen R.A., Stone C.J., Olshen R.A., Stone C.J., Stone C.J. Classification and regression trees. Chapman & Hall; New York: 1984. [Google Scholar]
- Carroll J.S., Liu X.S., Brodsky A.S., Li W., Meyer C.A., Szary A.J., Eeckhoute J., Shao W., Hestermann E.V., Geistlinger T.R., Liu X.S., Brodsky A.S., Li W., Meyer C.A., Szary A.J., Eeckhoute J., Shao W., Hestermann E.V., Geistlinger T.R., Brodsky A.S., Li W., Meyer C.A., Szary A.J., Eeckhoute J., Shao W., Hestermann E.V., Geistlinger T.R., Li W., Meyer C.A., Szary A.J., Eeckhoute J., Shao W., Hestermann E.V., Geistlinger T.R., Meyer C.A., Szary A.J., Eeckhoute J., Shao W., Hestermann E.V., Geistlinger T.R., Szary A.J., Eeckhoute J., Shao W., Hestermann E.V., Geistlinger T.R., Eeckhoute J., Shao W., Hestermann E.V., Geistlinger T.R., Shao W., Hestermann E.V., Geistlinger T.R., Hestermann E.V., Geistlinger T.R., Geistlinger T.R., et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Cheng A.S., Jin V.X., Fan M., Smith L.T., Liyanarachchi S., Yan P.S., Leu Y.W., Chan M.W., Plass C., Nephew K.P., Jin V.X., Fan M., Smith L.T., Liyanarachchi S., Yan P.S., Leu Y.W., Chan M.W., Plass C., Nephew K.P., Fan M., Smith L.T., Liyanarachchi S., Yan P.S., Leu Y.W., Chan M.W., Plass C., Nephew K.P., Smith L.T., Liyanarachchi S., Yan P.S., Leu Y.W., Chan M.W., Plass C., Nephew K.P., Liyanarachchi S., Yan P.S., Leu Y.W., Chan M.W., Plass C., Nephew K.P., Yan P.S., Leu Y.W., Chan M.W., Plass C., Nephew K.P., Leu Y.W., Chan M.W., Plass C., Nephew K.P., Chan M.W., Plass C., Nephew K.P., Plass C., Nephew K.P., Nephew K.P., et al. Combinatorial analysis of transcription factor partners reveals recruitment of c-Myc to estrogen receptor-α responsive promoters. Mol. Cell. 2006;21:393–404. doi: 10.1016/j.molcel.2005.12.016. [DOI] [PubMed] [Google Scholar]
- Crawford G.E., Davis S., Scacheri P.C., Renaud G., Halawi M.J., Erdos M.R., Green R., Meltzer P.S., Wolfsberg T.G., Collins F.S., Davis S., Scacheri P.C., Renaud G., Halawi M.J., Erdos M.R., Green R., Meltzer P.S., Wolfsberg T.G., Collins F.S., Scacheri P.C., Renaud G., Halawi M.J., Erdos M.R., Green R., Meltzer P.S., Wolfsberg T.G., Collins F.S., Renaud G., Halawi M.J., Erdos M.R., Green R., Meltzer P.S., Wolfsberg T.G., Collins F.S., Halawi M.J., Erdos M.R., Green R., Meltzer P.S., Wolfsberg T.G., Collins F.S., Erdos M.R., Green R., Meltzer P.S., Wolfsberg T.G., Collins F.S., Green R., Meltzer P.S., Wolfsberg T.G., Collins F.S., Meltzer P.S., Wolfsberg T.G., Collins F.S., Wolfsberg T.G., Collins F.S., Collins F.S. DNase-chip: A high-resolution method to identify DNase I hypersensitive sites using tiled microarrays. Nat. Methods. 2006;3:503–509. doi: 10.1038/NMETH888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das D., Nahle Z., Zhang M.Q., Nahle Z., Zhang M.Q., Zhang M.Q. Adaptively inferring human transcriptional subnetworks. Mol. Syst. Biol. 2006 doi: 10.1038/msb4100067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djordjevic M., Sengupta A.M., Shraiman B.I., Sengupta A.M., Shraiman B.I., Shraiman B.I. A biophysical approach to transcription factor binding site discovery. Genome Res. 2003;13:2381–2390. doi: 10.1101/gr.1271603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Down T.A., Hubbard T.J., Hubbard T.J. NestedMICA: Sensitive inference of over-represented motifs in nucleic acid sequence. Nucleic Acids Res. 2005;33:1445–1453. doi: 10.1093/nar/gki282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efron B. Bootstrap methods: Another look at the jackknife. Ann. Stat. 1979;7:1–26. [Google Scholar]
- Elnitski L., Jin V.X., Farnham P.J., Jones S.J.M., Jin V.X., Farnham P.J., Jones S.J.M., Farnham P.J., Jones S.J.M., Jones S.J.M. Locating mammalian transcription factor binding sites: A survey of computational and experimental techniques. Genome Res. 2006;16:1455–1464. doi: 10.1101/gr.4140006. [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium, The ENCODE (ENCyclopedia of DNA Elements) Project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution Int. J. Org. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Forwood J., Harley V., Jans D.A., Harley V., Jans D.A., Jans D.A. The C-terminal nuclear localization signal of the sex-determining region Y (SRY) high-mobility group domain mediates nuclear import through importin β1. J. Biol. Chem. 2001;276:46575–46582. doi: 10.1074/jbc.M101668200. [DOI] [PubMed] [Google Scholar]
- Friedman J.R., Fredericks W.J., Jensen D.E., Speicher D.W., Huang X.-P., Neilson E.G., Rauscher F.J., III, Fredericks W.J., Jensen D.E., Speicher D.W., Huang X.-P., Neilson E.G., Rauscher F.J., III, Jensen D.E., Speicher D.W., Huang X.-P., Neilson E.G., Rauscher F.J., III, Speicher D.W., Huang X.-P., Neilson E.G., Rauscher F.J., III, Huang X.-P., Neilson E.G., Rauscher F.J., III, Neilson E.G., Rauscher F.J., III, Rauscher F.J., III KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes & Dev. 1996;10:2067–2078. doi: 10.1101/gad.10.16.2067. [DOI] [PubMed] [Google Scholar]
- Gupta M., Liu J.S., Liu J.S. De novo cis-regulatory module elicitation for eukaryotic genomes. Proc. Natl. Acad. Sci. 2005;102:7079–7084. doi: 10.1073/pnas.0408743102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison C.T., Gordon D.B., Lee T.I., Rinaldi N.J., Macisaac K.D., Danford T.W., Hannett N.M., Tagne J.B., Reynolds D.B., Yoo J., Gordon D.B., Lee T.I., Rinaldi N.J., Macisaac K.D., Danford T.W., Hannett N.M., Tagne J.B., Reynolds D.B., Yoo J., Lee T.I., Rinaldi N.J., Macisaac K.D., Danford T.W., Hannett N.M., Tagne J.B., Reynolds D.B., Yoo J., Rinaldi N.J., Macisaac K.D., Danford T.W., Hannett N.M., Tagne J.B., Reynolds D.B., Yoo J., Macisaac K.D., Danford T.W., Hannett N.M., Tagne J.B., Reynolds D.B., Yoo J., Danford T.W., Hannett N.M., Tagne J.B., Reynolds D.B., Yoo J., Hannett N.M., Tagne J.B., Reynolds D.B., Yoo J., Tagne J.B., Reynolds D.B., Yoo J., Reynolds D.B., Yoo J., Yoo J., et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley V.R., Clarkson M.J., Argentaro A., Clarkson M.J., Argentaro A., Argentaro A. The molecular action and regulation of the testis-determining factors, SRY (sex-determining region on the Y chromosome) and SOX9 (SRY-related high-mobility group [HMG] box 9) Endocr. Rev. 2003;24:466–487. doi: 10.1210/er.2002-0025. [DOI] [PubMed] [Google Scholar]
- Harrow J., Denoeud F., Frankish A., Reymond A., Chen C., Chrast J., Lagarde J., Gilbert J.G.R., Storey R., Swarbreck D., Denoeud F., Frankish A., Reymond A., Chen C., Chrast J., Lagarde J., Gilbert J.G.R., Storey R., Swarbreck D., Frankish A., Reymond A., Chen C., Chrast J., Lagarde J., Gilbert J.G.R., Storey R., Swarbreck D., Reymond A., Chen C., Chrast J., Lagarde J., Gilbert J.G.R., Storey R., Swarbreck D., Chen C., Chrast J., Lagarde J., Gilbert J.G.R., Storey R., Swarbreck D., Chrast J., Lagarde J., Gilbert J.G.R., Storey R., Swarbreck D., Lagarde J., Gilbert J.G.R., Storey R., Swarbreck D., Gilbert J.G.R., Storey R., Swarbreck D., Storey R., Swarbreck D., Swarbreck D., et al. GENCODE: Producing a reference annotation for ENCODE. Genome Biol. 2006;7:S4. doi: 10.1186/gb-2006-7-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haverty P.M., Hansen U., Weng Z., Hansen U., Weng Z., Weng Z. Computational inference of transcriptional regulatory networks from expression profiling and transcription factor binding site identification. Nucleic Acids Res. 2004;32:179–188. doi: 10.1093/nar/gkh183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz G.Z., Stormo G.D., Stormo G.D. Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics. 1999;15:563–577. doi: 10.1093/bioinformatics/15.7.563. [DOI] [PubMed] [Google Scholar]
- Hong P., Liu X.S., Zhou Q., Lu X., Liu J.S., Wong W.H., Liu X.S., Zhou Q., Lu X., Liu J.S., Wong W.H., Zhou Q., Lu X., Liu J.S., Wong W.H., Lu X., Liu J.S., Wong W.H., Liu J.S., Wong W.H., Wong W.H. A boosting approach for motif modeling using ChIP-chip data. Bioinformatics. 2005a;21:2636–2643. doi: 10.1093/bioinformatics/bti402. [DOI] [PubMed] [Google Scholar]
- Hong P., Liu X.S., Zhou Q., Lu X., Liu J.S., Wong W.H., Liu X.S., Zhou Q., Lu X., Liu J.S., Wong W.H., Zhou Q., Lu X., Liu J.S., Wong W.H., Lu X., Liu J.S., Wong W.H., Liu J.S., Wong W.H., Wong W.H. A boosting approach for motif modeling using ChIP-chip data. Bioinformatics. 2005b;21:2636–2643. doi: 10.1093/bioinformatics/bti402. [DOI] [PubMed] [Google Scholar]
- Hossain A., Saunders G.F., Saunders G.F. The human sex-determining gene SRY is a direct target of WT1. J. Biol. Chem. 2001;276:16817–16823. doi: 10.1074/jbc.M009056200. [DOI] [PubMed] [Google Scholar]
- Ho-Sui S.J., Mortimer J., Arenillas D.J., Brumm J., Walsh C.J., Kennedy B.P., Wasserman W.W., Mortimer J., Arenillas D.J., Brumm J., Walsh C.J., Kennedy B.P., Wasserman W.W., Arenillas D.J., Brumm J., Walsh C.J., Kennedy B.P., Wasserman W.W., Brumm J., Walsh C.J., Kennedy B.P., Wasserman W.W., Walsh C.J., Kennedy B.P., Wasserman W.W., Kennedy B.P., Wasserman W.W., Wasserman W.W. oPOSSUM: Identification of over-represented transcription factor binding sites in co-expressed genes. Nucleic Acids Res. 2005;33:3154–3164. doi: 10.1093/nar/gki624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer V.R., Horak C.E., Scafe C.S., Botstein D., Snyder M., Brown P.O., Horak C.E., Scafe C.S., Botstein D., Snyder M., Brown P.O., Scafe C.S., Botstein D., Snyder M., Brown P.O., Botstein D., Snyder M., Brown P.O., Snyder M., Brown P.O., Brown P.O. Genomic binding sites of the yeast cell-cycle transcription factor SBF and MBF. Nature. 2001;409:533–538. doi: 10.1038/35054095. [DOI] [PubMed] [Google Scholar]
- Jin V., Rabinvich A., Squazzo S.L., Green R., Farnham P.J., Rabinvich A., Squazzo S.L., Green R., Farnham P.J., Squazzo S.L., Green R., Farnham P.J., Green R., Farnham P.J., Farnham P.J. A computational genomics approach to identify cis-regulatory modules from chromatin immunoprecipitation microarray data—A case study using E2F1. Genome Res. 2006;16:1585–1595. doi: 10.1101/gr.5520206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson W.E., Li W., Meyer C.A., Gottardo R., Carroll J.S., Brown M., Liu X.S., Li W., Meyer C.A., Gottardo R., Carroll J.S., Brown M., Liu X.S., Meyer C.A., Gottardo R., Carroll J.S., Brown M., Liu X.S., Gottardo R., Carroll J.S., Brown M., Liu X.S., Carroll J.S., Brown M., Liu X.S., Brown M., Liu X.S., Liu X.S. Model-based analysis of tiling arrays for ChIP-chip. Proc. Natl. Acad. Sci. 2006;103:12457–12462. doi: 10.1073/pnas.0601180103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karanam S., Moreno C.S., Moreno C.S. CONFAC: Automated application of comparative genomic promoter analysis to DNA microarray data sets. Nucleic Acids Res. 2004;32:W475–W484. doi: 10.1093/nar/gkh353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kel A.E., Gossling E., Reuter I., Cheremushkin E., Kel-Margoulis O.V., Wingender E., Gossling E., Reuter I., Cheremushkin E., Kel-Margoulis O.V., Wingender E., Reuter I., Cheremushkin E., Kel-Margoulis O.V., Wingender E., Cheremushkin E., Kel-Margoulis O.V., Wingender E., Kel-Margoulis O.V., Wingender E., Wingender E. MATCH: A tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 2003;31:3576–3579. doi: 10.1093/nar/gkg585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Shapiro D.J., Shapiro D.J. In simple synthetic promoters YY1-induced DNA bending is important in transcription activation and repression. Nucleic Acids Res. 1996;24:4341–4348. doi: 10.1093/nar/24.21.4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopman P., Bullejos M., Bowles J., Bullejos M., Bowles J., Bowles J. Regulation of male sexual development by Sry and Sox9. J. Exp. Zool. 2001;290:463–474. doi: 10.1002/jez.1089. [DOI] [PubMed] [Google Scholar]
- Krig S.R., Jin V.X., Bieda M.C., O’Geen H., Yaswen P., Green R., Farnham P.J., Jin V.X., Bieda M.C., O’Geen H., Yaswen P., Green R., Farnham P.J., Bieda M.C., O’Geen H., Yaswen P., Green R., Farnham P.J., O’Geen H., Yaswen P., Green R., Farnham P.J., Yaswen P., Green R., Farnham P.J., Green R., Farnham P.J., Farnham P.J. Identification of genes directly regulated by the oncogene ZNF217 using chromatin immunoprecipitation (ChIP)-chip assays. J. Biol. Chem. 2007;282:9703–9712. doi: 10.1074/jbc.M611752200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivan W., Wasserman W.W., Wasserman W.W. A predictive model for regulatory sequences directing liver-specific transcription. Genome Res. 2001;11:1559–1566. doi: 10.1101/gr.180601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence C.E., Altschul S.F., Boguski M.S., Liu J.S., Neuwald A.F., Wootton J.C., Altschul S.F., Boguski M.S., Liu J.S., Neuwald A.F., Wootton J.C., Boguski M.S., Liu J.S., Neuwald A.F., Wootton J.C., Liu J.S., Neuwald A.F., Wootton J.C., Neuwald A.F., Wootton J.C., Wootton J.C. Detecting subtle sequence signals: A Gibbs sampling strategy for multiple alignment. Science. 1993;262:208–214. doi: 10.1126/science.8211139. [DOI] [PubMed] [Google Scholar]
- Li H., Chen H., Bao L., Manly K.F., Chesler E.J., Lu L., Wang J., Zhou M., Williams R.W., Cui Y., Chen H., Bao L., Manly K.F., Chesler E.J., Lu L., Wang J., Zhou M., Williams R.W., Cui Y., Bao L., Manly K.F., Chesler E.J., Lu L., Wang J., Zhou M., Williams R.W., Cui Y., Manly K.F., Chesler E.J., Lu L., Wang J., Zhou M., Williams R.W., Cui Y., Chesler E.J., Lu L., Wang J., Zhou M., Williams R.W., Cui Y., Lu L., Wang J., Zhou M., Williams R.W., Cui Y., Wang J., Zhou M., Williams R.W., Cui Y., Zhou M., Williams R.W., Cui Y., Williams R.W., Cui Y., Cui Y. Integrative genetic analysis of transcription modules: Toward filling the gap between genetic loci and inherited traits. Hum. Mol. Genet. 2006;15:481–492. doi: 10.1093/hmg/ddi462. [DOI] [PubMed] [Google Scholar]
- Liu X.S., Brutlag D.L., Liu J.S., Brutlag D.L., Liu J.S., Liu J.S. An algorithm for finding protein–DNA binding sites with applications to chromatin-immunoprecipitation microarray experiments. Nat. Biotechnol. 2002;20:835–839. doi: 10.1038/nbt717. [DOI] [PubMed] [Google Scholar]
- Lockhart D., Dong H., Byrne M.C., Follettie M.T., Gallo M.V., Chee M.S., Mittmann M., Wang C., Kobayashi M., Horton H., Dong H., Byrne M.C., Follettie M.T., Gallo M.V., Chee M.S., Mittmann M., Wang C., Kobayashi M., Horton H., Byrne M.C., Follettie M.T., Gallo M.V., Chee M.S., Mittmann M., Wang C., Kobayashi M., Horton H., Follettie M.T., Gallo M.V., Chee M.S., Mittmann M., Wang C., Kobayashi M., Horton H., Gallo M.V., Chee M.S., Mittmann M., Wang C., Kobayashi M., Horton H., Chee M.S., Mittmann M., Wang C., Kobayashi M., Horton H., Mittmann M., Wang C., Kobayashi M., Horton H., Wang C., Kobayashi M., Horton H., Kobayashi M., Horton H., Horton H., et al. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat. Biotechnol. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- Loh Y.-H., Wu Q., Chew J.-L., Vega V.B., Zhang W., Chen X., Bourque G., George J., Leong B., Liu J., Wu Q., Chew J.-L., Vega V.B., Zhang W., Chen X., Bourque G., George J., Leong B., Liu J., Chew J.-L., Vega V.B., Zhang W., Chen X., Bourque G., George J., Leong B., Liu J., Vega V.B., Zhang W., Chen X., Bourque G., George J., Leong B., Liu J., Zhang W., Chen X., Bourque G., George J., Leong B., Liu J., Chen X., Bourque G., George J., Leong B., Liu J., Bourque G., George J., Leong B., Liu J., George J., Leong B., Liu J., Leong B., Liu J., Liu J., et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- Looman C., Abrink M., Mark C., Hellman L., Abrink M., Mark C., Hellman L., Mark C., Hellman L., Hellman L. KRAB zinc finger proteins: An analysis of the molecular mechanisms governing their increase in numbers and complexity during evolution. Mol. Biol. Evol. 2002;19:2118–2130. doi: 10.1093/oxfordjournals.molbev.a004037. [DOI] [PubMed] [Google Scholar]
- MacIsaac K.D., Gordon D.B., Nekludova L., Odom D.T., Schreiber J., Gifford D.K., Young R.A., Fraenkel E., Gordon D.B., Nekludova L., Odom D.T., Schreiber J., Gifford D.K., Young R.A., Fraenkel E., Nekludova L., Odom D.T., Schreiber J., Gifford D.K., Young R.A., Fraenkel E., Odom D.T., Schreiber J., Gifford D.K., Young R.A., Fraenkel E., Schreiber J., Gifford D.K., Young R.A., Fraenkel E., Gifford D.K., Young R.A., Fraenkel E., Young R.A., Fraenkel E., Fraenkel E. A hypothesis-based approach for identifying the binding specificity of regulatory proteins from chromatin immunoprecipitation data. Bioinformatics. 2006;22:423–429. doi: 10.1093/bioinformatics/bti815. [DOI] [PubMed] [Google Scholar]
- Nikolova G., Vilain E., Vilain E. Mechanisms of disease: Transcription factors in sex determination—Relevance to human disorders of sex development. Nat. Clin. Pract. Endocrinol. Metab. 2006;2:231–238. doi: 10.1038/ncpendmet0143. [DOI] [PubMed] [Google Scholar]
- O’Geen H., Nicolet C.M., Blahnik K., Green R., Farnham P.J., Nicolet C.M., Blahnik K., Green R., Farnham P.J., Blahnik K., Green R., Farnham P.J., Green R., Farnham P.J., Farnham P.J. Comparison of sample preparation methods for ChIP-chip assays. Biotechniques. 2006;41:577–580. doi: 10.2144/000112268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H.J., Lau Y.F., Lau Y.F. KRAB: A partner for SRY action on chromatin. Mol. Cell. Endocrinol. 2005;247:47–52. doi: 10.1016/j.mce.2005.12.011. [DOI] [PubMed] [Google Scholar]
- Oh H.J., Li Y., Lau Y.-F., Li Y., Lau Y.-F., Lau Y.-F. Sry associates with the heterochromatin protein 1 complex by interacting with a KRAB domain protein. Biol. Reprod. 2005;72:407–415. doi: 10.1095/biolreprod.104.034447. [DOI] [PubMed] [Google Scholar]
- Palaniswamy S.K., Jin V.X., Sun H., Davuluri R.V., Jin V.X., Sun H., Davuluri R.V., Sun H., Davuluri R.V., Davuluri R.V. OMGProm: An integrated resource of orthologous mammalian gene promoters. Bioinformatics. 2005;21:835–836. doi: 10.1093/bioinformatics/bti119. [DOI] [PubMed] [Google Scholar]
- Pavesi G., Mereghetti P., Mauri G., Pesole G., Mereghetti P., Mauri G., Pesole G., Mauri G., Pesole G., Pesole G. Weeder Web: Discovery of transcription factor binding sites in a set of sequences from co-regulated genes. Nucleic Acids Res. 2004;32:W199–W203. doi: 10.1093/nar/gkh465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen J.T., Moult J., Moult J. Genetic algorithms for protein structure prediction. Curr. Opin. Struct. Biol. 1996;6:227–231. doi: 10.1016/s0959-440x(96)80079-0. [DOI] [PubMed] [Google Scholar]
- Pesce M., Scholer H.R., Scholer H.R. Oct-4: Gatekeeper in the beginnings of mammalian development. Stem Cells. 2001;19:271–278. doi: 10.1634/stemcells.19-4-271. [DOI] [PubMed] [Google Scholar]
- Player A., Wang Y., Bhattacharya B., Rao M., Puri R.K., Kawasaki E.S., Wang Y., Bhattacharya B., Rao M., Puri R.K., Kawasaki E.S., Bhattacharya B., Rao M., Puri R.K., Kawasaki E.S., Rao M., Puri R.K., Kawasaki E.S., Puri R.K., Kawasaki E.S., Kawasaki E.S. Comparisons between transcriptional regulation and RNA expression in human embryonic stem cell lines. Stem Cells Dev. 2006;15:315–323. doi: 10.1089/scd.2006.15.315. [DOI] [PubMed] [Google Scholar]
- Poulat F., Barbara P.S., Desclozeaux M., Soullier S., Moniot B., Bonneaud N., Boizet B., Berta P., Barbara P.S., Desclozeaux M., Soullier S., Moniot B., Bonneaud N., Boizet B., Berta P., Desclozeaux M., Soullier S., Moniot B., Bonneaud N., Boizet B., Berta P., Soullier S., Moniot B., Bonneaud N., Boizet B., Berta P., Moniot B., Bonneaud N., Boizet B., Berta P., Bonneaud N., Boizet B., Berta P., Boizet B., Berta P., Berta P. The human testis determining factor SRY binds a nuclear factor containing PDZ protein interaction domains. J. Biol. Chem. 1997;272:7167–7172. doi: 10.1074/jbc.272.11.7167. [DOI] [PubMed] [Google Scholar]
- Ren B., Robert F., Wyrick J.J., Aparicio O., Jennings E.G., Simon I., Zeitlinger J., Schreiber J., Hannett N., Kanin E., Robert F., Wyrick J.J., Aparicio O., Jennings E.G., Simon I., Zeitlinger J., Schreiber J., Hannett N., Kanin E., Wyrick J.J., Aparicio O., Jennings E.G., Simon I., Zeitlinger J., Schreiber J., Hannett N., Kanin E., Aparicio O., Jennings E.G., Simon I., Zeitlinger J., Schreiber J., Hannett N., Kanin E., Jennings E.G., Simon I., Zeitlinger J., Schreiber J., Hannett N., Kanin E., Simon I., Zeitlinger J., Schreiber J., Hannett N., Kanin E., Zeitlinger J., Schreiber J., Hannett N., Kanin E., Schreiber J., Hannett N., Kanin E., Hannett N., Kanin E., Kanin E., et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- Robison K., McGuire A.M., Church G.M., McGuire A.M., Church G.M., Church G.M. A comprehensive library of DNA-binding site matrices for 55 proteins applied to the complete Escherichia coliK-12 genome. J. Mol. Biol. 1998;284:241–254. doi: 10.1006/jmbi.1998.2160. [DOI] [PubMed] [Google Scholar]
- Sandelin A., Alkema W., Engstrom P., Wasserman W.W., Lenhard B., Alkema W., Engstrom P., Wasserman W.W., Lenhard B., Engstrom P., Wasserman W.W., Lenhard B., Wasserman W.W., Lenhard B., Lenhard B. JASPAR: An open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004a;32:D91–D94. doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandelin A., Wasserman W.W., Lenhard B., Wasserman W.W., Lenhard B., Lenhard B. ConSite: Web-based prediction of regulatory elements using cross-species comparison. Nucleic Acids Res. 2004b;32:W249–W252. doi: 10.1093/nar/gkh372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schena M., Shalon D., Davis R.W., Brown P.O., Shalon D., Davis R.W., Brown P.O., Davis R.W., Brown P.O., Brown P.O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- Sharan R., Ovcharenko I., Ben-Hur A., Karp R.M., Ovcharenko I., Ben-Hur A., Karp R.M., Ben-Hur A., Karp R.M., Karp R.M. CREME: A framework for identifying cis-regulatory modules in human–mouse conserved segments. Bioinformatics. 2003;19:i283–i291. doi: 10.1093/bioinformatics/btg1039. [DOI] [PubMed] [Google Scholar]
- Sinha S. Discriminative motifs. J. Comput. Biol. 2003;10:599–615. doi: 10.1089/10665270360688219. [DOI] [PubMed] [Google Scholar]
- Smith A.D., Sumazin P., Das D., Zhang M.Q., Sumazin P., Das D., Zhang M.Q., Das D., Zhang M.Q., Zhang M.Q. Mining ChIP-chip data for transcription factor and cofactor binding sites. Bioinformatics. 2005;(Suppl 1) 21:i403–i412. doi: 10.1093/bioinformatics/bti1043. [DOI] [PubMed] [Google Scholar]
- Squazzo S.L., Komashko V.M., O’Geen H., Krig S., Jin V.X., Jang S.-W., Green R., Margueron R., Reinberg D., Farnham P.J., Komashko V.M., O’Geen H., Krig S., Jin V.X., Jang S.-W., Green R., Margueron R., Reinberg D., Farnham P.J., O’Geen H., Krig S., Jin V.X., Jang S.-W., Green R., Margueron R., Reinberg D., Farnham P.J., Krig S., Jin V.X., Jang S.-W., Green R., Margueron R., Reinberg D., Farnham P.J., Jin V.X., Jang S.-W., Green R., Margueron R., Reinberg D., Farnham P.J., Jang S.-W., Green R., Margueron R., Reinberg D., Farnham P.J., Green R., Margueron R., Reinberg D., Farnham P.J., Margueron R., Reinberg D., Farnham P.J., Reinberg D., Farnham P.J., Farnham P.J. Suz12 silences large regions of the genome in a cell type-specific manner. Genome Res. 2006;16:890–900. doi: 10.1101/gr.5306606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweitzer T.D., Hanover J.A., Hanover J.A. Calmodulin activates nuclear protein import: A link between signal transduction and nuclear transport. Proc. Natl. Acad. Sci. 1996;93:14574–14579. doi: 10.1073/pnas.93.25.14574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J., Higgins D.G., Gibson T.J., Gibson T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B., Henikoff S., Henikoff S. Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase. Nat. Biotechnol. 2000;18:424–428. doi: 10.1038/74487. [DOI] [PubMed] [Google Scholar]
- Wang W., Cherry J.M., Nochomovitz Y., Jolly E., Botstein D., Li H., Cherry J.M., Nochomovitz Y., Jolly E., Botstein D., Li H., Nochomovitz Y., Jolly E., Botstein D., Li H., Jolly E., Botstein D., Li H., Botstein D., Li H., Li H. Inference of combinatorial regulation in yeast transcriptional networks: A case study of sporulation. Proc. Natl. Acad. Sci. 2005;102:1998–2003. doi: 10.1073/pnas.0405537102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman W.W., Fickett J.W., Fickett J.W. Identification of regulatory regions which confer muscle-specific gene expression. J. Mol. Biol. 1998;278:167–181. doi: 10.1006/jmbi.1998.1700. [DOI] [PubMed] [Google Scholar]
- Wingender E., Chen X., Hehl R., Karas H., Matys V., Meinhardt T., Pruss M., Reuter I., Schacherer F., Chen X., Hehl R., Karas H., Matys V., Meinhardt T., Pruss M., Reuter I., Schacherer F., Hehl R., Karas H., Matys V., Meinhardt T., Pruss M., Reuter I., Schacherer F., Karas H., Matys V., Meinhardt T., Pruss M., Reuter I., Schacherer F., Matys V., Meinhardt T., Pruss M., Reuter I., Schacherer F., Meinhardt T., Pruss M., Reuter I., Schacherer F., Pruss M., Reuter I., Schacherer F., Reuter I., Schacherer F., Schacherer F. TRANSFAC: An integrated system for gene expression regulation. Nucleic Acids Res. 2000;28:316–319. doi: 10.1093/nar/28.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman C.T., Stormo G.D., Stormo G.D. ANN-Spec: A method for discovering transcription factor binding sites with improved specificity. Pac. Symp. Biocomput. 2000;2000:467–478. doi: 10.1142/9789814447331_0044. [DOI] [PubMed] [Google Scholar]
- Zhou Q., Wong W.H., Wong W.H. CisModule: De novo discovery of cis-regulatory modules by hierarchical mixture modeling. Proc. Natl. Acad. Sci. 2004;101:12114–12119. doi: 10.1073/pnas.0402858101. [DOI] [PMC free article] [PubMed] [Google Scholar]