Abstract
Identification of unconventional functional features such as fusion transcripts is a challenging task in the effort to annotate all functional DNA elements in the human genome. Paired-End diTag (PET) analysis possesses a unique capability to accurately and efficiently characterize the two ends of DNA fragments, which may have either normal or unusual compositions. This unique nature of PET analysis makes it an ideal tool for uncovering unconventional features residing in the human genome. Using the PET approach for comprehensive transcriptome analysis, we were able to identify fusion transcripts derived from genome rearrangements and actively expressed retrotransposed pseudogenes, which would be difficult to capture by other means. Here, we demonstrate this unique capability through the analysis of 865,000 individual transcripts in two types of cancer cells. In addition to the characterization of a large number of differentially expressed alternative 5′ and 3′ transcript variants and novel transcriptional units, we identified 70 fusion transcript candidates in this study. One was validated as the product of a fusion gene between BCAS4 and BCAS3 resulting from an amplification followed by a translocation event between the two loci, chr20q13 and chr17q23. Through an examination of PETs that mapped to multiple genomic locations, we identified 4055 retrotransposed loci in the human genome, of which at least three were found to be transcriptionally active. The PET mapping strategy presented here promises to be a useful tool in annotating the human genome, especially aberrations in human cancer genomes.
With the completion of the human genome project, attention has turned to the annotation of the human genome for functional contents, including gene-coding transcriptional units (TUs), cis-acting regulatory elements, and chromatin modifications that modulate chromosomal structure and gene expression (Kim et al. 2005; Harrow et al. 2006; Wei et al. 2006). A battery of technologies including high-throughput sequencing and genome tiling arrays have been engaged on the elucidation of these functional elements (The ENCODE Project Consortium 2004, 2007). Despite considerable success, current methodologies are nevertheless not well suited for high-throughput discovery of aberrant genomic features such as fusion transcripts derived from chromosomal structure variations (Eichler 2001; Zelent et al. 2004), mechanisms like trans-splicing (Mayer and Floeter-Winter 2005; Horiuchi and Aigaki 2006), transcription-induced chimerism (Akiva et al. 2006; Parra et al. 2006), and efficient identification of transcribed pseudogenes (Balakirev and Ayala 2003; Zheng et al. 2005).
In the past, cDNA sequencing approaches, including traditional sequencing-based cDNA analysis (full-length, EST, and short tag sequencing) (Adams et al. 1992; Velculescu et al. 1995; Carninci et al. 1997; Brenner et al. 2000), contributed an immense number of transcripts (Gerhard et al. 2004) but were limited by either huge operational cost and inefficiency (full-length cDNA) or limited information (SAGE tag). In contrast, hybridization-based approaches like DNA microarrays, in particular the most recent genome-wide tiling arrays (Bertone et al. 2004; Cheng et al. 2005), provide massively parallel approaches for the characterization of all expressed exons, and new promise for highly comprehensive transcriptome analysis. However, the exon data provided by tiling array have no inherent structural information for each characterized transcript; i.e., it is not straightforward to define the start and termination positions or the connectivity of individual transcript units. Furthermore, the tiling array approach suffers from cross hybridization noise when it is used to detect transcripts expressed in highly homologous genomic regions, and it is incapable of uncovering unconventional transcripts, such as fusion transcripts, multi-cistronic transcripts that give rise to new proteins with novel functions, and transcribed pseudogenes which have nucleotide sequences that are different from, but highly homologous to, their parental genes.
We recently developed the Gene Identification Signature analysis using Paired-End diTagging (GIS-PET) (Ng et al. 2005). In GIS-PET analysis, paired-end ditags from the two ends of each expressed transcript (18 bp from 5′ end and 18 bp from 3′ end) are extracted, concatenated, and subjected to sequencing analysis. We demonstrated the precision of GIS-PETs in demarcating transcript boundaries in mouse genome, inferring proximal promoter sites, and identifying novel genes or alternative variants. Besides the robustness of GIS-PET for comprehensive transcriptome analysis, the unique nature of paired-end ditags reveals the relationship of the two ends of any DNA fragment, which is particularly suitable for high-throughput and systematic identification of unconventional transcripts such as fusion transcripts expressed in mammalian genomes. Furthermore, the large volume of PET data and specificity of PET mapping allow us to comprehensively annotate retrotransposed loci (pseudogenes) and distinguish actively transcribed pseudogenes from their parental genes. Here, we describe our strategy for the comprehensive characterization of the human cancer cell transcriptome and demonstrate the established tag-to-genome mapping scheme of screening for novel unconventional transcripts with potential biological functions. In this study, we uncovered a total of 70 putative fusion transcripts in two cancer genomes, identified 653 duplicate gene regions and 4055 retrotransposed loci, and provided evidence for three actively transcribed pseudogenes. We believe that the approach presented in this study will be of value for the systematic characterization of unconventional features in human cancer genomes.
Results
Mapping the cancer cell PET sequences to the human genome
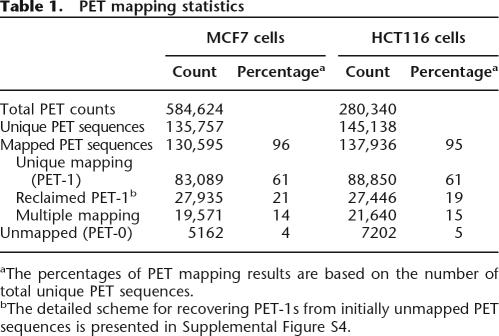
From the two cancer cell lines, MCF7 (breast cancer) and HCT116 (colon cancer), we generated 584,624 and 280,340 PET reads, which account for 135,757 and 145,138 non-redundant unique PET sequences, respectively (Table 1). Statistical analyses estimated that the numbers of transcripts captured by PETs represented ∼70% saturation of the two transcriptomes (Supplemental Information I and Fig. S1). Using standard mapping criteria (Ng et al. 2005), the majority (61%) of the PET sequences were mapped to unique single locations (thereafter referred to as PET-1) in the genome (an example of PET sequence mapping is shown in Supplemental Fig. S2). About 14%–15% of the PETs mapped to multiple locations in the genome and most likely represent transcripts expressed from genomic regions with high sequence homology such as duplicated gene families and pseudogene loci (Table 1). We also observed that ∼24%–25% of the PETs could not be mapped to the genome using the standard mapping criteria (referred to as PET-0). The main causes for the lack of mapping are sequencing errors, single nucleotide polymorphisms (SNPs), imperfection of the assembled reference genome as we described previously (Ng et al. 2005), and probably unconventional fusion transcripts derived from either trans-splicing or translocation (Supplemental Fig. S3). After a series of modified alignment analyses (details in Supplemental Information II), the majority (84% for MCF7 and 79% for HCT116) of these initially unmapped PETs were reclaimed as PETs that specifically map to the human genome (Supplemental Fig. S4) and which account for ∼20% of total PET sequences. Only 4%–5% of PET sequences (5162 for MCF7 and 7202 for HCT116) remained unmappable (Table 1). Collectively, these three categories of PET sequences (uniquely mapped, multiply-mapped, and unmapped PETs) represent the overall transcriptomes for these two cancer cell lines, and these transcriptomes are warrant for in-depth characterization.
Table 1.
PET mapping statistics
aThe percentages of PET mapping results are based on the number of total unique PET sequences.
bThe detailed scheme for recovering PET-1s from initially unmapped PET sequences is presented in Supplemental Figure S4.
Defining the transcriptome of cancer cells by uniquely mapped PET sequences
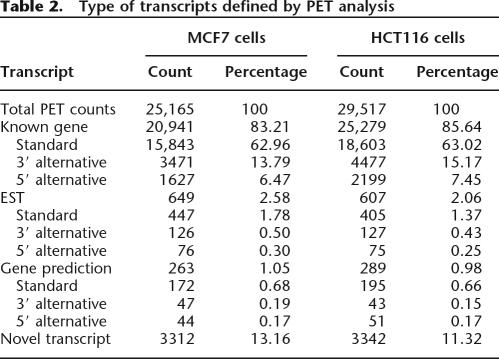
The uniquely mapped PETs were further grouped into individual transcripts if both 5′ and 3′ tag ends overlapped with each other (≥1 bp overlapping). The PET-defined transcripts that localized in the same regions but differed at either end by 1 kb or less were clustered further into transcript groups to represent a transcriptional unit (Supplemental Fig. S5). Through this clustering algorithm, the uniquely mapped PET-1 sequences including the reclaimed ones (111,024 for MCF7 and 116,296 for HCT116) were grouped into 25,165 and 29,517 transcrips in the two cell types. Most of the PET-defined TUs can be assigned to previously annotated gene features. In the MCF7 library, 20,941 (83.21%) PET-defined transcripts can be assigned to 9240 well-characterized known genes, while 3312 (13%) transcripts might be considered to encode novel genes because they covered genomic regions for which no solid transcriptional evidence (including known genes, RefSeq genes, Human mRNA from GenBank, and available ESTs) or gene predictions were available. Similarly in HCT116 cells, 25,279 (85.6%) transcripts were assigned to 8923 known genes and 3342 (11%) transcripts were considered novel (Table 2; Supplemental Fig. S6). Of these novel transcriptional units, 99 are located in the ENCODE regions and 43 have been validated by new data generated by ENCODE analysis (seven by GENCODE, 23 by Transfrag, and 13 by TARs). To further validate the remaining 56 regions identified only by GIS-PET data, we analyzed 17 PET-defined novel transcriptional units that have multiple PET counts and genomic span <500,000 bp by PCR from the full-length cDNA libraries. Of the 17 tested, 13 (76%) showed positive PCR results and were subsequently confirmed by sequencing analysis (Supplemental Table S1). This result suggests that the majority of novel transcripts identified by GIS-PET are bona fide transcripts.
Table 2.
Type of transcripts defined by PET analysis
To assess whether the full-length transcripts defined by GIS-PET data in this study were intact, we first examined the uniquely mapped PET sequences from the top 20 most abundant transcript clusters that matched well-characterized genes. Most of these genes have well-defined transcription start sites (TSS) and polyadenylation sites (PAS); therefore, they serve as good references for the evaluation of transcript intactness. Of the 61,569 and 23,704 PET-1 sequences assigned to the top 20 clusters in MCF7 and HCT116 cells, 98% and 93% of the PETs matched or spanned regions longer than the known 5′ and 3′ ends of the referenced transcripts (Supplemental Table S2). These results suggest that the vast majority of transcripts represented by GIS-PET sequences are intact. Therefore, when mapped specifically to the genome, these PET sequences can be used to accurately define transcription start and stop sites and applied to explore many interesting features of expressed genes and transcripts on the human genome.
Alternative 5′ transcription start sites and 3′ polyadenylation sites reflect the use of different promoters to regulate gene expression or transcript variants with different numbers or structures of exons. We observed significant variations in PET mapped 5′ and 3′ ends from previously annotated transcripts, which probably indicate a certain level of differential promoter usage that result in alternative TSS, as well as different polyA signals that result in alternative PAS. Because the majority of TSSs are scattered within a window of ±50 bp, we reason that a 200-bp difference between the PET-defined boundaries and previously annotated transcripts could be sufficient as a cutoff with high stringency to estimate the percentage of alternative TSS and PAS used by the genes in these two cancer cells. It was assessed that 86.4% (MCF7) and 86.8% (HCT116) of 5′ annotated PETs were within ±200 bp of TSS of their corresponding genes whereas 81.2% (MCF7) and 76.1% (HCT116) of 3′ annotated PETs were within ±200 bp of PAS. Therefore, ∼20% (±5) of the PET sequences did not map precisely to known transcripts and may represent transcript molecules with alternative 5′ TSS or 3′ PAS.
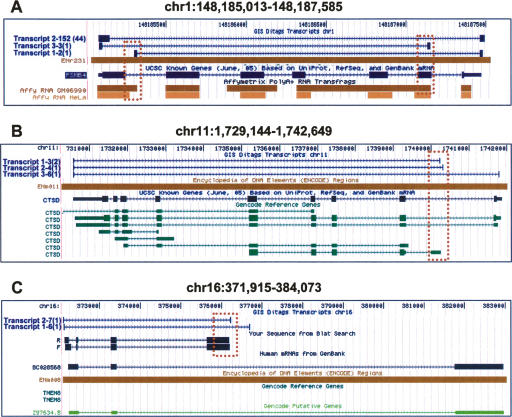
To be conservative and efficient, we defined PETs as 5′ TSS variants if their 5′ tags mapped to locations other than the first exon of their annotated transcript. We defined PETs as 3′ PAS variants if their 3′ tags mapped to locations other than the last exon of their annotated transcripts. From the 25,165 and 29,517 transcripts found in the two libraries, 1703 (MCF7) and 2274 (HCT116) 5′ alternative TSS transcripts and 3597 (MCF7) and 4604 (HCT116) 3′ alternative PAS transcripts were identified (Table 2). To determine the extent of overlap with TSSs found previously, we compared them with 30,964 alternative promoters identified through oligo-cap cDNA 5′ end sequencing (Kimura et al. 2006) and found that 315 (18.5%) and 396 (17.4%) were supported by these promoters found previously. Within the ENCODE regions, the majority of nonredundant (119/137; 86.9%) alternative TSS and alternative 3′ PAS (142/183; 77.6%) were validated by recent transcriptional evidence from GENCODE, transfrag, or TARs (Supplemental Table S1; examples in Fig. 1A,B). We conducted PCR analysis using PCR primers designed against ditags on the remaining 11 5′ alternative TSS and the 19 alternative 3′ PAS, and validated 10 of the tested TSS and 17 of the PAS (90%) variants (Supplemental Table S1; Fig. 1C; additional examples in Supplemental Fig. S7). Thus, GIS-PET uncovers transcript variants at 5′ and 3′ ends with high accuracy.
Figure 1.
Alternative 5′ TSS and 3′ PAS identified by GIS-PET analysis. Examples of 5′ and 3′ alternative TSS and PAS found in the ENCODE regions are shown. (A) Three different transcripts encoding PSMB4 were identified by GIS-PET in MCF7 cells. The top one (transcript 2; 152 copies) is the normal transcript identical to the PSMB4 in the known gene track. The 3′ PAS variant (middle transcript 3; three copies) and 5′ TSS variant (bottom transcript 1; two copies) are supported by transfrags detected by tiling array approaches. They are shown within the dotted box. (B) 5′ TSS variants of CTSD defined by three and four copies of PETs are supported by GENCODE reference gene in HCT116 cells. (C) A 3′ PAS variant of BC028568 detected by GIS-PET was confirmed by cDNA PCR and sequencing, shown as forward (F) and reverse (R) read.
Using this unique feature, we further explored differential promoter usages as indicated by 5′ alternative TSS identified from these two cell types. From a total of 6259 genes commonly expressed in both cells, 208 (3%) genes have alternative 5′ TSS transcripts predominantly expressed in one cell compared with the other cell type. Taking DNAJB4 as an example (Supplemental Fig. S8A), although about the same level of expression was detected in both of the cells (14 copies per million transcripts in HCT116 cells and 15 copies for MCF7), all transcripts of this gene in HCT116 are the standard form, while the TSS of this transcript in MCF7 were 25 kb upstream (chr1:78,156,944–78,195,014), probably due to the use of a bidirectional promoter shared with another gene, FUBP1 (chr1:78,124,189–78,156,774). MGAT1 is another example (Supplemental Fig. S8B). In HCT116 cells, there was only one type of transcript derived from the locus at chr5:180,150,207–180,162,458 with 39 copies per million transcripts. However, in MCF7, there were two variants of transcripts of this gene, one with five copies was the same as the transcript in HCT116 cells, while the other type (10 copies per million; chr5:180,169,756) was initiated from 19 kb upstream of the known transcription start site. Similarly, we also detected a few hundred genes with different 3′ PAS variants differentially expressed in either of these two cell types.
Identifying unconventional fusion transcripts from unpaired PET sequences
Fusion transcripts are unconventional and can be derived from either trans-splicing of separate pre-mRNA molecules (Mayer and Floeter-Winter 2005; Horiuchi and Aigaki 2006) or rearranged chromosomal regions that are genomic aberrations in which sections of two separate chromosomes were joined by translocation, deletion, or inversion (Supplemental Fig. S3B) (Masuda and Takahashi 2002; Saglio and Cilloni 2004). Fusion transcripts can generate distinct protein products as unique molecular signatures in specialized cell types (Sekiguchi et al. 2005; von Ahsen et al. 2005). In particular, fusion genes produced by chromosomal rearrangement are frequently involved in carcinogenesis (Mitelman et al. 1997). We suspect that some of the PET sequences in the PET-0 category were derived from fusion genes because the ditag sequences representing fusion transcripts would not be aligned in pairs along the human reference genome. All of the PET sequences in the PET-0 category can actually map to the human genome as separate 5′ tag and 3′ tag sequences, but the separately mapped tags cannot be paired by the standard pairing scheme used for conventional transcripts. Due to the short length of single tags, the individual 5′ and 3′ tags of these PET sequences would align to the reference genome at multiple locations, including the incorrect ones.
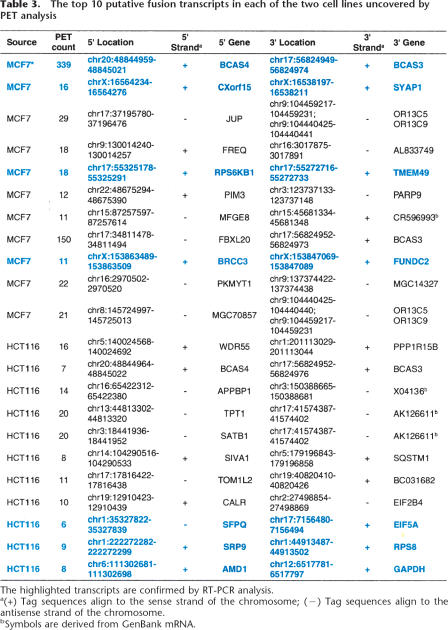
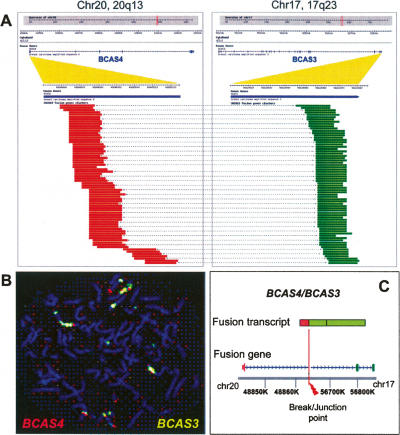
To identify correct PET sequences potentially representing fusion transcripts, we used a clustering strategy to group PET-0 sequences based on the mapping locations of the 5′ tags and 3′ tags separately. This strategy takes the advantage of the fact that multiple PET sequences representing the same transcripts will have slight shifts in the 5′ and 3′ boundaries along the genome sequence, and the mapping coordinates of 5′ tags or 3′ tags can be closely clustered. To a group of tags that have multiple alignment sites, the correct mapping sites are common to all the PET sequences derived from the same fusion transcripts, and the incorrect sites are scattered randomly. Therefore, we can distinguish true mapping sites from nonspecific ones by clustering. From the separate 5′ and 3′ mapping loci, putative fusion transcripts can be predicted. Out of the 5162 (MCF7) and 7202 (HCT116) PET-0 sequences, 777 and 750 clusters were grouped based on the 5′ tag and 3′ tag of PET mapping coordinates within 1 kb proximity, of which 173 and 119 clusters with ≥3 PET-0 sequences were considered of high confidence. We next looked for support from annotations of any known gene or EST data with start and stop sites that are <200 bp from these clustered 5′ and 3′ mapping coordinates. After manual inspection, 43 and 27 putative fusion transcripts were found from the MCF7 and HCT116 datasets, respectively. The 10 most abundant fusion transcripts for each of the two cell lines were listed in Table 3 and the complete list of the 70 fusion gene candidates is shown in Supplemental Table S5. The fusion gene candidate with the most abundant PET copies (339) in the MCF7 transcriptome showed mapping of the 5′ tag cluster (62 bp) to chr20:48,844,959–48,845,021 and 3′ tag cluster (25 bp) to chr17:56,824,949–56,824,974. Based on the genomic mapping coordinates, the 5′ region of this cluster is aligned to the 5′ end of BCAS4 on chromosome 20q13 and the 3′ region of this cluster is matched to the 3′ end of BCAS3 on chromosome 17q23 (Fig. 2A).
Table 3.
The top 10 putative transcripts in each of the two cell lines uncovered by PET analysis
The highlighted transcripts are confirmed by RT-PCR analysis.
a(+) Tag sequences align to the sense strand of the chromosome; (−) Tag sequences align to the antisense strand of the chromosome.
bSymbols are derived from GenBank mRNA.
Figure 2.
Validation of the fusion transcript BCAS4/BCAS3 found in MCF7 cells. (A) Three-hundred-thirty-nine copies of PET-0s were found to encode a putative BCAS4/BCAS3 fusion transcript. The top panels show the genomic regions for chr20q13 and chr17q23 and the lines below indicate the regions encoding BCAS4 and BCAS3 genes and their exon-intron structures. Portions of the first exon of BCAS4 and last exon of BCAS3 are expanded to show the PET mapping distributions along chromosome 20 between 48,845,002 and 48,845,295 (the 5′ end of BCAS4) and chromosome 17 between 56,824,953 and 56,824,980 (the 3′ end of BCAS3). (B) FISH analysis showed the amplification and colocalization of BCAS3 (green) and BCAS4 (red) genes. (C) Fusion transcript and inferred gene structures revealed by RT-PCR and sequencing analysis are shown. The exact breakpoint mapped by genomic PCR is displayed at the bottom.
We attempted to validate the top 11 most abundant fusion gene candidates from each of two cell lines by RT-PCR using the 5′ tag and 3′ tag information for PCR primer design. Of the 22 attempts (11 for each cell line), 17 (77%) showed positive results by PCR and seven (four from MCF7 cells and three from HCT116 cells) were further confirmed by sequencing analysis as genuine fusion transcripts (Table 3), including the known fusion gene BCAS4/BCAS3 that had been previously identified from MCF7 cells (Barlund et al. 2002). The other six fusion transcripts are CXorf15/SYAP1, RPS6KB1/TMEM49, BRCC3/FUNDC2, SFPQ/EIF5A, SRP9/RPS8, AMD1/GAPDH; and their detailed PCR results and cDNA sequences can be found in Supplemental Information IV. Because the fusion points of all seven fusion transcripts occurred at canonical exon splicing junctions, it is highly unlikely that these fusions were resulted from random cloning artifacts. Whether they are resulted from trans-splicing or genome translocation remains to be determined. Interestingly, some of these genes involved in the fusion events have been implicated in breast cancers either in chemotherapy response (SYAP1; Al-Dhaheri et al. 2006) or as oncogenic marker (RPS6KB1; van der Hage et al. 2004). We are currently investigating the clinical significance and prevalence of these fusion genes in different cancer types.
The cDNA product of the BCAS4/BCAS3 fusion transcript obtained by RT-PCR is identical with the fusion gene previously reported (Barlund et al. 2002). Although it has been suggested that this fusion transcript is a result of translocation between chr20 and chr17 on these locations, no cytogenetic evidence has been provided. Furthermore, the exact breakpoint junction and its molecular structure surrounding the translocation site have not been fully investigated at the genomic level. Therefore, we decided to characterize this fusion gene in great detail. Dual color fluorescence in situ hybridization (FISH) analysis by double la beling the metaphase chromosome from MCF7 cells with both BCAS3 and BCAS4 genomic DNA probes confirmed that these regions were not only amplified but also colocalized (Fig. 2B), confirming, therefore, the translocation events. To determine the precise translocation site, we used PCR to isolate the junction genomic segment from MCF7 genomic DNA. DNA sequencing analysis revealed that the exact breakpoint is at chr20:48, 863,986 for the BCAS4 locus, and chr17:56,642,488 for the BCAS3 locus (Fig. 2C). Interestingly, at the junction between the two breakpoints, there is a 448-bp (chr20:58,276,779–58,281,258) insertion that is located 9.4 mega bases away from the BCAS4 sites on chr20 based on the reference genome sequence (Supplemental Fig. S9). Collectively, these validation data conclude the fusion transcript BCAS4/BCAS3 is derived from the fusion gene BCAS4/BCAS3 that is the result of a chromosomal rearrangement between chr20 and chr17. It is intriguing to observe that 51 counts of PET sequences were detected for the full-length transcripts of BCAS4 in the MCF7 transcriptome. This observation suggests that not all 20q13 regions of chr20 in the MCF7 cell population were translocated. In contrast, no full-length BCAS3 transcript was detected by GIS-PET in MCF7 cells.
Taking these results together, we have demonstrated a use of GIS-PET analysis for the discovery of unconventional fusion transcripts, which would be difficult by other means such as conventional cDNA sequencing or tiling array hybridization approaches. These fusion transcripts can potentially produce new protein products uniquely to the cancer genomes analyzed here.
Characterization of duplicated gene families and retrotransposed gene loci
Another property of GIS-PET analysis is its ability to annotate genomic regions encoding transcripts that share high sequence homology, such as duplicated genes and retrotransposed loci with PETs that mapped to multiple locations in the genome. Duplicated genes (DG) usually have exon-intron structure and promoter regions (Fig. 3A), while the retrotransposed pseudogenes (RG) are cDNA sequences of parental genes subsequently reinserted back to genome at different genomic locations (Harrison and Gerstein 2002). They usually have features such as the presence of a polyA tail, lack of introns, and promoter elements. With random mutations accumulated over time, these loci have lost their transcription potential and become nonfunctional (processed pseudogenes) (Esnault et al. 2000). Duplicated genes and retrotransposed pseudogenes are abundant in mammalian genomes. The precise identification of these regions is important for accurate genome annotation and insight into the evolution of genes.
Figure 3.
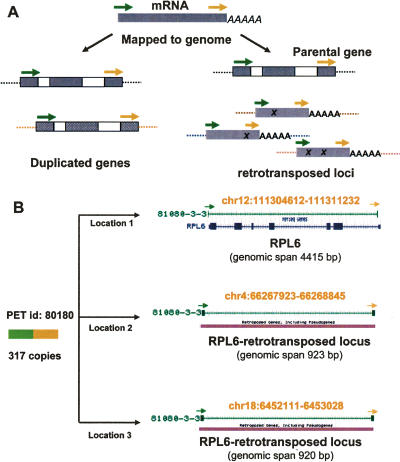
Gene duplication and retrotransposition mapped by GIS-PETs. (A) PETs derived from transcripts (mRNA) of duplicated gene family or from gene undergoing retrotransposition events map to multiple locations on the genome. (B) PET sequence with 317 copies derived from RPL6 was mapped specifically to three locations in the human genome hg17 assembly. The location on chromosome 12 is where the gene resides, while the other two locations on chromosome 4 and 18 are where RPL6 retrotransposed regions reside.
We first manually examined the multiple genomic locations mapped by same PET and found that most of them are of similar length and share a high degree of sequence identity with their corresponding mRNA sequences, indicating that they derived from either duplicated genes or retrotransposed genes. For example, PET sequence (GCTCTTTCCCATCTTGCATTAAATAGCT GACTACAA) mapped to three distinct locations (Fig. 3B): chr12:111,304,612–111,311,232 (RPL6; span 4415 bp), chr4:66,267,923–66,268,845 (span 923 bp), and chr18:6,452,111–6,453,028 (span 920 bp). When the latter two genomic sequences were aligned with the RPL6 mRNA sequence, they shared a 94% sequence identity over the entire 920-bp length. It was found that the first location encoded for the expressed RPL6 gene whereas the other two locations were retrotransposed pseudogene loci. Thus, by examining the genomic coordinates mapped by this category of PETs, we can not only locate the parental full-length transcripts but also annotate the genome with their corresponding duplicated gene regions and retrotransposed gene loci.
We next expanded the analysis to all PETs mapped to multiple locations. Genomic sequences from the multiple regions mapped by the same PET sequence were pairwise aligned by BLAT (Kent 2002). Of the 19,571 and 21,640 PETs with multiple mapping locations, >50% (10,181, 52% for MCF7, and 11,030, 51% for HCT116) map to multiple genomic locations that were highly homologous to each other, representing 3778 and 4200 genomic loci in MCF7 and HCT116 genomes, respectively. In the MCF7 genome, 468 duplicate gene loci and 3310 pseudogene loci were determined. Similarly, 446 duplicate genes and 3754 pseudogene loci were found in the HCT116 genome (Supplemental Fig. S10). From these two cell lines, a total of 653 duplicate gene loci of 126 genes and 4055 pseudogene loci of 526 parental genes were determined (Supplemental Table ST6).
We observed from these two datasets that high numbers of retrotransposed gene loci correlate well with the highly expressed housekeeping genes. For example, the gene that has the most retrotransposed gene copies (145 loci) is the ribosomal protein gene RPL21. In fact, 1580 out of 4055 (39%) identified retrotransposed gene loci were derived from ribosomal protein genes (Supplemental Table ST6). In addition, GAPDH, translation factors, and keratins contain high numbers of retrotransposed loci. Because the process of retrotransposition is considered to be highly random, we hypothesize that the number of inserted loci is proportional to mRNA copy number.
Retrotransposed genes have generally been discovered on the basis of sequence homology (Zhang et al. 2003). Because the prediction of retrotransposed loci was based on the sequence similarity with different thresholds and definitions, the numbers of human retrotransposed loci reported were inconsistent between different studies. By comparing the 4055 loci identified here with published datasets (7368 loci from Yale Pseudo track; 4416 loci from Vega Pseudogenes track for nine chromosomes; and 11,306 loci from Retroposed gene track of UCSC genome browser at http://genome.ucsc.edu), we found that the majority of retrotransposed loci identified in this study overlap with previously identified loci and >90% (3758 out of 4055) of our data can be supported by previous annotated loci (Supplemental Table S8). Among them, GIS-PET-identified retrotransposed loci share the highest overlap (3573/4055; 88%) with UCSC annotated track. Through the comparison, we identified 297 new retrotransposed loci in this study that were not found in other datasets.
Validating actively transcribed retrotransposed pseudogenes
Retrotransposed loci were believed to be nonfunctional. However, recent evidence shows that some retrotransposed genes are actively transcribed in specific cell types or conditions (Nguyen et al. 1991; Bard et al. 1995; Fujii et al. 1999; Olsen and Schechter 1999; Balakirev and Ayala 2003; Berger et al. 2005). More interestingly, the expressed pseudogenes may potentially play regulatory roles in specific biological processes (Korneev et al. 1999; Hirotsune et al. 2003). Because GIS-PET provides evidence of active transcription, PET-determined retrotransposed loci could be transcribed retrotransposed genes. One-hundred-seventeen nonredundant PETs only mapped to the retrotransposed loci but not to their parental genes, suggesting that the transcripts detected by PETs were derived from these loci. Indeed, 45 of these retrotransposed loci had supporting mRNA/EST evidence in public databases. We attempted to provide additional experimental evidence to validate the remaining loci by RT-PCR. Of the 72 candidates, 39 were tested by RT-PCR, and three were processed further for sequencing analysis. Based on sequence specificity, we validated that these three transcripts are from the three pseudogene loci: chr19:12,862,971–12,866,725 (RPS6), chr17:15,630,221– 15,632,178 (MEIS3), and chr1:27,335,644– 27,337,158 (ACTG1) (Supplemental Fig. 10). This limited study may suggest that there are more transcriptional activities associated with these “nonfunctional” loci than previously realized. Whether these transcriptional activities are biological relevant or simply reflect a certain low level of transcriptional noise requires further studies.
Discussion
In this study, we characterized the transcriptomes of two cancer cell lines (breast cancer and colon cancer) using GIS-PET analysis. Because of the large volume of transcript data for each transcriptome and the accuracy of PET sequences for the demarcation of transcription boundaries, we are able to identify significant numbers of putative novel genes and determine many alternative 5′ transcription start sites and 3′ termination sites. Furthermore, we demonstrated that this mapping process enabled identification of unconventional fusion transcripts and transcribed retrotransposed loci through the unique features of PET analysis.
Consistent with results generated by other technologies such as tiling array and RACEfrag within the the ENCODE Consortium (Kapranov et al. 2005; Gingeras 2006; The ENCODE Project Consortium 2007), GIS-PET showed that there are still significant numbers of novel transcripts yet to be characterized, and the human genome is transcribed more than we previously recognized. Furthermore, the large numbers of alternative TSS variants and cell-type specific promoter usage discovered here demonstrate the delicate regulation of transcriptional organization and emphasize that the human genome architecture is highly complex and dynamic. Thus, the complete catalogue of gene coding regions on human genome remains to be a big challenge for the ENCODE project.
One of the significant features exclusively provided by PET analysis is its efficiency in discovering unconventional fusion transcripts. Through this study, we have demonstrated a prototype scheme of using GIS-PET analysis as a screening pipeline for systematic identification of fusion genes resulting either from transcription-induced chimerism (Akiva et al. 2006; Parra et al. 2006), trans-splicing (Takahara et al. 2005), or genome rearrangement. Fusion transcripts are known to be regulated and have unique expression patterns and specific functions in certain biological systems (Maru 2001; Taki and Taniwaki 2006). Fusion transcripts resulting from chromosomal aberration are frequently observed in many hematological and solid tumors (Albertson et al. 2003; Mitelman et al. 2005) and the most well-known examples are the fusion oncoproteins such as BCR-ABL, PML-RARA, TEL/AML-1 (Mitelman et al. 2004). Among them, BCR-ABL was used successfully in the discovery of new diagnostic target and the development of effective drug for cancer therapy (Mauro et al. 2002). Giving the importance of chromosome rearrangements in the development of new concepts of prevention, diagnosis, and treatment of cancer, it is valuable to systematically categorize cancer genomes through comprehensive identification of all cancer-related abnormal fusion genes.
In the past, cancer signatures and structure variations were explored through various molecular or cytogenetic based approaches, such as SAGE-based digital karyotyping (Wang et al. 2002), chromosome banding (Mitelman et al. 1997), FISH (Tibiletti 2004), spectral karyotyping (SKY) (Schrock and Padilla-Nash 2000), single nucleotide polymorphism array (Dutt and Beroukhim 2007), and low resolution based array comparative genomic hybridization (CGH) (Kytola et al. 2000; Padilla-Nash et al. 2001). The information accumulated through these approaches has been integrated with the underlying human genome sequence sponsored by Cancer Chromosome Anatomy Project (CCAP) (Knutsen et al. 2005). Although some success has been achieved, the experimental approaches applied so far are neither comprehensive, only detecting copy number changes and deletion at low resolution, nor efficient for large scale and systematic characterization. Taking advantage of extensive EST data collections in public cDNA databases, computational efforts have also been attempted to identify fusion transcripts (Hahn et al. 2004). However, due to a lack of proper quality control and inconsistent standards of cDNA data accumulated over a decade from multiple sources, such excise of meta-transcriptome analyses has been inconclusive.
Recently, an approach similar to ours, called transcript End Sequence Profiling (tESP), has been reported for the identification of fusion transcripts associated with genome rearrangements in tumor genomes (Volik et al. 2006). Although, in theory the tESP strategy by sequencing the two ends of full-length cDNA clones can accurately identify prospective fusion transcripts, it is an impractical approach for identifying fusion transcripts through comprehensive screening of whole transcriptome, in which the majority (>90%) transcripts are conventional and most likely had been previously characterized ones, therefore, are considered as background for the purpose of obtaining fusion transcripts. In tESP analysis, two sequencing reads are required to generate paired ends for each full-length cDNA clone. In contrast, instead of directly sequencing full-length cDNA clones, GIS-PET extracts short paired ditags from the two ends of each full-length cDNA clone and then concatenates the ditags for efficient sequencing analysis. Each sequencing read in GIS-PET analysis generates an average of 15 paired-end ditags equivalent to 15 full-length transcripts (Ng et al. 2005). Therefore, GIS-PET analysis is 30-fold more efficient than tESP. In addition, by adapting the recently developed new sequencing technologies (Margulies et al. 2005; Shendure et al. 2005) on short reads (25–100 bp), we have further improved the sequencing efficiency of the tag-based GIS-PET analysis by another 100-fold (Ng et al. 2006). As tESP analysis is based on cDNA clones, it is difficult for it to adopt these new sequencing strategies. With our new capacity, a minimal one million PET sequence reads can be obtained by two 4-h sequencing runs for less than $10,000 cost. Giving that the number of transcripts in a mammalian cell is ∼200–300K (Jongeneel et al. 2003), the one million PET reads can provide near saturation coverage and superb sensitivity for the detection of fusion transcripts in a mixed cell populations, which is especially important in work with clinical biopsy tumor samples.
Using the position-based tag clustering algorithm described in this study, the fusion transcript identification provided by PET mapping is highly specific. This is supported by the high validation rate (seven out of 20) compared with previous reported results (Hahn et al. 2004; Volik et al. 2006). The chimeric BCAS4/3 fusion gene fully characterized in this study is one of the two fusion transcripts produced by imbalanced chromosomal translocation reported previously in MCF7 cells (Barlund et al. 2002; Hahn et al. 2004). The other one, TBL1XR1 (formerly IRA1)/RGS17, was identified through extensive search in the EST databases. Because the 17q23 locus amplification is found in 20% of primary breast tumors and the 20q13 locus amplification is found in 12%–39% of primary breast tumors (Kallioniemi et al. 1994; Muleris et al. 1994), it is likely that the BCAS4/3 fusion transcript found here was not an isolated event due to our cell culture condition and could be of clinical relevance. Because the GIS-PET sequencing approach offers a robust way to capture large numbers of fusion genes associated with chromosomal abnormalities, we can expect that the significance of such gene fusion for oncogenesis will be clarified in the near future.
Several efforts attempted to identify fusion transcripts in MCF7 cells. Hahn and colleagues established an informatics process to search for fusion transcripts among publicly available EST sequences (Hahn et al. 2004). They reported 237 fusion gene candidates from mixed of cancer and normal tissue samples and validated two fusion cDNAs (BCAS4/BCAS3; TBL1XR1/RGS17) in MCF7 cells. TBL1XR1/RGS17 fusion was also detected in our MCF7 cDNA library through PCR. However, we did not discover this fusion through PET mapping approach. The reason is because these corresponding two PETs (total counts 35) were of low sequence complexity and aligned to >100 different positions on the hg17 genome (for detail, see Supplemental Information IV). Therefore, they were excluded from our initial fusion gene analysis. Volik and colleagues uncovered 66 fusion genes by sequencing and mapping of pair-end reads from 5089 full-length cDNA clones (Volik et al. 2006). Of these, four were validated by PCR. Out of these 66 fusion genes reported by Volik and colleagues we did not find any of them overlapped with our 70 fusion candidates. To further verify the presence of the four confirmed genes found by Volik and colleagues in our MCF7 cDNA library, we carried out RT-PCR, and our PCR results did not yield any specific fragments from the MCF7 cells or even the MCF7 cDNA library. It is possible that the lack of overlap between these two studies is because of the massive rearrangements undergone in the MCF7 genome and the locations of the breakpoints, which are largely different between different labs and culturing conditions.
Another unique feature of GIS-PET analysis is identification of actively transcribed pseudogenes. In the human genome, the number of retrotransposed loci has been estimated to be ∼20,000 (Harrison et al. 2002). Although most of the pseudogenes are inactive, recent evidence has shown that some pseudogenes are expressed and they play active roles in regulating the expression of other genes either through translational inhibition (Korneev et al. 1999) or by affecting the stability of homologous mRNA (Hirotsune et al. 2003), and are not simply nonspecific transcriptional noise. It remains to be determined whether these observations are isolated incidences or prevalent mechanisms. The transcribed pseudogene loci identified so far were all through individual molecular cloning experiments (Yousef et al. 2004; Berger et al. 2005). No method has been reported for collection of such transcripts in a high-throughput, systematic manner. Due to the high degree of sequence identity between parental genes and pseudogenes, the array-based approach cannot offer sufficient hybridization specificity to distinguish between the expressed psuedogene loci as opposed to noise resulting from cross hybridization. In contrast, GIS-PET analysis offers an unmatchable solution with specificity at nucleotide level and efficiency of whole transcriptome coverage for comprehensive identification of active pseudogenes. In this study, we have demonstrated that PET sequences are sufficient to distinguish the nucleotide difference between the 117 transcribed pseudogene loci and their parental genes. The availability of GIS-PET data would facilitate the identification of transcribed pseudogenes and enable the research community to examine their functional relevance in many different biological systems.
We have been continually improving the specificity and capacity of GIS-PET analysis. One direction is to increase the tag length while maintaining its superb efficiency. The new GIS-PET protocol now can generate PETs with tag lengths of more than 50 bp (Y. Ruan, unpubl.). With the increased specificity of PET mappings to the genome for demarcation of transcription boundaries, the intrinsic efficiency of PETs for whole genome analysis, and the unique capability of PETs to recognize unconventional fusion transcripts and transcribed retrotransposed loci, we expect that the GIS-PET analysis will be a very valuable platform technology for decoding the genes and transcript elements of the entire human genome.
Methods
Cell lines, growth condition, and RNA preparation
Two human cancer cell lines were used for GIS-PET analysis. HCT116 is a human colorectal cancer cell line (ATCC no. CCL-247) and MCF7 is a human breast cancer cell line (ATCC no. HTB-22). The cells were grown under standard culture conditions and harvested at log phase. Two-hundred micrograms of total RNA and polyA+ RNA (10 μg) were prepared by TRIzol method and oligo-d(T) using standard molecular biology procedures.
GIS-PET library construction
Full-length cDNA libraries were made by a modified biotinylated cap-trapper approach (Carninci et al. 1997). Purified plasmid prepared from the full-length cDNA library was digested with MmeI, end-polished with T4 DNA polymerase, and the resulting plasmids containing a pair of end tags from each terminus of the original cDNA insert were self-ligated. They were then transformed to form a transitional single-PET library. Plasmid DNA extracted from this library was digested with BamHI to release the 50-bp paired-end ditags (PETs). The PETs were concatenated and cloned into the BamHI-cut pZErO-1 to form the final GIS-PET library for sequencing analysis (Ng et al. 2005).
PET sequencing and mapping
In MCF7 library, 584,624 PET sequences were extracted from 74,537 high quality (QV ≥ 20) sequence reads. In HCT116 library, 280,340 PET sequences were extracted from 53,758 sequence reads. Each PET sequence was split into its 5′ tag and 3′ tag components, and the tags were searched independently for matches in the human genome assembly hg17 in compressed suffix array (CSA) format. The mapped tags were then paired based on the mapping locations of their 5′ and 3′ signatures if they were on the same chromosome, in the correct order, orientation (5′ → 3′), and within appropriate genomic distance (one million base pairs) (Chiu et al. 2006).
PET-0 processing pipeline
First, the unmappable PET-0s from the default mapping were clustered with PET-1s and grouped if they shared two consecutive regions of at least length 12 and 10 nucleotides in the 5′ ends and 3′ ends, respectively. The PET-0s that could be clustered with PET-1s were reassigned back to PET-1 and further classified into clusters as either transcripts with known gene support or potential novel transcripts (no known genes are surrounding the mapping locations). Next, the remaining PET-0s were clustered on the 5′ and 3′ ends based on the same criteria (minimal 12 and 10 nucleotides in the 5′ and 3′ ends, respectively). These clusters were remapped to the genome using the consensus sequences. The PET clusters were considered mapped if the 5′ and 3′ matches shared a stretch of consecutive nucleotides of at least 17 and 15 bases in length with the hg17 genome, and have a span of at most one million bases. The remaining PET clusters with only one end (either 5′ or 3′) mapped to genome were further mapped and paired by allowing one base variation (mismatch, deletion, and insertion) into the other end. This process is known as End-Guided Mapping. All the PET-0s that cannot be paired with the “End-Guided Mapping” method were blasted against a list containing the first and last 100 nucleotide sequences from a set of GenBank mRNA transcripts downloaded from NCBI.
Fusion transcript analysis
The PET clusters that failed to map to the genome with the PET-0 processing pipeline were selected for potential fusion gene identification. The 5′ and 3′ consensus mapping coordinates from these PET clusters were grouped separately if their distances from each other were within 1-kb proximity. For each cluster grouped by 5′ mapping location, we extracted its 3′ mapping location and grouped them using the same criterion and vice versa. Only the clusters with both of their 5′ and 3′ mapping coordinates grouped together were considered putative candidates. Clusters containing more than three PET-0 sequences were collected and their 5′ and 3′ tags were blasted against a list containing 200 nucleotides of the starts and ends of EST and mRNA sequences. The final clusters were selected if they shared sequence homology with the ends from mRNA or EST and if they passed manual curation.
BCAS3/BCAS4 RT-PCR validation
Two sets of PCR primers (see below) were designed from PET sequences and internal regions from BCAS3 and BCAS4 mRNA to RT-PCR amplify fusion cDNA from MCF7 mRNA. Forward primer 1 (from BCAS4 5′ tag), 5′-GCGGGGCTCCGAGGCCCGGG; Forward primer 2 (47 bp 3′ of forward primer 1), 5′-CCTCCTGA TGCTGCTCGT; Reverse primer (from BCAS3 3′ tag), 5′-CTGCAGCGTGATTTATTGGA.
FISH hybridization
Three BAC clones (BCAS3: RP11-1081E4 and RP11-805G4; BCAS4: RP11-474E9) were used for the FISH analysis. BAC DNA was prepared following the alkaline lysis method using Qiagen DNA preparation protocol with Qia-TIP 500 columns. Two micrograms of BAC DNA was labeled by nick translation using fluorescin 12-dUTP and Texas red 5-dUTP (Perkin Elmer). For each hybridization, 100 ng of labeled BAC clone probe was mixed in 15 μL of hybridization mixture along with a 10× excess of unlabeled human Cot-1 DNA (Invitrogen). FISH hybridization was performed by codenaturation of probe on Thermobrite (Statspin) for 2 min at 80°C followed by incubation at 37°C overnight. Post-hybridization washes were performed using 2× SSC at 45°C twice for 5 min each followed by two washes in 0.5× SSC/0.1% Tween 20 at 45°C for 5 min each. Slides were rinsed in distilled water and 100% ethanol. Twenty microliters DAPI was added as counterstain and stored at 4°C. Fluorescent images were captured using a CCD camera mounted on NIKON-80i fluorescence microscope. Captured images were processed using ISIS (in situ imaging system) (Metasytems, GMBH) software.
Transcribed retrotransposed loci identification
To select retrotransposed loci with transcriptional evidence, we examined the mapping locations of PET-1s (PETs mapped to specific genomic locations). First, the first G of the 5′ end and the AA tail at the 3′ end of each PET were removed. Then, we extracted the genomic sequences of the retrotransposed locus mapped by the PET-1 as the query sequence and the parental gene locus as the subject sequence. Pairwise alignment was done between the query sequence and subject sequence using BLAT (using ≥80% sequence similarity and ≥0.6 completeness as cutoff values) to determine the differences between these two sequences. To select highly reliable PET-1s derived from the transcribed loci, we followed the criterion that the PET-1 sequences aligned to the retrotransposed loci must have at least 1 bp different from the parental expressed gene in the 5′ (18 bp) or 3′ (18 bp) sequence regions. The difference could be a mismatch, deletion, or insertion. All the mismatch differences were verified against the SNPdb, and the known SNPs were removed.
Acknowledgments
We thank Mr. H. Thoreau and the Genome Technology & Biology Group at the Genome Institute of Singapore for HTP sequencing support, and Ms. Melissa Jane Fullwood for critical reading of the manuscript. We also thank The ENCODE Project Consortium for making their data publicly available, especially the Genes and Transcripts subgroup (GENCODE, Yale and Affymetrix) for their transcript data. This work is supported by the Agency for Science, Technology, and Research (A*STAR) of Singapore and the NIH ENCODE grant 1R01HG003521-01.
Footnotes
[Supplemental material is available online at www.genome.org. DNA sequences reported in this study are available to public through UCSC genome browser at http://genome.ucsc.edu track name GIS-PET RNA and http://t2g.bii.a-star.edu.sg.]
Article is online at http://www.genome.org/cgi/doi/10.1101/gr.6018607
References
- Adams M.D., Dubnick M., Kerlavage A.R., Moreno R., Kelley J.M., Utterback T.R., Nagle J.W., Fields C., Venter J.C., Dubnick M., Kerlavage A.R., Moreno R., Kelley J.M., Utterback T.R., Nagle J.W., Fields C., Venter J.C., Kerlavage A.R., Moreno R., Kelley J.M., Utterback T.R., Nagle J.W., Fields C., Venter J.C., Moreno R., Kelley J.M., Utterback T.R., Nagle J.W., Fields C., Venter J.C., Kelley J.M., Utterback T.R., Nagle J.W., Fields C., Venter J.C., Utterback T.R., Nagle J.W., Fields C., Venter J.C., Nagle J.W., Fields C., Venter J.C., Fields C., Venter J.C., Venter J.C. Sequence identification of 2,375 human brain genes. Nature. 1992;355:632–634. doi: 10.1038/355632a0. [DOI] [PubMed] [Google Scholar]
- Akiva P., Toporik A., Edelheit S., Peretz Y., Diber A., Shemesh R., Novik A., Sorek R., Toporik A., Edelheit S., Peretz Y., Diber A., Shemesh R., Novik A., Sorek R., Edelheit S., Peretz Y., Diber A., Shemesh R., Novik A., Sorek R., Peretz Y., Diber A., Shemesh R., Novik A., Sorek R., Diber A., Shemesh R., Novik A., Sorek R., Shemesh R., Novik A., Sorek R., Novik A., Sorek R., Sorek R. Transcription-mediated gene fusion in the human genome. Genome Res. 2006;16:30–36. doi: 10.1101/gr.4137606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertson D.G., Collins C., McCormick F., Gray J.W., Collins C., McCormick F., Gray J.W., McCormick F., Gray J.W., Gray J.W. Chromosome aberrations in solid tumors. Nat. Genet. 2003;34:369–376. doi: 10.1038/ng1215. [DOI] [PubMed] [Google Scholar]
- Al-Dhaheri M.H., Shah Y.M., Basrur V., Pind S., Rowan B.G., Shah Y.M., Basrur V., Pind S., Rowan B.G., Basrur V., Pind S., Rowan B.G., Pind S., Rowan B.G., Rowan B.G. Identification of novel proteins induced by estradiol, 4-hydroxytamoxifen and acolbifene in T47D breast cancer cells. Steroids. 2006;71:966–978. doi: 10.1016/j.steroids.2006.07.006. [DOI] [PubMed] [Google Scholar]
- Balakirev E.S., Ayala F.J., Ayala F.J. Pseudogenes: Are they “junk” or functional DNA? Annu. Rev. Genet. 2003;37:123–151. doi: 10.1146/annurev.genet.37.040103.103949. [DOI] [PubMed] [Google Scholar]
- Bard J.A., Nawoschik S.P., O'Dowd B.F., George S.R., Branchek T.A., Weinshank R.L., Nawoschik S.P., O'Dowd B.F., George S.R., Branchek T.A., Weinshank R.L., O'Dowd B.F., George S.R., Branchek T.A., Weinshank R.L., George S.R., Branchek T.A., Weinshank R.L., Branchek T.A., Weinshank R.L., Weinshank R.L. The human serotonin 5-hydroxytryptamine1D receptor pseudogene is transcribed. Gene. 1995;153:295–296. doi: 10.1016/0378-1119(94)00693-m. [DOI] [PubMed] [Google Scholar]
- Barlund M., Monni O., Weaver J.D., Kauraniemi P., Sauter G., Heiskanen M., Kallioniemi O.P., Kallioniemi A., Monni O., Weaver J.D., Kauraniemi P., Sauter G., Heiskanen M., Kallioniemi O.P., Kallioniemi A., Weaver J.D., Kauraniemi P., Sauter G., Heiskanen M., Kallioniemi O.P., Kallioniemi A., Kauraniemi P., Sauter G., Heiskanen M., Kallioniemi O.P., Kallioniemi A., Sauter G., Heiskanen M., Kallioniemi O.P., Kallioniemi A., Heiskanen M., Kallioniemi O.P., Kallioniemi A., Kallioniemi O.P., Kallioniemi A., Kallioniemi A. Cloning of BCAS3 (17q23) and BCAS4 (20q13) genes that undergo amplification, overexpression, and fusion in breast cancer. Genes Chromosomes Cancer. 2002;35:311–317. doi: 10.1002/gcc.10121. [DOI] [PubMed] [Google Scholar]
- Berger I.R., Buschbeck M., Bange J., Ullrich A., Buschbeck M., Bange J., Ullrich A., Bange J., Ullrich A., Ullrich A. Identification of a transcriptionally active hVH-5 pseudogene on 10q22.2. Cancer Genet. Cytogenet. 2005;159:155–159. doi: 10.1016/j.cancergencyto.2004.10.010. [DOI] [PubMed] [Google Scholar]
- Bertone P., Stolc V., Royce T.E., Rozowsky J.S., Urban A.E., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Stolc V., Royce T.E., Rozowsky J.S., Urban A.E., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Royce T.E., Rozowsky J.S., Urban A.E., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Rozowsky J.S., Urban A.E., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Urban A.E., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Tongprasit W., Samanta M., Weissman S., Samanta M., Weissman S., Weissman S., et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–2246. doi: 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- Brenner S., Johnson M., Bridgham J., Golda G., Lloyd D.H., Johnson D., Luo S., McCurdy S., Foy M., Ewan M., Johnson M., Bridgham J., Golda G., Lloyd D.H., Johnson D., Luo S., McCurdy S., Foy M., Ewan M., Bridgham J., Golda G., Lloyd D.H., Johnson D., Luo S., McCurdy S., Foy M., Ewan M., Golda G., Lloyd D.H., Johnson D., Luo S., McCurdy S., Foy M., Ewan M., Lloyd D.H., Johnson D., Luo S., McCurdy S., Foy M., Ewan M., Johnson D., Luo S., McCurdy S., Foy M., Ewan M., Luo S., McCurdy S., Foy M., Ewan M., McCurdy S., Foy M., Ewan M., Foy M., Ewan M., Ewan M., et al. Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat. Biotechnol. 2000;18:630–634. doi: 10.1038/76469. [DOI] [PubMed] [Google Scholar]
- Carninci P., Westover A., Nishiyama Y., Ohsumi T., Itoh M., Nagaoka S., Sasaki N., Okazaki Y., Muramatsu M., Schneider C., Westover A., Nishiyama Y., Ohsumi T., Itoh M., Nagaoka S., Sasaki N., Okazaki Y., Muramatsu M., Schneider C., Nishiyama Y., Ohsumi T., Itoh M., Nagaoka S., Sasaki N., Okazaki Y., Muramatsu M., Schneider C., Ohsumi T., Itoh M., Nagaoka S., Sasaki N., Okazaki Y., Muramatsu M., Schneider C., Itoh M., Nagaoka S., Sasaki N., Okazaki Y., Muramatsu M., Schneider C., Nagaoka S., Sasaki N., Okazaki Y., Muramatsu M., Schneider C., Sasaki N., Okazaki Y., Muramatsu M., Schneider C., Okazaki Y., Muramatsu M., Schneider C., Muramatsu M., Schneider C., Schneider C., et al. High efficiency selection of full-length cDNA by improved biotinylated cap trapper. DNA Res. 1997;4:61–66. doi: 10.1093/dnares/4.1.61. [DOI] [PubMed] [Google Scholar]
- Cheng J., Kapranov P., Drenkow J., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Kapranov P., Drenkow J., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Drenkow J., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Patel S., Long J., Stern D., Tammana H., Helt G., Long J., Stern D., Tammana H., Helt G., Stern D., Tammana H., Helt G., Tammana H., Helt G., Helt G., et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- Chiu K.P., Wong C.H., Chen Q., Ariyaratne P., Ooi H.S., Wei C.L., Sung W.K., Ruan Y., Wong C.H., Chen Q., Ariyaratne P., Ooi H.S., Wei C.L., Sung W.K., Ruan Y., Chen Q., Ariyaratne P., Ooi H.S., Wei C.L., Sung W.K., Ruan Y., Ariyaratne P., Ooi H.S., Wei C.L., Sung W.K., Ruan Y., Ooi H.S., Wei C.L., Sung W.K., Ruan Y., Wei C.L., Sung W.K., Ruan Y., Sung W.K., Ruan Y., Ruan Y. PET-Tool: A software suite for comprehensive processing and managing of Paired-End diTag (PET) sequence data. BMC Bioinformatics. 2006;7:390. doi: 10.1186/1471-2105-7-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutt A., Beroukhim R., Beroukhim R. Single nucleotide polymorphism array analysis of cancer. Curr. Opin. Oncol. 2007;19:43–49. doi: 10.1097/CCO.0b013e328011a8c1. [DOI] [PubMed] [Google Scholar]
- Eichler E.E. Recent duplication, domain accretion and the dynamic mutation of the human genome. Trends Genet. 2001;17:661–669. doi: 10.1016/s0168-9525(01)02492-1. [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium, The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium, Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007 doi: 10.1038/nature05874. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnault C., Maestre J., Heidmann T., Maestre J., Heidmann T., Heidmann T. Human LINE retrotransposons generate processed pseudogenes. Nat. Genet. 2000;24:363–367. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
- Fujii G.H., Morimoto A.M., Berson A.E., Bolen J.B., Morimoto A.M., Berson A.E., Bolen J.B., Berson A.E., Bolen J.B., Bolen J.B. Transcriptional analysis of the PTEN/MMAC1 pseudogene, psiPTEN. Oncogene. 1999;18:1765–1769. doi: 10.1038/sj.onc.1202492. [DOI] [PubMed] [Google Scholar]
- Gerhard D.S., Wagner L., Feingold E.A., Shenmen C.M., Grouse L.H., Schuler G., Klein S.L., Old S., Rasooly R., Good P., Wagner L., Feingold E.A., Shenmen C.M., Grouse L.H., Schuler G., Klein S.L., Old S., Rasooly R., Good P., Feingold E.A., Shenmen C.M., Grouse L.H., Schuler G., Klein S.L., Old S., Rasooly R., Good P., Shenmen C.M., Grouse L.H., Schuler G., Klein S.L., Old S., Rasooly R., Good P., Grouse L.H., Schuler G., Klein S.L., Old S., Rasooly R., Good P., Schuler G., Klein S.L., Old S., Rasooly R., Good P., Klein S.L., Old S., Rasooly R., Good P., Old S., Rasooly R., Good P., Rasooly R., Good P., Good P., et al. The status, quality, and expansion of the NIH full-length cDNA project: The Mammalian Gene Collection (MGC) Genome Res. 2004;14:2121–2127. doi: 10.1101/gr.2596504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingeras T.R. The multitasking genome. Nat. Genet. 2006;38:608–609. doi: 10.1038/ng0606-608. [DOI] [PubMed] [Google Scholar]
- Hahn Y., Bera T.K., Gehlhaus K., Kirsch I.R., Pastan I.H., Lee B., Bera T.K., Gehlhaus K., Kirsch I.R., Pastan I.H., Lee B., Gehlhaus K., Kirsch I.R., Pastan I.H., Lee B., Kirsch I.R., Pastan I.H., Lee B., Pastan I.H., Lee B., Lee B. Finding fusion genes resulting from chromosome rearrangement by analyzing the expressed sequence databases. Proc. Natl. Acad. Sci. 2004;101:13257–13261. doi: 10.1073/pnas.0405490101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison P.M., Gerstein M., Gerstein M. Studying genomes through the aeons: Protein families, pseudogenes and proteome evolution. J. Mol. Biol. 2002;318:1155–1174. doi: 10.1016/s0022-2836(02)00109-2. [DOI] [PubMed] [Google Scholar]
- Harrison P.M., Hegyi H., Balasubramanian S., Luscombe N.M., Bertone P., Echols N., Johnson T., Gerstein M., Hegyi H., Balasubramanian S., Luscombe N.M., Bertone P., Echols N., Johnson T., Gerstein M., Balasubramanian S., Luscombe N.M., Bertone P., Echols N., Johnson T., Gerstein M., Luscombe N.M., Bertone P., Echols N., Johnson T., Gerstein M., Bertone P., Echols N., Johnson T., Gerstein M., Echols N., Johnson T., Gerstein M., Johnson T., Gerstein M., Gerstein M. Molecular fossils in the human genome: Identification and analysis of the pseudogenes in chromosomes 21 and 22. Genome Res. 2002;12:272–280. doi: 10.1101/gr.207102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J., Denoeud F., Frankish A., Reymond A., Chen C.K., Chrast J., Lagarde J., Gilbert J.G., Storey R., Swarbreck D., Denoeud F., Frankish A., Reymond A., Chen C.K., Chrast J., Lagarde J., Gilbert J.G., Storey R., Swarbreck D., Frankish A., Reymond A., Chen C.K., Chrast J., Lagarde J., Gilbert J.G., Storey R., Swarbreck D., Reymond A., Chen C.K., Chrast J., Lagarde J., Gilbert J.G., Storey R., Swarbreck D., Chen C.K., Chrast J., Lagarde J., Gilbert J.G., Storey R., Swarbreck D., Chrast J., Lagarde J., Gilbert J.G., Storey R., Swarbreck D., Lagarde J., Gilbert J.G., Storey R., Swarbreck D., Gilbert J.G., Storey R., Swarbreck D., Storey R., Swarbreck D., Swarbreck D., et al. GENCODE: Producing a reference annotation for ENCODE. Genome Biol. 2006;7:S41–S49. doi: 10.1186/gb-2006-7-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirotsune S., Yoshida N., Chen A., Garrett L., Sugiyama F., Takahashi S., Yagami K., Wynshaw-Boris A., Yoshiki A., Yoshida N., Chen A., Garrett L., Sugiyama F., Takahashi S., Yagami K., Wynshaw-Boris A., Yoshiki A., Chen A., Garrett L., Sugiyama F., Takahashi S., Yagami K., Wynshaw-Boris A., Yoshiki A., Garrett L., Sugiyama F., Takahashi S., Yagami K., Wynshaw-Boris A., Yoshiki A., Sugiyama F., Takahashi S., Yagami K., Wynshaw-Boris A., Yoshiki A., Takahashi S., Yagami K., Wynshaw-Boris A., Yoshiki A., Yagami K., Wynshaw-Boris A., Yoshiki A., Wynshaw-Boris A., Yoshiki A., Yoshiki A. An expressed pseudogene regulates the messenger-RNA stability of its homologous coding gene. Nature. 2003;423:91–96. doi: 10.1038/nature01535. [DOI] [PubMed] [Google Scholar]
- Horiuchi T., Aigaki T., Aigaki T. Alternative trans-splicing: A novel mode of pre-mRNA processing. Biol. Cell. 2006;98:135–140. doi: 10.1042/BC20050002. [DOI] [PubMed] [Google Scholar]
- Jongeneel C.V., Iseli C., Stevenson B.J., Riggins G.J., Lal A., Mackay A., Harris R.A., O’Hare M.J., Neville A.M., Simpson A.J., Iseli C., Stevenson B.J., Riggins G.J., Lal A., Mackay A., Harris R.A., O’Hare M.J., Neville A.M., Simpson A.J., Stevenson B.J., Riggins G.J., Lal A., Mackay A., Harris R.A., O’Hare M.J., Neville A.M., Simpson A.J., Riggins G.J., Lal A., Mackay A., Harris R.A., O’Hare M.J., Neville A.M., Simpson A.J., Lal A., Mackay A., Harris R.A., O’Hare M.J., Neville A.M., Simpson A.J., Mackay A., Harris R.A., O’Hare M.J., Neville A.M., Simpson A.J., Harris R.A., O’Hare M.J., Neville A.M., Simpson A.J., O’Hare M.J., Neville A.M., Simpson A.J., Neville A.M., Simpson A.J., Simpson A.J., et al. Comprehensive sampling of gene expression in human cell lines with massively parallel signature sequencing. Proc. Natl. Acad. Sci. 2003;100:4702–4705. doi: 10.1073/pnas.0831040100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallioniemi A., Kallioniemi O.P., Piper J., Tanner M., Stokke T., Chen L., Smith H.S., Pinkel D., Gray J.W., Waldman F.M., Kallioniemi O.P., Piper J., Tanner M., Stokke T., Chen L., Smith H.S., Pinkel D., Gray J.W., Waldman F.M., Piper J., Tanner M., Stokke T., Chen L., Smith H.S., Pinkel D., Gray J.W., Waldman F.M., Tanner M., Stokke T., Chen L., Smith H.S., Pinkel D., Gray J.W., Waldman F.M., Stokke T., Chen L., Smith H.S., Pinkel D., Gray J.W., Waldman F.M., Chen L., Smith H.S., Pinkel D., Gray J.W., Waldman F.M., Smith H.S., Pinkel D., Gray J.W., Waldman F.M., Pinkel D., Gray J.W., Waldman F.M., Gray J.W., Waldman F.M., Waldman F.M. Detection and mapping of amplified DNA sequences in breast cancer by comparative genomic hybridization. Proc. Natl. Acad. Sci. 1994;91:2156–2160. doi: 10.1073/pnas.91.6.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P., Drenkow J., Cheng J., Long J., Helt G., Dike S., Gingeras T.R., Drenkow J., Cheng J., Long J., Helt G., Dike S., Gingeras T.R., Cheng J., Long J., Helt G., Dike S., Gingeras T.R., Long J., Helt G., Dike S., Gingeras T.R., Helt G., Dike S., Gingeras T.R., Dike S., Gingeras T.R., Gingeras T.R. Examples of the complex architecture of the human transcriptome revealed by RACE and high-density tiling arrays. Genome Res. 2005;15:987–997. doi: 10.1101/gr.3455305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W.J. BLAT—The BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.H., Barrera L.O., Zheng M., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Barrera L.O., Zheng M., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Zheng M., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Richmond T.A., Wu Y., Green R.D., Ren B., Wu Y., Green R.D., Ren B., Green R.D., Ren B., Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K., Wakamatsu A., Suzuki Y., Ota T., Nishikawa T., Yamashita R., Yamamoto J., Sekine M., Tsuritani K., Wakaguri H., Wakamatsu A., Suzuki Y., Ota T., Nishikawa T., Yamashita R., Yamamoto J., Sekine M., Tsuritani K., Wakaguri H., Suzuki Y., Ota T., Nishikawa T., Yamashita R., Yamamoto J., Sekine M., Tsuritani K., Wakaguri H., Ota T., Nishikawa T., Yamashita R., Yamamoto J., Sekine M., Tsuritani K., Wakaguri H., Nishikawa T., Yamashita R., Yamamoto J., Sekine M., Tsuritani K., Wakaguri H., Yamashita R., Yamamoto J., Sekine M., Tsuritani K., Wakaguri H., Yamamoto J., Sekine M., Tsuritani K., Wakaguri H., Sekine M., Tsuritani K., Wakaguri H., Tsuritani K., Wakaguri H., Wakaguri H., et al. Diversification of transcriptional modulation: Large-scale identification and characterization of putative alternative promoters of human genes. Genome Res. 2006;16:55–65. doi: 10.1101/gr.4039406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutsen T., Gobu V., Knaus R., Padilla-Nash H., Augustus M., Strausberg R.L., Kirsch I.R., Sirotkin K., Ried T., Gobu V., Knaus R., Padilla-Nash H., Augustus M., Strausberg R.L., Kirsch I.R., Sirotkin K., Ried T., Knaus R., Padilla-Nash H., Augustus M., Strausberg R.L., Kirsch I.R., Sirotkin K., Ried T., Padilla-Nash H., Augustus M., Strausberg R.L., Kirsch I.R., Sirotkin K., Ried T., Augustus M., Strausberg R.L., Kirsch I.R., Sirotkin K., Ried T., Strausberg R.L., Kirsch I.R., Sirotkin K., Ried T., Kirsch I.R., Sirotkin K., Ried T., Sirotkin K., Ried T., Ried T. The interactive online SKY/M-FISH & CGH database and the Entrez cancer chromosomes search database: Linkage of chromosomal aberrations with the genome sequence. Genes Chromosomes Cancer. 2005;44:52–64. doi: 10.1002/gcc.20224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korneev S.A., Park J.H., O’Shea M., Park J.H., O’Shea M., O’Shea M. Neuronal expression of neural nitric oxide synthase (nNOS) protein is suppressed by an antisense RNA transcribed from an NOS pseudogene. J. Neurosci. 1999;19:7711–7720. doi: 10.1523/JNEUROSCI.19-18-07711.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kytola S., Rummukainen J., Nordgren A., Karhu R., Farnebo F., Isola J., Larsson C., Rummukainen J., Nordgren A., Karhu R., Farnebo F., Isola J., Larsson C., Nordgren A., Karhu R., Farnebo F., Isola J., Larsson C., Karhu R., Farnebo F., Isola J., Larsson C., Farnebo F., Isola J., Larsson C., Isola J., Larsson C., Larsson C. Chromosomal alterations in 15 breast cancer cell lines by comparative genomic hybridization and spectral karyotyping. Genes Chromosomes Cancer. 2000;28:308–317. doi: 10.1002/1098-2264(200007)28:3<308::aid-gcc9>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Margulies M., Egholm M., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braveman M.S., Chen Y.J., Chen Z., Egholm M., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braveman M.S., Chen Y.J., Chen Z., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braveman M.S., Chen Y.J., Chen Z., Attiya S., Bader J.S., Bemben L.A., Berka J., Braveman M.S., Chen Y.J., Chen Z., Bader J.S., Bemben L.A., Berka J., Braveman M.S., Chen Y.J., Chen Z., Bemben L.A., Berka J., Braveman M.S., Chen Y.J., Chen Z., Berka J., Braveman M.S., Chen Y.J., Chen Z., Braveman M.S., Chen Y.J., Chen Z., Chen Y.J., Chen Z., Chen Z., et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maru Y. Molecular biology of chronic myeloid leukemia. Int. J. Hematol. 2001;73:308–322. doi: 10.1007/BF02981955. [DOI] [PubMed] [Google Scholar]
- Masuda A., Takahashi T., Takahashi T. Chromosome instability in human lung cancers: Possible underlying mechanisms and potential consequences in the pathogenesis. Oncogene. 2002;21:6884–6897. doi: 10.1038/sj.onc.1205566. [DOI] [PubMed] [Google Scholar]
- Mauro M.J., O’Dwyer M., Heinrich M.C., Druker B.J., O’Dwyer M., Heinrich M.C., Druker B.J., Heinrich M.C., Druker B.J., Druker B.J. STI571: A paradigm of new agents for cancer therapeutics. J. Clin. Oncol. 2002;20:325–334. doi: 10.1200/JCO.2002.20.1.325. [DOI] [PubMed] [Google Scholar]
- Mayer M.G., Floeter-Winter L.M., Floeter-Winter L.M. Pre-mRNA trans-splicing: From kinetoplastids to mammals, an easy language for life diversity. Mem. Inst. Oswaldo Cruz. 2005;100:501–513. doi: 10.1590/s0074-02762005000500010. [DOI] [PubMed] [Google Scholar]
- Mitelman F., Mertens F., Johansson B., Mertens F., Johansson B., Johansson B. A breakpoint map of recurrent chromosomal rearrangements in human neoplasia. Nat. Genet. 1997;15:417–474. doi: 10.1038/ng0497supp-417. [DOI] [PubMed] [Google Scholar]
- Mitelman F., Johansson B., Mertens F., Johansson B., Mertens F., Mertens F. Fusion genes and rearranged genes as a linear function of chromosome aberrations in cancer. Nat. Genet. 2004;36:331–334. doi: 10.1038/ng1335. [DOI] [PubMed] [Google Scholar]
- Mitelman F., Mertens F., Johansson B., Mertens F., Johansson B., Johansson B. Prevalence estimates of recurrent balanced cytogenetic aberrations and gene fusions in unselected patients with neoplastic disorders. Genes Chromosomes Cancer. 2005;43:350–366. doi: 10.1002/gcc.20212. [DOI] [PubMed] [Google Scholar]
- Muleris M., Almeida A., Gerbault-Seureau M., Malfoy B., Dutrillaux B., Almeida A., Gerbault-Seureau M., Malfoy B., Dutrillaux B., Gerbault-Seureau M., Malfoy B., Dutrillaux B., Malfoy B., Dutrillaux B., Dutrillaux B. Detection of DNA amplification in 17 primary breast carcinomas with homogeneously staining regions by a modified comparative genomic hybridization technique. Genes Chromosomes Cancer. 1994;10:160–170. doi: 10.1002/gcc.2870100303. [DOI] [PubMed] [Google Scholar]
- Ng P., Wei C.L., Sung W.K., Chiu K.P., Lipovich L., Ang C.C., Gupta S., Shahab A., Ridwan A., Wong C.H., Wei C.L., Sung W.K., Chiu K.P., Lipovich L., Ang C.C., Gupta S., Shahab A., Ridwan A., Wong C.H., Sung W.K., Chiu K.P., Lipovich L., Ang C.C., Gupta S., Shahab A., Ridwan A., Wong C.H., Chiu K.P., Lipovich L., Ang C.C., Gupta S., Shahab A., Ridwan A., Wong C.H., Lipovich L., Ang C.C., Gupta S., Shahab A., Ridwan A., Wong C.H., Ang C.C., Gupta S., Shahab A., Ridwan A., Wong C.H., Gupta S., Shahab A., Ridwan A., Wong C.H., Shahab A., Ridwan A., Wong C.H., Ridwan A., Wong C.H., Wong C.H., et al. Gene identification signature (GIS) analysis for transcriptome characterization and genome annotation. Nat. Methods. 2005;2:105–111. doi: 10.1038/nmeth733. [DOI] [PubMed] [Google Scholar]
- Ng P., Tan J.J., Ooi H.S., Lee Y.L., Chiu K.P., Fullwood M.J., Srinivasan K.G., Perbost C., Du L., Sung W.K., Tan J.J., Ooi H.S., Lee Y.L., Chiu K.P., Fullwood M.J., Srinivasan K.G., Perbost C., Du L., Sung W.K., Ooi H.S., Lee Y.L., Chiu K.P., Fullwood M.J., Srinivasan K.G., Perbost C., Du L., Sung W.K., Lee Y.L., Chiu K.P., Fullwood M.J., Srinivasan K.G., Perbost C., Du L., Sung W.K., Chiu K.P., Fullwood M.J., Srinivasan K.G., Perbost C., Du L., Sung W.K., Fullwood M.J., Srinivasan K.G., Perbost C., Du L., Sung W.K., Srinivasan K.G., Perbost C., Du L., Sung W.K., Perbost C., Du L., Sung W.K., Du L., Sung W.K., Sung W.K., et al. Multiplex sequencing of paired-end ditags (MS-PET): A strategy for the ultra-high-throughput analysis of transcriptomes and genomes. Nucleic Acids Res. 2006;34:e84. doi: 10.1093/nar/gkl444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T., Sunahara R., Marchese A., Van Tol H.H., Seeman P., O’Dowd B.F., Sunahara R., Marchese A., Van Tol H.H., Seeman P., O’Dowd B.F., Marchese A., Van Tol H.H., Seeman P., O’Dowd B.F., Van Tol H.H., Seeman P., O’Dowd B.F., Seeman P., O’Dowd B.F., O’Dowd B.F. Transcription of a human dopamine D5 pseudogene. Biochem. Biophys. Res. Commun. 1991;181:16–21. doi: 10.1016/s0006-291x(05)81375-0. [DOI] [PubMed] [Google Scholar]
- Olsen M.A., Schechter L.E., Schechter L.E. Cloning, mRNA localization and evolutionary conservation of a human 5-HT7 receptor pseudogene. Gene. 1999;227:63–69. doi: 10.1016/s0378-1119(98)00588-5. [DOI] [PubMed] [Google Scholar]
- Padilla-Nash H.M., Heselmeyer-Haddad K., Wangsa D., Zhang H., Ghadimi B.M., Macville M., Augustus M., Schrock E., Hilgenfeld E., Ried T., Heselmeyer-Haddad K., Wangsa D., Zhang H., Ghadimi B.M., Macville M., Augustus M., Schrock E., Hilgenfeld E., Ried T., Wangsa D., Zhang H., Ghadimi B.M., Macville M., Augustus M., Schrock E., Hilgenfeld E., Ried T., Zhang H., Ghadimi B.M., Macville M., Augustus M., Schrock E., Hilgenfeld E., Ried T., Ghadimi B.M., Macville M., Augustus M., Schrock E., Hilgenfeld E., Ried T., Macville M., Augustus M., Schrock E., Hilgenfeld E., Ried T., Augustus M., Schrock E., Hilgenfeld E., Ried T., Schrock E., Hilgenfeld E., Ried T., Hilgenfeld E., Ried T., Ried T. Jumping translocations are common in solid tumor cell lines and result in recurrent fusions of whole chromosome arms. Genes Chromosomes Cancer. 2001;30:349–363. doi: 10.1002/gcc.1101. [DOI] [PubMed] [Google Scholar]
- Parra G., Reymond A., Dabbouseh N., Dermitzakis E.T., Castelo R., Thomson T.M., Antonarakis S.E., Guigo R., Reymond A., Dabbouseh N., Dermitzakis E.T., Castelo R., Thomson T.M., Antonarakis S.E., Guigo R., Dabbouseh N., Dermitzakis E.T., Castelo R., Thomson T.M., Antonarakis S.E., Guigo R., Dermitzakis E.T., Castelo R., Thomson T.M., Antonarakis S.E., Guigo R., Castelo R., Thomson T.M., Antonarakis S.E., Guigo R., Thomson T.M., Antonarakis S.E., Guigo R., Antonarakis S.E., Guigo R., Guigo R. Tandem chimerism as a means to increase protein complexity in the human genome. Genome Res. 2006;16:37–44. doi: 10.1101/gr.4145906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saglio G., Cilloni D., Cilloni D. Abl: The prototype of oncogenic fusion proteins. Cell. Mol. Life Sci. 2004;61:2897–2911. doi: 10.1007/s00018-004-4271-0. [DOI] [PubMed] [Google Scholar]
- Schrock E., Padilla-Nash H., Padilla-Nash H. Spectral karyotyping and multicolor fluorescence in situ hybridization reveal new tumor-specific chromosomal aberrations. Semin. Hematol. 2000;37:334–347. doi: 10.1016/s0037-1963(00)90014-3. [DOI] [PubMed] [Google Scholar]
- Sekiguchi N., Watanabe T., Kobayashi Y., Inokuchi C., Kim S.W., Yokota Y., Tanimoto K., Matsuno Y., Tobinai K., Watanabe T., Kobayashi Y., Inokuchi C., Kim S.W., Yokota Y., Tanimoto K., Matsuno Y., Tobinai K., Kobayashi Y., Inokuchi C., Kim S.W., Yokota Y., Tanimoto K., Matsuno Y., Tobinai K., Inokuchi C., Kim S.W., Yokota Y., Tanimoto K., Matsuno Y., Tobinai K., Kim S.W., Yokota Y., Tanimoto K., Matsuno Y., Tobinai K., Yokota Y., Tanimoto K., Matsuno Y., Tobinai K., Tanimoto K., Matsuno Y., Tobinai K., Matsuno Y., Tobinai K., Tobinai K. The application of molecular analyses for primary granulocytic sarcoma with a specific chromosomal translocation. Int. J. Hematol. 2005;82:210–214. doi: 10.1532/IJH97.04151. [DOI] [PubMed] [Google Scholar]
- Shendure J., Porreca G.J., Reppas N.B., Lin X., McCutcheon J.P., Rosenbaum A.M., Wang M.D., Zhang K., Mitra R.D., Church G.M., Porreca G.J., Reppas N.B., Lin X., McCutcheon J.P., Rosenbaum A.M., Wang M.D., Zhang K., Mitra R.D., Church G.M., Reppas N.B., Lin X., McCutcheon J.P., Rosenbaum A.M., Wang M.D., Zhang K., Mitra R.D., Church G.M., Lin X., McCutcheon J.P., Rosenbaum A.M., Wang M.D., Zhang K., Mitra R.D., Church G.M., McCutcheon J.P., Rosenbaum A.M., Wang M.D., Zhang K., Mitra R.D., Church G.M., Rosenbaum A.M., Wang M.D., Zhang K., Mitra R.D., Church G.M., Wang M.D., Zhang K., Mitra R.D., Church G.M., Zhang K., Mitra R.D., Church G.M., Mitra R.D., Church G.M., Church G.M. Accurate multiplex polony sequencing of an evolved bacterial genome. Science. 2005;309:1728–1732. doi: 10.1126/science.1117389. [DOI] [PubMed] [Google Scholar]
- Takahara T., Tasic B., Maniatis T., Akanuma H., Yanagisawa S., Tasic B., Maniatis T., Akanuma H., Yanagisawa S., Maniatis T., Akanuma H., Yanagisawa S., Akanuma H., Yanagisawa S., Yanagisawa S. Delay in synthesis of the 3′ splice site promotes trans-splicing of the preceding 5′ splice site. Mol. Cell. 2005;18:245–251. doi: 10.1016/j.molcel.2005.03.018. [DOI] [PubMed] [Google Scholar]
- Taki T., Taniwaki M., Taniwaki M. Chromosomal translocations in cancer and their relevance for therapy. Curr. Opin. Oncol. 2006;18:62–68. doi: 10.1097/01.cco.0000198972.69541.04. [DOI] [PubMed] [Google Scholar]
- Tibiletti M.G. Specificity of interphase fluorescence in situ hybridization for detection of chromosome aberrations in tumor pathology. Cancer Genet. Cytogenet. 2004;155:143–148. doi: 10.1016/j.cancergencyto.2004.03.005. [DOI] [PubMed] [Google Scholar]
- van der Hage J.A., van den Broek L.J., Legrand C., Clahsen P.C., Bosch C.J., Robanus-Maandag E.C., de van Vel C.J., de van Vijver M.J., van den Broek L.J., Legrand C., Clahsen P.C., Bosch C.J., Robanus-Maandag E.C., de van Vel C.J., de van Vijver M.J., Legrand C., Clahsen P.C., Bosch C.J., Robanus-Maandag E.C., de van Vel C.J., de van Vijver M.J., Clahsen P.C., Bosch C.J., Robanus-Maandag E.C., de van Vel C.J., de van Vijver M.J., Bosch C.J., Robanus-Maandag E.C., de van Vel C.J., de van Vijver M.J., Robanus-Maandag E.C., de van Vel C.J., de van Vijver M.J., de van Vel C.J., de van Vijver M.J., de van Vijver M.J. Overexpression of P70 S6 kinase protein is associated with increased risk of locoregional recurrence in node-negative premenopausal early breast cancer patients. Br. J. Cancer. 2004;90:1543–1550. doi: 10.1038/sj.bjc.6601741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velculescu V.E., Zhang L., Vogelstein B., Kinzler K.W., Zhang L., Vogelstein B., Kinzler K.W., Vogelstein B., Kinzler K.W., Kinzler K.W. Serial analysis of gene expression. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- Volik S., Raphael B.J., Huang G., Stratton M.R., Bignel G., Murnane J., Brebner J.H., Bajsarowicz K., Paris P.L., Tao Q., Raphael B.J., Huang G., Stratton M.R., Bignel G., Murnane J., Brebner J.H., Bajsarowicz K., Paris P.L., Tao Q., Huang G., Stratton M.R., Bignel G., Murnane J., Brebner J.H., Bajsarowicz K., Paris P.L., Tao Q., Stratton M.R., Bignel G., Murnane J., Brebner J.H., Bajsarowicz K., Paris P.L., Tao Q., Bignel G., Murnane J., Brebner J.H., Bajsarowicz K., Paris P.L., Tao Q., Murnane J., Brebner J.H., Bajsarowicz K., Paris P.L., Tao Q., Brebner J.H., Bajsarowicz K., Paris P.L., Tao Q., Bajsarowicz K., Paris P.L., Tao Q., Paris P.L., Tao Q., Tao Q., et al. Decoding the fine-scale structure of a breast cancer genome and transcriptome. Genome Res. 2006;16:394–404. doi: 10.1101/gr.4247306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Ahsen I., Rogalla P., Bullerdiek J., Rogalla P., Bullerdiek J., Bullerdiek J. Expression patterns of the LPP-HMGA2 fusion transcript in pulmonary chondroid hamartomas with t(3;12)(q27 approximately 28;q14 approximately 15) Cancer Genet. Cytogenet. 2005;163:68–70. doi: 10.1016/j.cancergencyto.2005.02.023. [DOI] [PubMed] [Google Scholar]
- Wang T.L., Maierhofer C., Speicher M.R., Lengauer C., Vogelstein B., Kinzler K.W., Velculescu V.E., Maierhofer C., Speicher M.R., Lengauer C., Vogelstein B., Kinzler K.W., Velculescu V.E., Speicher M.R., Lengauer C., Vogelstein B., Kinzler K.W., Velculescu V.E., Lengauer C., Vogelstein B., Kinzler K.W., Velculescu V.E., Vogelstein B., Kinzler K.W., Velculescu V.E., Kinzler K.W., Velculescu V.E., Velculescu V.E. Digital karyotyping. Proc. Natl. Acad. Sci. 2002;99:16156–16161. doi: 10.1073/pnas.202610899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C.L., Wu Q., Vega V.B., Chiu K.P., Ng P., Zhang T., Shahab A., Yong H.C., Fu Y., Weng Z., Wu Q., Vega V.B., Chiu K.P., Ng P., Zhang T., Shahab A., Yong H.C., Fu Y., Weng Z., Vega V.B., Chiu K.P., Ng P., Zhang T., Shahab A., Yong H.C., Fu Y., Weng Z., Chiu K.P., Ng P., Zhang T., Shahab A., Yong H.C., Fu Y., Weng Z., Ng P., Zhang T., Shahab A., Yong H.C., Fu Y., Weng Z., Zhang T., Shahab A., Yong H.C., Fu Y., Weng Z., Shahab A., Yong H.C., Fu Y., Weng Z., Yong H.C., Fu Y., Weng Z., Fu Y., Weng Z., Weng Z., et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- Yousef G.M., Borgono C.A., Michael I.P., Diamandis E.P., Borgono C.A., Michael I.P., Diamandis E.P., Michael I.P., Diamandis E.P., Diamandis E.P. Cloning of a kallikrein pseudogene. Clin. Biochem. 2004;37:961–967. doi: 10.1016/j.clinbiochem.2004.07.012. [DOI] [PubMed] [Google Scholar]
- Zelent A., Greaves M., Enver T., Greaves M., Enver T., Enver T. Role of the TEL-AML1 fusion gene in the molecular pathogenesis of childhood acute lymphoblastic leukaemia. Oncogene. 2004;23:4275–4283. doi: 10.1038/sj.onc.1207672. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Harrison P.M., Liu Y., Gerstein M., Harrison P.M., Liu Y., Gerstein M., Liu Y., Gerstein M., Gerstein M. Millions of years of evolution preserved: A comprehensive catalog of the processed pseudogenes in the human genome. Genome Res. 2003;13:2541–2558. doi: 10.1101/gr.1429003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D., Zhang Z., Harrison P.M., Karro J., Carriero N., Gerstein M., Zhang Z., Harrison P.M., Karro J., Carriero N., Gerstein M., Harrison P.M., Karro J., Carriero N., Gerstein M., Karro J., Carriero N., Gerstein M., Carriero N., Gerstein M., Gerstein M. Integrated pseudogene annotation for human chromosome 22: Evidence for transcription. J. Mol. Biol. 2005;349:27–45. doi: 10.1016/j.jmb.2005.02.072. [DOI] [PubMed] [Google Scholar]